Abstract
Heterozygosity–fitness correlations (HFCs) have been used to understand the complex interactions between inbreeding, genetic diversity and evolution. Although frequently reported for decades, evidence for HFCs was often based on underpowered studies or inappropriate methods, and hence their underlying mechanisms are still under debate. Here, we used 6100 genome-wide single nucleotide polymorphisms (SNPs) to test for general and local effect HFCs in maritime pine (Pinus pinaster Ait.), an iconic Mediterranean forest tree. Survival was used as a fitness proxy, and HFCs were assessed at a four-site common garden under contrasting environmental conditions (total of 16 288 trees). We found no significant correlations between genome-wide heterozygosity and fitness at any location, despite variation in inbreeding explaining a substantial proportion of the total variance for survival. However, four SNPs (including two non-synonymous mutations) were involved in significant associations with survival, in particular in the common gardens with higher environmental stress, as shown by a novel heterozygosity–fitness association test at the species-wide level. Fitness effects of SNPs involved in significant HFCs were stable across maritime pine gene pools naturally growing in distinct environments. These results led us to dismiss the general effect hypothesis and suggested a significant role of heterozygosity in specific candidate genes for increasing fitness in maritime pine. Our study highlights the importance of considering the species evolutionary and demographic history and different spatial scales and testing environments when assessing and interpreting HFCs.
Keywords: genetic variation, adaptation, survival, single nucleotide polymorphism, maritime pine
1. Introduction
It is well known that a decrease in heterozygosity, for example due to mating between relatives or genetic drift after population bottlenecks, can lead to a concomitant reduction in fitness. This is believed to be caused by the expression of recessive deleterious alleles and the lower occurrence of beneficial overdominance effects [1–4]. However, understanding how heterozygosity, as evaluated by molecular markers, correlates with fitness (heterozygosity–fitness correlations or HFCs) at different spatial scales remains a key issue in the study of the complex interaction between genetics, ecology and evolution [5–7]. The mechanisms underlying HFCs are still hotly debated, since the hypotheses proposed so far involve alternative causative agents and have different evolutionary consequences [7,8].
Three main hypotheses have been proposed to explain HFCs (reviewed in [3,7]). The first two explain HFCs in terms of heterozygote advantage at particular loci: (i) the direct effect hypothesis, in which a molecular marker itself has a functional influence on a fitness trait; and (ii) the local effect hypothesis, which assumes HFCs to be caused by heterozygosity in genes that are in linkage disequilibrium with the studied molecular markers. Finally, (iii) the general effect hypothesis assumes that molecular marker heterozygosity reflects genome-wide heterozygosity, which in turn is correlated with the individual inbreeding level and the expression of deleterious recessive alleles. Direct, local and general effects are not mutually exclusive [9].
Numerous organisms show evidence for direct/local fitness effects (e.g. cattle [10] and fish [11]), general fitness effects (e.g. amphibians [12]) or both (e.g. birds [13], humans [14] and plants; see [15] for a review). HFCs for specific loci rather than for genome-wide heterozygosity tend to predominate in the literature. However, it has been recently argued that expected effects are small and hard to detect, and that inappropriate statistical testing (e.g. single-locus regression instead of multilocus models incorporating specific effects for each locus) may have biased the results towards the predominance of the direct and local effect hypotheses [7]. Meta-analyses have shown that HFCs, although positive overall, are weak (explaining around 1–5% of the variance of fitness traits in studies with microsatellites), far from universal and context-dependent [5,7,16,17]. Several authors have also pointed out a possible publication bias towards reporting positive, significant HFCs [7,17,18] (but see [19]). Furthermore, HFC studies could have also been hindered by the choice of fitness traits and molecular markers, incomplete knowledge on demographic history and population structure for target species, and lack of statistical power [7,18]. For example, under current theory, only life-history traits involving many different loci under directional dominance, such as survival or reproductive success, are expected to correlate with heterozygosity, while many studies have focused on less appropriate growth or behavioural traits [7,17].
Another highly controversial issue is the degree to which heterozygosity measured at a small number of molecular markers captures whole-genome heterozygosity [9,19,20], challenging the support for the general effect hypothesis in studies from the ‘pre-genomics era’. Moreover, for marker loci to be able to estimate general inbreeding, the studied population must have gone through some mechanism generating variation in inbreeding, such as consanguineous matings, genetic drift or a recent bottleneck [7,21]. Rates of both local and genome-wide recombination are also relevant, as they influence the effect of selection on neighbouring neutral sites through linkage (e.g. [22]) and the long-term maintenance of diversity [23]. Finally, the type and genome location of molecular markers used to test for HFCs have great importance, as fitness effects are expected to be different depending on whether the markers are neutral or potentially functional, and local or widespread in the genome [4,13,21,24,25]. In this context, genome-wide single nucleotide polymorphisms (SNPs) may help to overcome the inherent limitations traditionally associated to the study of HFCs [18,26,27]. Here, we used a large SNP dataset (6100 SNPs), including both putatively neutral control SNPs and potentially functional polymorphisms from candidate genes, to address HFCs in maritime pine (Pinus pinaster Ait.), an outcrossing conifer with a fragmented but widespread distribution in southwestern Europe and northern Africa.
Outcrossing species with long generation times, such as forest trees, can accumulate substantial genetic load [28] and are thus suitable for detecting general HFCs. They also often have high levels of standing genetic variation for adaptive genes (e.g. [29] for Mediterranean pines), which makes them ideal to identify loci involved in direct or local HFCs too. Moreover, forest trees, given their typically high population differentiation over extensive distribution ranges, can also be useful to study HFCs at different spatial scales. However, to date, only scarce data exist on HFCs for forest tree species [15], and the few available studies were based on only a handful of molecular markers (typically less than 20). These studies reported disparate results, from positive HFCs [30] to negative [31] or null correlations [32], or even positive and negative HFCs within different populations of the same species [33].
In this study, individual HFCs were assessed for distinct maritime pine populations and gene pools in a multisite common garden, thus providing an ideal scenario to test whether HFCs remain stable or vary across the species range or under distinct environmental conditions [15]. HFCs considering distinct selection pressures have seldom been tested or suffered from limitations due to reduced environmental variability or lack of replication [12] (but see [34]). Using survival as a fitness proxy, we tested for both general effects (correlating neutral SNP heterozygosity and fitness data) and local effects (as revealed by a novel heterozygosity–fitness association test). The interpretation of our results in the light of the relatively well-known evolutionary history of maritime pine and the use of complementary analytical approaches at different spatial scales provided comprehensive insights into the mechanisms underlying HFCs in this outcrossing, long-lived forest tree.
2. Material and methods
(a). Study species
Maritime pine (P. pinaster Ait.) is a long-lived, outcrossing conifer, native to the occidental Mediterranean Basin and the European Atlantic front (figure 1). This species has great ecological and economic importance, being an iconic tree of Mediterranean landscapes as well as the main commercial softwood source in southern Europe. It grows in a variety of substrates, from siliceous to calcareous, in elevations ranging from seashore to over 2000 m.a.s.l., and under different climate regimes [35]. In the Mediterranean area, maritime pine is found in scattered populations, from the coast to the Iberian central plateau. In contrast, Atlantic populations, which grow in a moist-temperate climate with low seasonality, are large and continuous, partly due to afforestation.
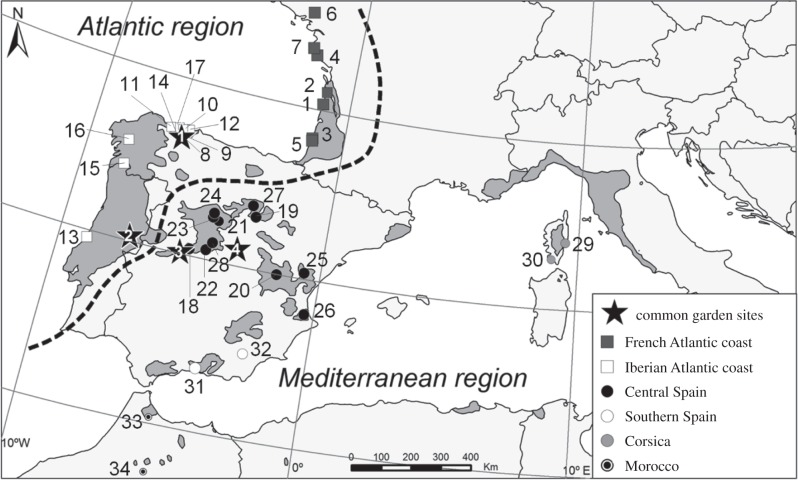
Figure 1.
Location of the four common gardens (stars) and the 34 source populations used to test for HFCs in P. pinaster. Populations are represented by numbers; see key in table 1. Symbols represent different gene pools as indicated in the legend. The size of the symbol is proportional to population mean heterozygosity (Ho) for 6100 SNPs. Shaded grey areas represent the species's natural distribution range.
Maritime pine survived the last glaciations in multiple refugia located in southeastern Spain, northern Africa, southern Italy and the Atlantic coast of Portugal [36], where it presumably overcame severe bottlenecks (see fig. 3 in [29]; see also [37]). Based on molecular markers, six extant gene pools have been described in the species (French Atlantic coast, Iberian Atlantic coast, central Spain, southern Spain, Corsica and Morocco [38,39]; figure 1), which also showed evidence of genetic differentiation for adaptive traits [38,40]. High molecular and phenotypic variability among gene pools, together with population adaptive divergence, make maritime pine particularly interesting for HFCs studies at different spatial scales.
(b). Plant material
Open-pollinated seeds were collected from 34 natural populations selected across maritime pine natural range. The sampling included representations of the six gene pools previously described using neutral molecular markers (see above and figure 1). Seedlings were grown in nursery and each individual was propagated vegetatively by cuttings [41]. These cuttings (1–28 genotypes per population; table 1) were used to establish a common garden experiment (total of 16 288 trees) at four locations that covered the species's climatic breadth (figure 1). Common gardens in Asturias and Portugal are located in the Atlantic region, with high annual rainfall (1160 and 985 mm, respectively) and mild temperatures (11.3 and 14.2°C, respectively). Nevertheless, Portugal shows less than half the summer rainfall than Asturias, indicating higher seasonality (63 mm and 152 mm, respectively). By contrast, trials in Madrid and Cáceres are located in continental areas under Mediterranean influence (annual rainfall of 443 and 898 mm, and mean temperature of 13.7 and 15.5°C, respectively), with large seasonal temperature oscillations and a marked summer drought. Apart from its climate, clay soils make Cáceres a worse site than Madrid for maritime pine survival and growth. Each of the four replicated trials comprised 4072 trees, with 509 genotypes and eight ramets per genotype, set in a randomized complete block design. This clonal common garden design allowed multiple survival measurements for each genotype and accurate performance evaluation.
Table 1.
Heterozygosity and inbreeding for 34 P. pinaster populations evaluated in four common gardens with contrasting environments (Asturias, Portugal, Madrid and Cáceres; figure 1). Numbers in brackets after population names denote population location in figure 1. N, number of individuals (genotypes); Ho, observed heterozygosity based on 6100 SNPs; g2, a multilocus estimator of ID; s(g2), inbreeding coefficient for the population estimated from marker data [42]; s(g2) values were not computed for the three populations with fewer than five genotypes.
| gene pool | population | N | Ho | g2 | s(g2) |
|---|---|---|---|---|---|
| French Atlantic coast | Hourtin (1) | 26 | 0.274 | 0.003 | 0.013 |
| Leverdon (2) | 26 | 0.260 | 0.023 | 0.083 | |
| Mimizan (3) | 18 | 0.269 | 0.013 | 0.050 | |
| Olonne sur Mer (4) | 23 | 0.266 | 0.024 | 0.086 | |
| Petrocq (5) | 22 | 0.269 | 0.013 | 0.048 | |
| Pleucadec (6) | 20 | 0.278 | 0.004 | 0.014 | |
| St-Jean des Monts (7) | 26 | 0.266 | 0.015 | 0.056 | |
| overall | 161 | 0.268 | 0.014 | 0.051 | |
| Iberian Atlantic coast | Alto de la Llama (8) | 9 | 0.245 | 0.006 | 0.025 |
| Armayán (9) | 8 | 0.236 | 0.001 | 0.003 | |
| Cadavedo (10) | 10 | 0.215 | 0.033 | 0.113 | |
| Castropol (11) | 10 | 0.221 | 0.014 | 0.053 | |
| Lamuño (12) | 9 | 0.235 | 0.001 | 0.005 | |
| Leiria (13) | 23 | 0.258 | 0.001 | 0.005 | |
| Puerto de Vega (14) | 8 | 0.219 | 0.019 | 0.069 | |
| San Cipriano (15) | 8 | 0.249 | 0.006 | 0.022 | |
| Sergude (16) | 21 | 0.246 | 0.006 | 0.023 | |
| Sierra de Barcia (17) | 8 | 0.225 | 0.022 | 0.079 | |
| overall | 114 | 0.239 | 0.012 | 0.045 | |
| Central Spain | Arenas de S. Pedro (18) | 17 | 0.264 | 0.002 | 0.009 |
| Bayubas de Abajo (19) | 18 | 0.277 | 0.000 | 0.000 | |
| Boniches (20) | 9 | 0.267 | 0.004 | 0.015 | |
| Carbonero (21) | 5 | 0.259 | n.a. | n.a. | |
| Cenicientos (22) | 9 | 0.271 | 0.005 | 0.019 | |
| Coca (23) | 17 | 0.270 | 0.003 | 0.011 | |
| Cuellar (24) | 28 | 0.270 | 0.002 | 0.008 | |
| Olba (25) | 21 | 0.265 | 0.016 | 0.058 | |
| Quatretonda (26) | 17 | 0.266 | 0.021 | 0.076 | |
| San Leonardo (27) | 14 | 0.267 | 0.004 | 0.016 | |
| Valdemaqueda (28) | 12 | 0.269 | 0.000 | 0.001 | |
| overall | 167 | 0.268 | 0.006 | 0.021 | |
| Corsica | Pinia (29) | 9 | 0.225 | 0.002 | 0.010 |
| Pineta (30) | 12 | 0.205 | 0.036 | 0.121 | |
| overall | 21 | 0.217 | 0.013 | 0.049 | |
| southern Spain | Cómpeta (31) | 4 | 0.270 | n.a. | n.a. |
| Oria (32) | 26 | 0.261 | 0.004 | 0.014 | |
| overall | 30 | 0.262 | 0.003 | 0.013 | |
| Morocco | Madisouka (33) | 1 | 0.185 | n.a. | n.a. |
| Tamrabta (34) | 15 | 0.175 | 0.008 | 0.032 | |
| overall | 16 | 0.176 | 0.009 | 0.035 | |
| species | 509 | 0.256 | 0.014 | 0.053 |
(c). Molecular markers
Illumina Infinium technology was used to genotype 8949 SNPs. After standard quality filters (based on genotype clustering scores, SNP call frequency and Hardy–Weinberg equilibrium tests) and careful visual inspection, 6100 polymorphic SNPs from 3511 distinct amplicons/unigenes with lower than 10% missing data were kept for this study. The successfully scored SNP set included 2661 control SNPs (see [43,44] for details). These SNPs were randomly chosen from transcriptome sequence data originated from cDNA libraries of different tissues without any prior experimental treatment and are thus expected to represent neutral or quasi-neutral variants. The remaining 3439 SNPs were obtained from mixed sources (see table S3 in [44]), including potentially functional markers from candidate genes (e.g. 18 SNPs associated with climate adaptation [39] and 17 SNPs associated with serotiny, a fire-response adaptive trait [45]). SNPs were also selected to provide wide genome coverage and low linkage with other SNPs from the same amplicon/unigene. Thus, they provide independent genome-wide samplings of the genome. Further details on SNP selection, assay construction, DNA extraction and SNP typing methods are provided in [44].
(d). Fitness and heterozygosity estimates
(i). Fitness
Tree survival was scored as a binary variable (0/1) 3 years after plantation and used as a fitness proxy [7,46]. Survival best linear unbiased predictors (BLUPs) for each population and genotype were estimated by restricted maximum likelihood (REML), as implemented in ASREML [47]. A general linear mixed model was fitted for each common garden, including block as a fixed term, and population and clone within population as random factors (for details, see electronic supplementary material, appendix SA). This way, we obtained one survival BLUP for each genotype, subsequently used as the individual fitness estimate for HFCs. These fitness estimates represent the genetic merit of each genotype for survival under the testing conditions of each common garden and have reduced environmental noise, thus increasing the power to detect HFCs. To avoid spurious HFCs due to population structure when merging genotypes from different populations [7], genotype survival estimates did not include population effects (i.e. population mean estimates were subtracted from the genotype survival estimates), as if all trees formed a unique population.
(ii). Heterozygosity
Individual multilocus observed heterozygosity (Ho) was determined as:
where m is the number of loci successfully scored, and Hij is a binary variable that takes the value of 1 if the jth locus is heterozygous within the ith individual, and 0 otherwise [14]. We preferred multilocus observed heterozygosity (Ho) over other standardized measures of marker heterozygosity (e.g. adaptive distance or internal relatedness) because Ho is, in general, more robust [7] (see [17] for a discussion). To combine data from different populations for HFC analyses, individual genotype heterozygosity estimates were standardized with respect to the population mean, in a similar way to the survival data [7]. An inbreeding coefficient (s) based on g2, an estimate of identity disequilibrium (ID), was also computed for each population using RMES software [42]. The procedure estimates s from g2 (the covariance in heterozygosity standardized by average heterozygosity), which depends only on the mean and variance of inbreeding in the population, and not on locus-specific characteristics (for details, see [42]).
(e). Heterozygosity–fitness correlations
(i). General fitness effects
To test for general fitness effects using individual multilocus heterozygosity, the molecular markers assayed must carry information about genome-wide levels of heterozygosity and inbreeding [20]. Thus, prior to the analysis of general effect HFCs, we used two methods based on ID to assess the ability of the SNP markers used to capture individual inbreeding levels. First, we ran a heterozygosity–heterozygosity correlation test [9,48] with 1000 randomizations of the marker partition, using Rhh package [48] in R v. 3.1.2 [49]. This test is based in comparing random partitions of the marker set. If independent subsets of loci are able to produce correlated heterozygosity estimates, then we could assume that the marker set has enough power to capture genome-wide information. Second, we computed g2, species-wide and for each population and gene pool, running 1000 iterations in RMES [42]. The parameter g2 is an estimator of ID that uses all markers simultaneously and is thus expected to be only affected by demographic history and not by the particular set of markers used [7,18]. Then, we assessed the expected power of our SNP marker set to detect general-effect HFCs using formulae from Miller et al. [27] (eqn 5).
To test for the general effect hypothesis, the HFCs' significance level was assessed with standard Pearson correlation coefficients (r) between individual heterozygosity based on 6100 SNPs and survival estimates (BLUPs), after Bonferroni corrections for multiple testing. These coefficients were first computed species-wide considering all individuals (genotypes) as if they were part of a single population (i.e. after subtracting population effects, see above). Maritime pine has a strong population genetic structure resulting from its complex evolutionary history and high level of fragmentation (see Plant material). Thus, as HFCs' significance and sign may vary among distinct geographical groups, HFCs were also studied at different spatial levels: (i) the gene pool level, which also considered individual data without population effects; and (ii) the population level, where HFCs were determined considering only the genotypes tested in each population (see table 1 for details on sample sizes). Three populations with five or fewer genotypes (Madisouka, Carbonero and Cómpeta) were excluded for these later analyses. As phenotypic correlations across common gardens were low (see Results), general HFCs were tested separately for each one of the four common gardens. Finally, the expected correlation between heterozygosity and inbreeding (f) and between inbreeding and fitness, as gauged by survival BLUPs, were computed following Szulkin et al. [7].
(ii). Local fitness effects
Local fitness effects were tested using a novel association test between heterozygosity and fitness. As for the case of general HFCs, an independent test was conducted for each common garden. SNPs were coded as 0 if the locus was heterozygous and as −1 otherwise. Heterozygote effects were computed using a Bayesian mixed linear model that imputes missing data and fits simultaneously additive SNP effects and population structure [50], as implemented in BAMD software v. 3.5 [51] (see electronic supplementary material, appendix SB). The kinship matrix was inferred from the full SNP dataset in SpaGeDi v. 1.3 [52]. Negative genetic covariances between individuals were set to zero as in the study by Yu et al. [53]. To avoid false positives and control for confounding associations between marker heterozygosity and survival, individual assignment probabilities to each gene pool were included in the model as covariates (i.e. the Q matrix, as obtained from STRUCTURE software v. 2.2 [54]; details on STRUCTURE runs are provided in electronic supplementary material, table S1). A total of 500 000 iterations were performed on BAMD and the last 200 000 were kept to estimate the distribution of additive SNP effects (γ). Then, a one-sided test (α = 0.05) was used to identify significant local HFCs (i.e. those with γ > 0). Finally, fitness effects of SNPs involved in significant HFCs were also computed separately for trees belonging to the two gene pools with higher sample sizes, French Atlantic Coast (n = 135) and central Spain (n = 168), which represent two highly contrasted growing conditions (Atlantic versus Mediterranean) within the species range. In this case, the distributions of additive SNP effects were used to identify significant differences (i.e. non-overlapping 95% confidence intervals, CIs) with SNP effects on fitness at the species-wide scale.
3. Results
(a). Fitness and heterozygosity estimates
Tree survival ranged from 8% to 95% across test sites (average of 8% in Cáceres, 25% in Madrid, 68% in Portugal and 95% in Asturias; electronic supplementary material, table S2), evidencing highly contrasted levels of environmental stress at each site. Site-to-site correlations for individual genotype survival were not significant (see electronic supplementary material, table S3). Multilocus observed heterozygosity (Ho) ranged from 0.085 to 0.316, with mean of 0.256 ± 0.033 (table 1). Populations from Mediterranean and Atlantic regions did not show significant differences in heterozygosity, with mean of 0.268 for both the French Atlantic Coast and central Spain gene pools (table 1). Inbreeding coefficients (s) ranged from 0 to 0.121 (table 1; electronic supplementary material, figure S1), and neither showed any remarkable differences across the six gene pools or between the Mediterranean and Atlantic regions.
(b). Heterozygosity–fitness correlations
The multilocus heterozygosity–heterozygosity mean correlation coefficient was 0.95 (95% CIs: 0.94–0.96). Such a highly positive and significant value indicates that our SNP markers are suitable to detect general effect HFCs in this species, if present. Accordingly, the g2 estimator of ID differed significantly from zero species-wide (g2 = 0.014, s.d. = 0.003, p < 0.001) and also for the Atlantic and Mediterranean regions separately (g2 = 0.015, s.d. = 0.004, p < 0.001, and g2 = 0.013, s.d. = 0.004, p < 0.001, respectively; see table 1 for population estimates). The expected power to detect HFCs according to Miller et al. [27] was very high (r2 = 0.97). But correlations between heterozygosity (Ho) and fitness, as evaluated by survival BLUPs, were not significant (after Bonferroni corrections) at any of the spatial levels tested (i.e. species, gene pool or population) in any of the four common gardens (see electronic supplementary material, table S4). The same results were obtained when only the 2661 control SNPs were used (see electronic supplementary material, table S5). Among all computed correlations, 55% were positive and 45% were negative, and no particular sign bias was observed. Variation in inbreeding among individuals accounted for 22–51% of the total variance in survival, depending on the common garden. However, heterozygosity was only moderately correlated with inbreeding (r2 = 0.17; see also electronic supplementary material, figure S1).
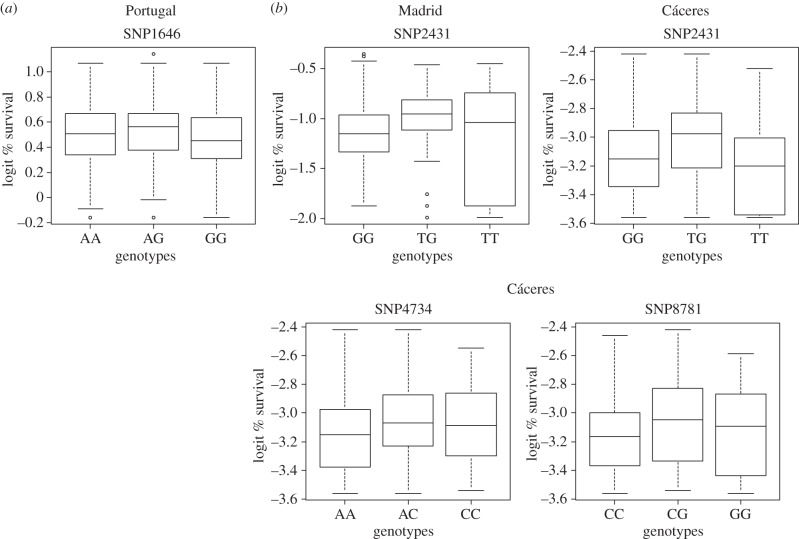
Using a heterozygosity–fitness association test, we found that four SNPs (SNP1646, SNP2431, SNP4734 and SNP8781; see [44] for notation), two of them non-synonymous (SNP1646 and SNP2431), were involved in local effect HFCs (table 2 and figure 2). Interestingly, one of the non-synonymous mutations (SNP2431) was significantly associated with survival in two common gardens (Madrid and Cáceres). Most of the significant positive associations (four out of five, involving three SNPs out of four) were found in the common gardens with higher environmental stress: one in Madrid and three in Cáceres, the common garden with highest mortality. Fitness effects of SNPs involved in HFCs were stable across maritime pine gene pools naturally growing in distinct Atlantic and Mediterranean environments (i.e. they had overlapping 95% CIs with species-wide SNP effect estimates on fitness; table 2).
Table 2.
Additive SNP effects (γ) on fitness and one-sided test for local HFCs (γ > 0) based on SNP effect distributions obtained using a Bayesian heterozygosity–fitness association test at the rangewide geographical scale. Additive SNP effects and a two-sided test for significant differences with species-wide SNP effect estimates are also shown for two maritime pine gene pools growing under highly contrasted environments (Atlantic versus Mediterranean climate); n.s., not significant (i.e. overlapping 95% CI). non-syn, non-synonymous; na, unknown/data unavailable.
| regional SNP effects (γ) |
||||||
|---|---|---|---|---|---|---|
| SNP codea | SNP typeb | annotation (BLASTx) | accession number | SNP effects (γ) [p-values] | French Atlantic Coast | central Spain |
| Portugal trial | ||||||
| SNP1646 | [A/G]non-syn | V-type proton ATPase | 501311935 | 0.04144 [0.0053] |
0.02918 n.s. |
0.04673 n.s. |
| Madrid trial | ||||||
| SNP2431 | [T/G]non-syn | Expansin 2 | 1485845574 | 0.05451 [0.0125] |
0.04153 n.s. |
0.02739 n.s. |
| Cáceres trial | ||||||
| SNP2431 | [T/G]non-syn | Expansin 2 | 1485845574 | 0.04435 [0.0342] |
0.03969 n.s. |
0.02922 n.s. |
| SNP4734 | [A/C]na | Serine-type endopeptidase | 501314366 | 0.04923 [0.0071] |
0.03989 n.s. |
0.01850 n.s. |
| SNP8781 | [C/G]na | GPCR-type G protein 1 | 1516987379 | 0.05752 [0.0020] |
0.05905 n.s. |
0.04158 n.s. |
aSee notation in [44].
bAs provided by NCBI's dbSNP.
Figure 2.
Box-plots for significant local HFCs at the species-wide geographical scale. (a) Common gardens under mild Atlantic environmental conditions (Portugal, there were not any significant HFC in Asturias). (b) Common gardens under harsh Mediterranean environmental conditions (Madrid and Cáceres). Boxes denote the interquartile range and horizontal lines within boxes the phenotypic means (percentage survival in logit scale).
4. Discussion
HFCs were examined in the keystone tree maritime pine, under contrasted environments, using 6100 genome-wide SNP markers. Variation in inbreeding explained a substantial proportion of the total variance in survival, but no significant correlation was found between genome-wide heterozygosity and fitness at any spatial level (i.e. species, gene pool or population). However, heterozygosity in four SNPs was positively associated with survival, in particular in the common gardens with higher environmental stress, as shown by an association test. Local HFCs were stable across maritime pine gene pools naturally growing under Mediterranean and Atlantic climates. These results led us to dismiss a global relationship between heterozygosity and fitness in maritime pine (i.e. general effects). Alternatively, we suggest that not enough variance in inbreeding depression and demographic history may have decoupled heterozygosity and inbreeding.
(a). Lack of general fitness effects
Previous studies in conifers found no clear HFC patterns [32,33], but their results may have been swayed by a lack of statistical power, due to a reduced number of markers, inappropriate analytical methods, and the confounding effects of environment and population structure [7,55]. It can even be argued that previous HFC studies in forest trees were all unwittingly looking for local effects, as they used a very small number of markers that were poor estimators of genome-wide heterozygosity [9]. Our multisite clonal common garden design (with several hundreds of individuals from across the species's natural range, planted in contrasting environments), our use of a high number of SNP markers with wide heterozygosity range and the performance of analyses at different spatial levels (accounting for population structure) should have overcome previous drawbacks, maximizing the chance to detect HFCs in the species, if present [1,7,46]. The expected power to detect HFCs [27] was very high, and this is one of the first studies reporting significant g2 values (but see [56]) and a high heterozygosity–heterozygosity correlation [18,57]. These results endorse the greater ability of a high number of SNP markers (in our case 6100), despite incomplete coverage of the genome, for the estimation of genome-wide heterozygosity, as recently suggested by Hoffman et al. [26].
Common gardens covered a wide range of environments, from very harsh in Cáceres to very mild in Asturias, as evidenced by survival rates ranging from 8% to 95%, which allowed us to assess HFCs in highly contrasted growing conditions. The correlation between individual heterozygosity and fitness is expected to be particularly strong under high stress conditions [13,56,57] (but see [1,24]). This is because the deleterious effects of inbreeding (i.e. inbreeding depression) tend to increase under environmental stressful conditions, as the increased variance in fitness in the inbred population allows natural selection to act against the least fit genotypes, reducing the cumulative loss of fitness predicted to occur in more benign environments [58]. However, we did not find significant correlations between genome-wide multilocus heterozygosity and fitness under any testing environment, including those with low survival.
This lack of correlation could be explained by maritime pine mating system and evolutionary history. Forest trees and maritime pine in particular are mostly outcrossing, with little variation in outcrossing rates across populations, and show low levels of correlated paternity [59] and high heterozygosity [39]. As a mainly outbreeding species, maritime pine may not have enough variance in inbreeding as to result in significant general effect HFCs. However, when compared with other Pinus species, maritime pine has reduced inbreeding depression (although still significant), in particular for fecundity traits and general vigour [60]. Evidence of historical demographic bottlenecks in the species [29,37] suggests that some genetic load may have been purged during maritime pine survival in isolated glacial refugia [2,28,61,62], contributing to the observed lack of general effect HFCs [5,17,28,63]. This hypothesis is supported by a relatively low correlation between heterozygosity and inbreeding (r2 = 0.17), which renders HFCs non-significant despite a substantial proportion of the total variance in survival being explained by variation in inbreeding (22–51%, depending on common garden). Similar low or non-significant HFCs despite relatively high association between inbreeding and fitness have been reported for both plants and animals by Szulkin et al. [7].
Agricultural hybrids where two homozygous inbred lines are crossed to produce heterozygote F1 individuals with outstanding performance (i.e. heterosis) can be considered an extreme case of general HFCs (e.g. in maize or rice [64]). However, naturally occurring differences in individual heterozygosity are normally much lower and, as we showed in this study, did not lead to significant fitness differences in maritime pine. To our knowledge, efforts to produce high-performance forest trees by crossing double haploid lines in breeding programmes have been generally unsuccessful [65].
(b). Significant local fitness effects
Heterozygosity at four SNP markers was positively associated with fitness, in particular in the two common gardens with higher mortality (Madrid and Cáceres), suggesting that heterozygosity at specific loci can confer selective advantage to maritime pine under stressful environmental conditions. Apart from including a large set of control SNPs, our SNP assay was enriched for polymorphisms from candidate genes, which could have fostered the identification of local HFCs, despite the huge size of the pine genome (approx. 20–25 Gbp). These local HFCs were stable across maritime pine gene pools naturally growing under Atlantic and Mediterranean climates, suggesting broad, species-wide adaptation processes. Individuals with increased heterozygosity at candidate genes may well possess the necessary diversity of alleles required to adequately cope with environmental stochasticity [17]. Changing selection pressures (i.e. drought, extreme temperatures) could favour one allele or the other, alternately, along the extensive reproductive period and long life of trees (e.g. wet or dry years or decades). It has been recently argued that heterozygote advantage should be very common during adaptation if selection is stabilizing and at least some mutations are large enough to overshoot the optimum [66]. Although there is scarce experimental proof of particular loci showing heterozygote advantage [67], it has been suggested that single alleles can confer general stress resistance for some types of stress [10,58]. For example, heterozygote advantage might include some important cases of recent adaptation under strong selection (e.g. mutants in livestock and companion animals, warfarin resistance in rats; see [67] for discussion and more examples). However, it is generally not possible to differentiate between heterozygote advantage and other types of balancing selection [67].
Additive SNP effects in significant local HFCs were modest, ranging from 0.041 to 0.058 (table 2), as would be expected for highly polygenic fitness traits such as survival. Integrated life-history traits (e.g. survival, reproductive success) involve many different loci, all of which are targets for deleterious recessive mutations. This genetic architecture favours the expression of HFCs [7]. However, the expected effect sizes for those traits are very small, as they have a more complex polygenic architecture than morphological or physiological traits [17,68]. Hence, it is not surprising that previous studies based on microsatellites and invoking local effect HFCs explained very little variance of the studied traits [7,17]. Recently, García-Navas et al. [56] found heterozygosity at a single microsatellite locus to explain a disproportionately large variation in offspring size in great tits (37%). Although they suggest a possible heterozygote advantage for this locus, they also argue that this result could be biased by an underlying general inbreeding effect. It is interesting to note, however, the much stronger mean effect sizes found for plants than for animals [17]. Anyhow, because purging of genetic load and decay of linkage disequilibrium are slow processes, effect sizes tend to be also smaller in populations that underwent historical bottlenecks, as is the case of maritime pine, than in those that suffered more recent ones [17].
Remarkably, a non-synonymous polymorphism, SNP2431, was involved in HFCs for the two Mediterranean common gardens with high environmental stress (Madrid and Cáceres). This SNP is located in a candidate gene encoding an expansin, which belongs to a family of closely related plant cell wall proteins involved in cell growth [69,70]. The heterozygote advantage observed at this locus may then be explained by direct selection, as it appears to play an important functional role in plant growth and development [71]. Some previous studies have also reported significant HFCs for markers located in genes of known function, but not for neutral markers [25,72]. However, most of the local HFCs found in this study were located in poorly annotated genes and it is likely that they are in linkage with causative polymorphisms rather than being the causative polymorphisms themselves (i.e. they can be considered local but not direct HFCs). Thus, further work is needed to validate or identify causative polymorphisms and fully understand the functional basis of the significant HFCs found in our study.
5. Conclusion
Our study suggests that genome-wide heterozygosity has little impact on maritime pine fitness, even though variation in inbreeding seems to be associated with survival in the species. However, a novel application of a genetic association test allowed us to identify the heterozygosity of some putatively adaptive markers to be associated with survival in maritime pine, revealing that local fitness effects may drive HFCs in this species. Nevertheless, because our SNP assay was enriched for polymorphisms from candidate genes, this assertion must be taken with caution. HFCs were stable across distinct maritime gene pools pointing to broad, species-wide effects on fitness. Moreover, significant HFCs were found in common gardens under harsh Mediterranean climatic conditions, suggesting that heterozygosity at these loci is more important for survival under increased selection pressures. Thus, heterozygosity in specific candidate genes may hold a relevant role for survival of forest tree populations under warmer and drier climates, as those expected under current climate change. Our study highlights the utmost importance of integrating knowledge on species evolutionary and demographic history across different spatial scales and testing environments in the estimation and interpretation of HFCs in future studies.
Acknowledgements
We thank Prof. John Pannell for critical reading and discussion of a previous version of the manuscript. Thanks are extended to A. Saldaña, F. del Caño, E. Ballesteros and D. Barba for field assistance and to J. Fernández for help with figure 1. Data used in this research are part of the Spanish Network of Genetic Trials (GENFORED, http://www.genfored.es). We thank all persons and institutions linked to the establishment and maintenance of field trials used in this study. The language in the article has been revised by a scientific editor, P. C. Grant.
Data accessibility
SNP data were deposited in the Dryad repository at http://dx.doi.org/10.5061/dryad.8d6k1. Survival data have been deposited in GENFORED, the Spanish Network of Genetic Trials (http://www.genfored.es).
Authors' contributions
I.R.-Q. collected field data, carried out the statistical analyses and drafted the manuscript. J.P.J.-C., R.A. and S.C.G.-M. conceived and designed the study. S.C.G.-M. and R.A. coordinated the study, helped with statistical analysis and helped to draft the manuscript. L.S.-B. participated in data analysis. I.R.-Q., D.G. and S.C.G.-M. contributed to the SNP assay design and molecular laboratory work. J.M. helped with field data collection. G.G.V. and S.C.G.-M. produced the genotyping data. All authors contributed to manuscript discussion and review, and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This study was funded by the Spanish Ministry of Economy and Competitiveness through projects RTA2010-00120-C02-02 (CLONAPIN), CGL2011-30182-C02-01 (AdapCon) and AGL2012-40151-C03-02 (FENOPIN), and by the European Research Area-Net BiodivERsA (TipTree project, ANR-12-EBID-0003), which included the French National Research Agency (ANR) as national funder (part of the 2012 BiodivERsA call for research proposals). I.R.-Q. acknowledges a PhD scholarship (FPI-INIA) from the Spanish National Institute for Agricultural and Food Research and Technology (INIA), and D.G. and J.P.J.-C. ‘Ramón y Cajal’ and ‘Juan de la Cierva’ fellowships, respectively, from the Spanish Ministry of Science and Innovation. S.C.G.-M. thanks the support of a senior Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (PIEF-GA-2012-328146). G.G.V. was supported by a grant by the Italian MIUR project ‘Biodiversitalia’ (RBAP10A2T4).
References
- 1.Waller DM, Dole J, Bersch AJ. 2008. Effects of stress and phenotypic variation on inbreeding depression in Brassica rapa. Evolution 62, 917–931. ( 10.1111/j.1558-5646.2008.00325.x) [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. ( 10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 3.Hansson B, Westerberg L. 2002. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 11, 2467–2474. ( 10.1046/j.1365-294X.2002.01644.x) [DOI] [PubMed] [Google Scholar]
- 4.Olano-Marin J, Mueller JC, Kempenaers B. 2011. Heterozygosity and survival in blue tits (Cyanistes caeruleus): contrasting effects of presumably functional and neutral loci. Mol. Ecol. 20, 4028–4041. ( 10.1111/j.1365-294X.2011.05177.x) [DOI] [PubMed] [Google Scholar]
- 5.Reed DH, Frankham R. 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. ( 10.1046/j.1523-1739.2003.01236.x) [DOI] [Google Scholar]
- 6.Luquet E, David P, Lena JP, Joly P, Konecny L, Dufresnes C, Perrin N, Plenet S. 2011. Heterozygosity–fitness correlations among wild populations of European tree frog (Hyla arborea) detect fixation load. Mol. Ecol. 20, 1877–1887. ( 10.1111/j.1365-294X.2011.05061.x) [DOI] [PubMed] [Google Scholar]
- 7.Szulkin M, Bierne N, David P. 2010. Heterozygosity–fitness correlations: a time for reappraisal. Evolution 64, 1202–1217. ( 10.1111/j.1558-5646.2010.00966.x) [DOI] [PubMed] [Google Scholar]
- 8.Mueller JC, Hermisson J, Olano-Marin J, Hansson B, Kempenaers B. 2011. Linking genetic mechanisms of heterozygosity–fitness correlations to footprints of selection at single loci. Evol. Ecol. 25, 1–11. ( 10.1007/s10682-010-9377-2) [DOI] [Google Scholar]
- 9.Balloux F, Amos W, Coulson T. 2004. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 13, 3021–3031. ( 10.1111/j.1365-294X.2004.02318.x) [DOI] [PubMed] [Google Scholar]
- 10.Amos W, Acevedo-Whitehouse K. 2009. A new test for genotype-fitness associations reveals a single microsatellite allele that strongly predicts the nature of tuberculosis infections in wild boar. Mol. Ecol. Resour. 9, 1102–1111. ( 10.1111/j.1755-0998.2009.02560.x) [DOI] [PubMed] [Google Scholar]
- 11.Lieutenant-Gosselin M, Bernatchez L. 2006. Local heterozygosity–fitness correlations with global positive effects on fitness in threespine stickleback. Evolution 60, 1658–1668. ( 10.1111/j.0014-3820.2006.tb00510.x) [DOI] [PubMed] [Google Scholar]
- 12.Lesbarrères D, Primmer CR, Laurila A, Merilä J. 2005. Environmental and population dependency of genetic variability–fitness correlations in Rana temporaria. Mol. Ecol. 14, 311–323. ( 10.1111/j.1365-294X.2004.02394.x) [DOI] [PubMed] [Google Scholar]
- 13.Voegeli B, Saladin V, Wegmann M, Richner H. 2013. Heterozygosity is linked to the costs of immunity in nestling great tits (Parus major). Ecol. Evol. 3, 4815–4827. ( 10.1002/ece3.854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindaraju DR, Larson MG, Yin X, Benjamin EJ, Rao MB, Vasan RS. 2009. Association between SNP heterozygosity and quantitative traits in the Framingham Heart Study. Ann. Hum. Genet. 73, 465–473. ( 10.1111/j.1469-1809.2009.00514.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leimu R, Mutikainen P, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 94, 942–952. ( 10.1111/j.1365-2745.2006.01150.x) [DOI] [Google Scholar]
- 16.Coltman DW, Slate J. 2003. Microsatellite measures of inbreeding: a meta-analysis. Evolution 57, 971–983. ( 10.1111/j.0014-3820.2003.tb00309.x) [DOI] [PubMed] [Google Scholar]
- 17.Chapman JR, Nakagawa S, Coltman D, Slate J, Sheldon B. 2009. A quantitative review of heterozygosity–fitness correlations in animal populations. Mol. Ecol. 18, 2746–2765. ( 10.1111/j.1365-294X.2009.04247.x) [DOI] [PubMed] [Google Scholar]
- 18.Miller JM, Coltman DW. 2014. Assessment of identity disequilibrium and its relation to empirical heterozygosity fitness correlations: a meta-analysis. Mol. Ecol. 23, 1899–1909. ( 10.1111/mec.12707) [DOI] [PubMed] [Google Scholar]
- 19.Slate J, David P, Dodds KG, Veenvliet BA, Glass BC, Broad TE, McEwan JC. 2004. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity 93, 255–265. ( 10.1038/sj.hdy.6800485) [DOI] [PubMed] [Google Scholar]
- 20.De Woody YD, De Woody JA. 2005. On the estimation of genome-wide heterozygosity using molecular markers. Heredity 96, 85–88. ( 10.1093/jhered/esi017) [DOI] [PubMed] [Google Scholar]
- 21.Agudo R, Carrete M, Alcaide M, Rico C, Hiraldo F, Donazar JA. 2012. Genetic diversity at neutral and adaptive loci determines individual fitness in a long-lived territorial bird. Proc. R. Soc. B 279, 3241–3249. ( 10.1098/rspb.2011.2606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbett-Detig RB, Hartl DL, Sackton TB. 2015. Natural selection constrains neutral diversity across a wide range of species. PLOS Biol. 13, e1002112 ( 10.1371/journal.pbio.1002112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaramillo-Correa JP, Verdú M, González-Martínez SC. 2010. The contribution of recombination to heterozygosity differs among plant evolutionary lineages and life-forms. BMC Evol. Biol. 10, 22 ( 10.1186/1471-2148-10-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szulkin M, David P. 2011. Negative heterozygosity–fitness correlations observed with microsatellites located in functional areas of the genome. Mol. Ecol. 20, 3949–3952. ( 10.1111/j.1365-294X.2011.05277.x) [DOI] [PubMed] [Google Scholar]
- 25.Da Silva A, et al. 2009. Heterozygosity–fitness correlations revealed by neutral and candidate gene markers in roe deer from a long-term study. Evolution 63, 403–417. ( 10.1111/j.1558-5646.2008.00542.x) [DOI] [PubMed] [Google Scholar]
- 26.Hoffman JI, Simpson F, David P, Rijks JM, Kuiken T, Thorne MAS, Lacy RC, Dasmahapatra KK. 2014. High-throughput sequencing reveals inbreeding depression in a natural population. Proc. Natl Acad. Sci. USA 111, 3775–3780. ( 10.1073/pnas.1318945111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JM, Malenfant RM, David P, Davis CS, Poissant J, Hogg JT, Festa-Bianchet M, Coltman DW. 2014. Estimating genome-wide heterozygosity: effects of demographic history and marker type. Heredity 112, 240–247. ( 10.1038/hdy.2013.99) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byers DL, Waller DM. 1999. Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu. Rev. Ecol. Syst. 30, 479–513. ( 10.1146/annurev.ecolsys.30.1.479) [DOI] [Google Scholar]
- 29.Grivet D, Sebastiani F, Alía R, Bataillon T, Torre S, Zabal-Aguirre M, Vendramin GG, González-Martínez SC. 2011. Molecular footprints of local adaptation in two Mediterranean conifers. Mol. Biol. Evol. 28, 101–116. ( 10.1093/molbev/msq190) [DOI] [PubMed] [Google Scholar]
- 30.Mosseler A, Major JE, Rajora OP. 2003. Old-growth red spruce forests as reservoirs of genetic diversity and reproductive fitness. Theor. Appl. Genet. 106, 931–937. ( 10.1007/s00122-002-1156-1) [DOI] [PubMed] [Google Scholar]
- 31.Abrahamsson S. 2011. Genetic dissection of quantitative traits in Scots pine. PhD thesis, Swedish University of Agricultural Sciences, Umeå, Sweden.
- 32.Savolainen O, Hedrick P. 1995. Heterozygosity and fitness: no association in Scots pine. Genetics 140, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bush RM, Smouse PE, Ledig FT. 1987. The fitness consequences of multiple-locus heterozygosity: the relationship between heterozygosity and growth rate in pitch pine (Pinus rigida Mill.). Evolution 41, 787–798. ( 10.2307/2408888) [DOI] [PubMed] [Google Scholar]
- 34.Richardson JL, Urban MC, Bolnick DI, Skelly DK. 2014. Microgeographic adaptation and the spatial scale of evolution. Trends Ecol. Evol. 29, 165–176. ( 10.1016/j.tree.2014.01.002) [DOI] [PubMed] [Google Scholar]
- 35.Alía R, Martín S. 2003. EUFORGEN: technical guidelines for genetic conservation and use for Maritime pine (Pinus pinaster). Rome, Italy: International Plant Genetic Resources Institute. [Google Scholar]
- 36.Burban C, Petit RJ. 2003. Phylogeography of maritime pine inferred with organelle markers having contrasted inheritance. Mol. Ecol. 12, 1487–1495. ( 10.1046/j.1365-294X.2003.01817.x) [DOI] [PubMed] [Google Scholar]
- 37.Naydenov KD, Alexandrov A, Matevski V, Vasilevski K, Naydenov MK, Gyuleva V, Carcaillet C, Wahid N, Kamary S. 2014. Range-wide genetic structure of maritime pine predates the last glacial maximum: evidence from nuclear DNA. Hereditas 151, 1–13. ( 10.1111/j.1601-5223.2013.00027.x) [DOI] [PubMed] [Google Scholar]
- 38.Santos-del-Blanco L, Climent J, González-Martínez SC, Pannell JR. 2012. Genetic differentiation for size at first reproduction through male versus female functions in the widespread Mediterranean tree Pinus pinaster. Ann. Bot. 110, 1449–1460. ( 10.1093/aob/mcs210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaramillo-Correa JP, et al. 2015. Molecular proxies for climate maladaptation in a long-lived tree (Pinus pinaster Aiton, Pinaceae). Genetics 199, 793–807. ( 10.5061/dryad.fb436.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Martínez SC, Alía R, Gil L. 2002. Population genetic structure in a Mediterranean pine (Pinus pinaster Ait.): a comparison of allozyme markers and quantitative traits. Heredity 89, 199–206. ( 10.1038/sj.hdy.6800114) [DOI] [PubMed] [Google Scholar]
- 41.Majada J, Martínez-Alonso C, Feito I, Kidelman A, Aranda I, Alía R. 2011. Mini-cuttings: an effective technique for the propagation of Pinus pinaster Ait. New For. 41, 399–412. ( 10.1007/s11056-010-9232-x) [DOI] [Google Scholar]
- 42.David P, Pujol B, Viard F, Castella V, Goudet J. 2007. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 16, 2474–2487. ( 10.1111/j.1365-294X.2007.03330.x) [DOI] [PubMed] [Google Scholar]
- 43.Chancerel E, et al. 2013. High-density linkage mapping in a pine tree reveals a genomic region associated with inbreeding depression and provides clues to the extent and distribution of meiotic recombination. BMC Biol. 11, 50 ( 10.1186/1741-7007-11-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plomion C, et al. In press. High-density SNP assay development for genetic analysis in maritime pine (Pinus pinaster). Mol. Ecol. Resour. ( 10.1111/1755-0998.12464) [DOI] [PubMed] [Google Scholar]
- 45.Budde KB, Heuertz M, Hernández-Serrano A, Pausas JG, Vendramin GG, Verdú M, González-Martínez SC. 2014. In situ genetic association for serotiny, a fire-related trait, in Mediterranean maritime pine (Pinus pinaster). New Phytol. 201, 230–241. ( 10.1111/nph.12483) [DOI] [PubMed] [Google Scholar]
- 46.David P. 1998. Heterozygosity–fitness correlations: new perspectives on old problems. Heredity 80, 531–537. ( 10.1046/j.1365-2540.1998.00393.x) [DOI] [PubMed] [Google Scholar]
- 47.Gilmour AR, Gogel BJ, Cullis BR, Thompson R. 2009. ASREML User Guide Release 3.0. Hemel Hempstead, UK: VSN International. [Google Scholar]
- 48.Alho JS, Välimäki K, Merilä J. 2010. Rhh: an R extension for estimating multilocus heterozygosity and heterozygosity–heterozygosity correlation. Mol. Ecol. Resour. 10, 720–722. ( 10.1111/j.1755-0998.2010.02830.x) [DOI] [PubMed] [Google Scholar]
- 49.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Fundation for Statistical Computing. [Google Scholar]
- 50.Quesada T, et al. 2010. Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.). Genetics 186, 677–686. ( 10.1534/genetics.110.117549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Gopal V, Li X, Davis JM, Casella G. 2012. Simultaneous SNP identification in association studies with missing data. Ann. Appl. Stat. 6, 432–456. ( 10.1214/11-AOAS516) [DOI] [Google Scholar]
- 52.Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. ( 10.1046/j.1471-8278) [DOI] [Google Scholar]
- 53.Yu J, et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. ( 10.1038/ng1702) [DOI] [PubMed] [Google Scholar]
- 54.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox CW, Reed DH. 2011. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution 65, 246–258. ( 10.1111/j.1558-5646.2010.01108.x) [DOI] [PubMed] [Google Scholar]
- 56.García-Navas V, Cáliz-Campal C, Ferrer ES, Sanz JJ, Ortego J. 2014. Heterozygosity at a single locus explains a large proportion of variation in two fitness-related traits in great tits: a general or a local effect? J. Evol. Biol. 27, 2807–2819. ( 10.1111/jeb.12539) [DOI] [PubMed] [Google Scholar]
- 57.Grueber CE, Waters JM, Jamieson IG. 2011. The imprecision of heterozygosity–fitness correlations hinders the detection of inbreeding and inbreeding depression in a threatened species. Mol. Ecol. 20, 67–79. ( 10.1111/j.1365-294X.2010.04930.x) [DOI] [PubMed] [Google Scholar]
- 58.Armbruster P, Reed DH. 2005. Inbreeding depression in benign and stressful environments. Heredity 95, 235–242. ( 10.1038/sj.hdy.6800721) [DOI] [PubMed] [Google Scholar]
- 59.De Lucas AI, Robledo-Arnuncio JJ, Hidalgo E, González-Martínez SC. 2008. Mating system and pollen gene flow in Mediterranean maritime pine. Heredity 100, 390–399. ( 10.1038/sj.hdy.6801090) [DOI] [PubMed] [Google Scholar]
- 60.Durel CE, Bertin P, Kremer A. 1996. Relationship between inbreeding depression and inbreeding coefficient in maritime pine (Pinus pinaster). Theor. Appl. Genet. 92, 347–356. ( 10.1007/s001220050134) [DOI] [PubMed] [Google Scholar]
- 61.Richardson DS, Komdeur J, Burke T. 2004. Inbreeding in the Seychelles warbler: environment-dependent maternal effects. Evolution 58, 2037–2048. ( 10.1111/j.0014-3820.2004.tb00488.x) [DOI] [PubMed] [Google Scholar]
- 62.Bucci G, González-Martínez SC, Le Provost G, Plomion C, Ribeiro MM, Sebastiani F, Alía R, Vendramin GG. 2007. Range-wide phylogeography and gene zones in Pinus pinaster Ait. revealed by chloroplast microsatellite markers. Mol. Ecol. 16, 2137–2153. ( 10.1111/j.1365-294X.2007.03275.x) [DOI] [PubMed] [Google Scholar]
- 63.Reed DH, Frankham R. 2001. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55, 1095–1103. ( 10.1554/0014-3820%282001%29055%5B1095%3AHCCAMA%5D2.0.CO%3B2) [DOI] [PubMed] [Google Scholar]
- 64.Tester M, Langridge P. 2010. Breeding technologies to increase crop production in a changing world. Science 327, 818–822. ( 10.1126/science.1183700) [DOI] [PubMed] [Google Scholar]
- 65.Andersen SB. 2005. Haploids in the improvement of woody species. In Haploids in crop improvement II (eds Palmer CED, Keller WA, Kasha K), pp. 243–257. Berlin, Germany: Springer. [Google Scholar]
- 66.Sellis D, Callahan BJ, Petrov DA, Messer PW. 2011. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl Acad. Sci. USA 108, 20 666–20 671. ( 10.1073/pnas.1114573108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hedrick PW. 2012. What is the evidence for heterozygote advantage selection? Trends Ecol. Evol. 27, 698–704. ( 10.1016/j.tree.2012.08.012) [DOI] [PubMed] [Google Scholar]
- 68.Merilä J, Sheldon BC. 1999. Genetic architecture of fitness and non-fitness traits: empirical patterns and development of ideas. Heredity 83, 103–109. ( 10.1046/j.1365-2540.1999.00585.x) [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Jones L, McQueen-Mason S. 2003. Expansins and cell growth. Curr. Opin. Plant Biol. 6, 603–610. ( 10.1016/j.pbi.2003.09.003) [DOI] [PubMed] [Google Scholar]
- 70.Cosgrove DJ. 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. ( 10.1038/nrm1746) [DOI] [PubMed] [Google Scholar]
- 71.Muller B, et al. 2007. Association of specific expansins with growth in maize leaves is maintained under environmental, genetic, and developmental sources of variation. Plant Physiol. 143, 278–290. ( 10.1104/pp.106.087494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luikart G, Pilgrim K, Visty J, Ezenwa VO, Schwartz MK. 2008. Candidate gene microsatellite variation is associated with parasitism in wild bighorn sheep. Biol. Lett. 4, 228–231. ( 10.1098/rsbl.2007.0633) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SNP data were deposited in the Dryad repository at http://dx.doi.org/10.5061/dryad.8d6k1. Survival data have been deposited in GENFORED, the Spanish Network of Genetic Trials (http://www.genfored.es).