Abstract
Self-fertilization (selfing) favours reproductive success when mate availability is low, but renders populations more vulnerable to environmental change by reducing genetic variability. A mixed-breeding strategy (alternating selfing and outcrossing) may allow species to balance these needs, but requires a system for regulating sexual identity. We explored the role of DNA methylation as a regulatory system for sex-ratio modulation in the mixed-mating fish Kryptolebias marmoratus. We found a significant interaction between sexual identity (male or hermaphrodite), temperature and methylation patterns when two selfing lines were exposed to different temperatures during development. We also identified several genes differentially methylated in males and hermaphrodites that represent candidates for the temperature-mediated sex regulation in K. marmoratus. We conclude that an epigenetic mechanism regulated by temperature modulates sexual identity in this selfing species, providing a potentially widespread mechanism by which environmental change may influence selfing rates. We also suggest that K. marmoratus, with naturally inbred populations, represents a good vertebrate model for epigenetic studies.
Keywords: selfing, mate availability, mixed mating, inbreeding, DNA methylation, mangrove killifish
1. Introduction
Self-fertilization (selfing) has been an evolutionary enigma ever since Darwin [1]. Selfing is a particularly cost-effective breeding mode compared with outcrossing (bi-parental reproduction) [2]. The ability of hermaphrodites to reproduce without a mate avoids the cost and uncertainty of gonochoric reproduction, and should be particularly advantageous when mate availability is low [3]. Conversely, outcrossing preserves populations' allelic diversity and heterozygosity, helps to confer resilience against future epidemiological or environmental challenges. Many hermaphrodite animals and plants that are able to self-fertilize seem to balance the contrasting needs for reproductive assurance (favouring selfing) and allelic diversity (favoured by outcrossing) by operating a mixed-mating strategy where selfing and outcrossing occur at variable rates [4]. Thus, in mixed-mating species the main genetic consequences of selfing (i.e. accumulation of deleterious mutations, reduced rate of adaptation, inbreeding depression and loss of heterozygosity) can be counteracted by the relative advantages of outcrossing, such as greater offspring plasticity and genetic variability [5]. Species with a mixed-mating model could thereby circumvent the issue of inbreeding depression as a critical evolutionary constraint of self-fertilization [6]. However, selfing rates are very variable in natural populations, and non-genetic and ecological factors seem to influence them [7]. Among the non-genetic factors influencing selfing, mate availability and the need for reproductive assurance have received particular attention [4,8], although the regulatory mechanism remains unclear [9]. Mate availability plays an important role in regulating selfing; in cestodes and nematodes, low population density induces delayed selfing [10,11], whereas stress alters both sex ratios and outcrossing rates [12]. Yet the mechanisms that modulate the sex ratios of mixed-mating species in the face of changes to population structure and the environment remain elusive.
Epigenetic modifications (such as DNA methylation or histone modification) are influenced by fluctuations in the environment and mediate changes in gene expression that contribute to phenotypic diversity in populations without altering the underlying genetic code [13]. Changes in temperature can alter DNA methylation in plants at specific genomic loci [14], and in animals, dietary regimes have been shown to affect DNA methylation and therefore expression of specific genes [15]. Epigenetic modifications thereby can create phenotypic variability in asexual and selfing organisms that, without involving DNA mutations, allows them to adapt to the environmental change [16]. In addition, there is a relationship between inbreeding depression and DNA methylation that suggests that epigenetic modulation may influence the magnitude of environment-dependent inbreeding depression [17,18], and therefore could contribute to polymorphism in selfing rates [19]. We therefore hypothesized that in species with mixed mating, epigenetic control systems could play an important role in determining sex ratios, and hence the balance between selfing and outcrossing rates. Kryptolebias marmoratus (the mangrove killifish) is an androdioecious fish with a mixed-mating reproductive strategy that is moderated by temperature [20]. Here, we compare sex ratios and methylation patterns in isogenic lines under different environmental conditions (incubation temperatures) to test the hypothesis that DNA methylation plays a role in determining temperature-dependent sex ratios, and ultimately influences selfing rates. In this and other androdioecious species, hermaphrodites coexist with males, with the latter being usually present at low frequencies. Hermaphrodites are able to self-fertilize but cannot outcross, so the frequency of males is critical in determining outcrossing rates. The ability of K. marmoratus to self-fertilize makes it easy to control for genetic variation when studying epigenetic effects. Its natural populations live under highly variable environmental conditions and, although they predominantly consist of highly homozygous selfing hermaphrodites, the presence of males has been observed in some populations at proportions varying from 1% to 20% [21]. Factors determining the frequency of males in natural populations of K. marmoratus are not well known, but the proportion of males can be manipulated by modifying egg incubation temperature (lower incubation temperature increases male percentage) [22]. Although selfing is the predominant mode of reproduction, outcrossing is common in the populations with the highest proportion of males, resulting in high individual heterozygosity and low population linkage disequilibria that contrast with the patterns of high homozygosity characteristic of populations with the lowest percentage of males [21]. We have previously demonstrated that outcrossing with males increases genetic diversity and parasite resistance in the offspring compared with selfed progeny [23,24], suggesting a potential adaptive value of maintaining males for outcrossing. In addition, males show a behavioural preference for hermaphrodites that are genetically dissimilar to them, a potential mechanism to increase genetic diversity in this species through outcrossing [25].
2. Material and methods
(a). Sex ratios of Kryptolebias marmoratus at different incubation temperatures
The study fish had two different selfing backgrounds: the R strain was derived originally from Belize in the early 1990s, and the DAN strain was also derived from Belize but in the early 2000s [26]. These strains were chosen due to known differences in their levels of genotypic diversity (i.e. DAN strain has higher levels of individual and within-strain heterozygosity than R) [24]. A total of 240 eggs (30 from each strain and temperature) were collected on the day of oviposition and incubated at four different temperatures between 18°C and 25°C, within the natural temperature range experienced by this species. Eggs were allowed to develop for 750 degree-days before being hatched by dechorionation. Newly hatched fry were then reared under standard growing conditions of 25°C and a 12 L : 12 D cycle. Fish were euthanized according to Home Office Schedule 1 methods 60 days post-hatching, and fish heads were removed and preserved in ethanol for brain tissue dissection. Sexual identity (male or hermaphrodite) was determined by histological examination of the gonads. Logistic regression was used to analyse variation in sex ratios (proportion of males) in relation to selfing strain and temperature.
(b). Genome-wide methylation analysis
Genome-wide DNA methylation was assessed in 79 fish from both genetic strains incubated at the two temperature extremes (18°C and 25°C). Total genomic DNA was extracted from whole brains, and analysed for differential patterns of DNA methylation using a modification of the methylation-sensitive amplified fragment length polymorphism (MS-AFLP) method [27]. A single organ was chosen in order to minimize tissue-specific differences in methylation. Genomic DNA was cleaved using restriction enzymes HpaII + EcoRI or restriction enzymes MspI + EcoRI in two separate reactions. HpaII and MspI vary in their sensitivity to methylation. Comparison of the two restriction profiles for each individual allowed assessment of the methylation state of the restriction sites in K. marmoratus. The four possible types of variation were pooled in methylated and non-methylated restriction sites [28]. For the selective amplification step, the combination of primers used was HpaII + CA (5′-GATGAGTCTAGAACGGCA-3′)/EcoRI + AAA (5′-GACTGCGTACCAATTCAAA). Fragments were run on an ABI PRISM 3100 (Applied Biosystems) and resultant profiles were analysed using GeneMapper v. 4.0 (Applied Biosystems). Only fragments larger than 100 bp in size were considered to reduce the potential impact of size homoplasy [29]. Singleton observations were excluded from the dataset (i.e. markers with only one non-consensus sample).
We used hierarchical analyses of molecular variance (AMOVA) in GenAlEx v. 6 [30] on both enzyme combinations to compare genetic and epigenetic variances among selfing lineages (ΦRT), among sex and incubation temperature groups within selfing lineages (ΦPR), and among individuals (ΦPT). We also performed independent hierarchical AMOVAs to compare variances with the individuals grouped by sex or incubation temperatures separately. Groups with fewer than five individuals were not considered. We used 9999 permutations to estimate statistical significance, adjusting for multiple comparisons by the sequential Bonferroni method. Differences in the presence/absence MS-AFLP profiles were also explored using principal coordinate analysis (PCA) in GenAlEx v. 6 [30]. PCA was first carried out to compare methylation-sensitive (HpaII/EcoRI) and methylation-insensitive (MspI/EcoRI) fragment presence/absence profiles. The variance of the scores for the first two coordinates of profiles from HpaII/EcoRI and MspI/EcoRI was calculated and 95% CI were determined using jackknifing [31]. PCA was then used to analyse epigenetic variation between groups (separated by sex, temperature and lineage) from the combined presence/absence profiles from HpaII enzyme combinations. In addition, to assess the statistical significance in the differences of principal coordinate scores between genetic strains, sexes and temperatures, generalized linear models (GLMs) were implemented on the scores of the first two coordinates, with individual as random effect. The initial models (where 1st or 2nd coordinate score was the dependent variable) used all factors (sex, temperature and genotype, where genotype represented the MspI scores of the first principal component) as independent variables plus all two-term interactions. Model selection was then carried out based on the Akaike information criterion corrected for small sample size [32]. All statistical modelling was carried out using R v. 2.11.0 with the MuMIn package [33].
(c). Detection of differentially methylated regions using methyl-capture sequencing (MethylCap-seq)
Genomic DNA was isolated from a male (18°C incubation) and a hermaphrodite (25°C incubation) from the R selfing line, sheared to 200–400 bp by sonication and enriched for methylated DNA using MethylMiner Methylated DNA Enrichment Kit (Invitrogen Inc., Carlsbad, CA, USA). Following methyl capture, the recovered DNA was diluted to 0.2 ng µl−1 and used to create a Nextera XT library (Illumina Inc.) for each sample. Libraries were uniquely indexed for multiplexing on the Illumina MiSeq NGS platform. Insert size was 200–500 bp. Libraries were pooled at 10 nM and diluted to 8 pM for sequencing. The sequencing run produced paired-end 2 × 150 bp reads that were automatically trimmed to remove adaptors. DNA was also isolated from a single R hermaphrodite and sequenced in a similar way to provide a reference draft sequence for the alignment of the methyl-captured DNA. As the genome sequence for K. marmoratus is not yet available, resulting sequences were aligned to the published genome from tilapia (Oreochromis niloticus; www.ensembl.org/Oreochromis_niloticus). Read information was extracted from BAM files and methylation levels across the genome were extrapolated from the sequencing coverage for each region (i.e. more coverage indicates higher level of methylation).
Two approaches with different stringencies were used to determine the significance of the observed methylation differences. First, differentially methylated regions (DMRs) were calculated for each window by simple subtraction of each (normalized) sample count. Differential methylation was deemed significant when the calculated difference in methylation between both samples was 2 (p < 0.01) or 3 standard deviations away from the mean (p < 0.001). Loci counts were calculated based on 1 857 793 500 bp non-overlapping sliding windows covering the entire aligned genome. Read counts were normalized to window size (reads per base) to account for windows smaller than 500 bp. Windows with no counts for either sample (819 840) were discarded. Normalized counts for both samples were plotted to identify the correlation between the loci count from each sample. Second, an additional analysis with a higher level of stringency was used to identify significant differentially methylated regions. Analysis was carried out using a model-based peak-calling algorithm previously used to analyse methyl capture data [33]. Model-based analysis of ChIP-Seq (MACS [34]) was run on both samples, using default parameters, p-value and q-value cut-offs, and an effective genome size of 9.27 × 108. DMRs resulting from both approaches were inspected for the presence of K. marmoratus sex-determining related sequences [35] aligned to tilapia with a minimum BLAST bit score of 192, in particular: cyp19a (DQ339107.1), ER alpha (AB251458.1), ER beta (AB251457), aromatase B (AB251459), Sox9a (DQ683739.1), Sox9b (DQ683740.1), Sox9c (DQ683741.1), figalpha (DQ683743.1), dmrt1 (DQ683742.1), foxl2 (DQ683738.1) and GnRHR (DQ996268.2).
(d). Validation of putative DMRs using high-resolution melting analysis
We then used methylation-sensitive high-resolution melting (MS-HRM) analysis to validate the observed differences in methylation between males and hermaphrodites in three of the genes: cyp19a, Sox9a and dmrt1. For this analysis, DNA from 17 individuals from the R strain—males (five) and hermaphrodites (five) incubated at low temperature (18°C), and males (two) and hermaphrodites (five) incubated at high temperature (25°C)—was extracted, and sodium bisulfite treated using the EZ DNA Methylation-Gold kit (Zymo Research) according to the manufacturer's instructions. Converted DNA concentration was then assessed using a NanoDrop 1000 spectrophotometer with the RNA setting. PCRs for cyp19a and dmrt1 were carried out using primers designed for Japanese flounder as described by Shao et al. [36]. PCRs for Sox9a were carried out using primers designed for alligator by Parrott et al. [37]. PCR conditions for the Sox9a and dmrt1 promoters were as follows: 5 min at 95°C; five cycles of 20 s at 95°C, 15 s at 55°C, 40 s at 68°C; 30 cycles of 15 s at 95°C, 30 s at 53°C and 40 s at 68°C. The cyp19a promoter was analysed by nested PCR with external cycle as follows: 95°C for 5 min, followed by seven cycles of 10 s at 95°C, 60 s at 55°C, 40 s at 72; 35 cycles at 95°C for 30 s, 52°C for 30 s, 72°C for 30 s and final extension at 72°C for an additional 10 min period. Nested PCR conditions for cyp19a were: 5 min at 95°C; 40 cycles of 30 s at 95°C, 60 s at 50°C, 60 s at 72°C. After PCR, resultant fragments were directly subjected to the same HRM conditions. In brief, melting curve analysis was conducted on the Rotor-Gene 6000 v. 1.7 software (Qiagen, UK) using the Cycling A-Green channel. During HRM, temperature was increased from 50 to 90°C in 0.1°C incremental steps, with each step held for 2 s. Using Rotor-Gene 6000 software, melting curves were normalized by calculation of the ‘line of best fit’ between two normalization regions selected before and after the major decrease in fluorescence (representing the ‘fragment melting’). Comparisons were made between sex/temperature DNA samples in terms of Tm or by a combination of Tm and altered curve shape.
(e). Analysis of expression of differentially methylated sex determination genes
A total of 17 fish were selected for RNA extraction: males (five) and hermaphrodites (five) incubated at low temperature, and males (two) and hermaphrodites (five) incubated at high temperature. RNA was extracted with the ISOLATE II RNA Mini Kit (Bioline UK). Fish were euthanized following Schedule 1 using benzocaine; the head of each fish was removed and transferred into 350 µl lysis buffer RLY, and kept on ice. Samples were homogenized using a micro pestle, 3.5 µl of β-mercaptoethanol were added and the rest of the extraction followed the manufacturer's instructions. RNA was eluted in 60 µl of RNase-free water and 2 µl were used for quantification using the NanoDrop 2000. In order to validate the methylation results, the expression profiles of two genes (sox9a and cyp19a) were analysed by reverse transcription quantitative PCR (RT-qPCR) using the primers and TM conditions detailed in [35]. The reactions were performed in duplicate per sample using the SensiFAST SYBR No-ROX One Step kit (Bioline Ltd, UK) in a Bio Rad CFX 96 Real-Time System. Reaction mixes of 20 µl were prepared following the manufacturer's instructions with 2 µl of extracted RNA. 18S rRNA was used as standard. PCR efficiency was estimated by linear regression analysis of the logarithm of SYBR Green fluorescence versus cycle number. We then used Pfaffl's method [38] to estimate the ratio of gene expression of each sample relative to that of the most common phenotype, used as calibrator (hermaphrodites incubated at high temperature). Normalization was carried out against the 18S rRNA housekeeping gene. In addition, for each experimental group we estimated the ΔCt of each gene normalized relative to the standard and then compared them using a Kruskal–Wallis test [39] in Systat v. 11.
3. Results
(a). Sex ratios
In the DAN strain, the proportion of males ranged from 0% at 25°C to 83.33% at 18°C. By contrast, the R strain yielded 6.66% males at 25°C but only 50.00% at 18°C. Logistic regression showed both genetic strain (p = 0.003) and temperature (p < 0.001) to be significant predictors of the probability of being male (electronic supplementary material, table S1). We also found a significant interaction (p = 0.006) between temperature and strain (figure 1).
Figure 1.
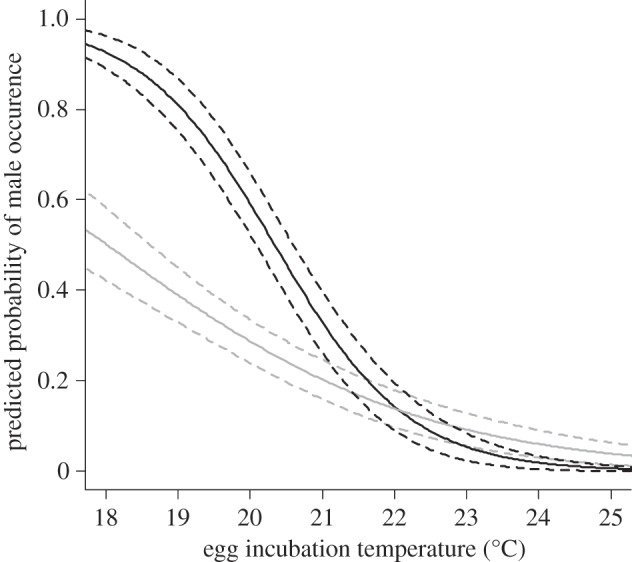
Logistic regression model predicting the probability of producing primary males across a range of egg incubation temperatures. Black lines represent DAN strain and grey lines represent R strain. Dotted lines indicate the 95% CI bands.
(b). Genome-wide analysis of temperature induced methylation changes using MS-AFLP
The MS-AFLP analysis of the 77 samples (from two strains at four incubation temperatures: DAN: 13 hermaphrodites at 25°C and 5 at 18°C, 14 males at 18°C; R: 13 hermaphrodites at 25°C and 15 at 18°C, 4 males at 25°C and 13 at 25°C) yielded 105 scorable polymorphic fragments, 91 generated by MspI (restriction enzyme insensitive to methylation) and 76 by HpaII (sensitive to methylation) (63 common to both enzymes; electronic supplementary material, table S2 and figure S1). AMOVA analysis of the MspI profiles indicated that 89.29% of the variation was explained by differences between genetic strains (ΦRT = 0.893, p = 0.001), 0.12% by differences between groups defined by incubation temperature and sex (ΦPR = 0.011, p = 0.357), and 10.58% by individual differences (ΦPT = 0.894, p = 0.001) (electronic supplementary material, table S3). By contrast, for HpaII profiles, 44.16% of the variation was explained by differences between genetic strains (ΦRT = 0.442, p = 0.001), 2.88% by differences between temperature/sex groups (ΦPR = 0.052, p = 0.018) and 52.97% by individual differences (ΦPT = 0.470, p = 0.001), implying a methylation basis for the differences between sex and temperature groups as well as more epigenetic than genetic individual differences. HpaII AMOVAs grouping the fish by either sex or temperature within genetic strains indicated that 2.07% and 3.63% of the variance was explained by differences between sexes (ΦPR = 0.037, p = 0.034) and temperature (ΦPR = 0.064, p = 0.015), respectively (electronic supplementary material, table S3). To evaluate whether genetic strain, egg incubation temperature and/or sex produced different epigenetic profiles, PCA was used to describe and visualize the variation contained in the polymorphic MS-AFLP loci. The first two coordinates of the PCA explained 47.47% and 15.58% of variation in MS-AFLP profiles, respectively (electronic supplementary material, figure S2a). General linear modelling selection showed that for the coordinate 1 PCA scores, genotype (p < 0.001) and sex (p = 0.014) had a significant influence with an additional significant interaction between sex and genotype (p = 0.007) (table 1; electronic supplementary material, figure S2). For coordinate 2 scores, only temperature was included in the final GLM (p = 0.045).
Table 1.
Results of GLMs evaluating the effect of genotype, sex and temperature on principal coordinate analysis (PCA) scores. PCA scores derived from variation in methylation epigenotypes between samples (R and DAN males and hermaphrodites incubated at high and low temperatures), based on the presence/absence scores of 63 polymorphic MS-AFLP markers. Significant values shown in bold. Genotype represents the scores of the first principal component of the MspI PCA analysis.
| parameter | d.f. | F | p-value |
|---|---|---|---|
| coordinate 1 | |||
| genotype | 1 | 38.922 | <0.001 |
| sex | 1 | 2.512 | 0.014 |
| temperature | 1 | 0.802 | 0.425 |
| genotype × sex | 1 | 2.796 | 0.007 |
| temperature × sex | 1 | −1.620 | 0.108 |
| error | 71 | ||
| coordinate 2 | |||
| temperature | 1 | −2.045 | 0.045 |
| error | 75 | ||
(c). Characterization of putative DMRs associated to sex determination using MethylCap-seq
After sequencing and subsequent alignment, the coverage of the read mappings across the tilapia genome was calculated from a samtools ‘pileup’ file: 96.5% of the genome was covered by less than 1 read, and approximately 1% of the genome had an average coverage of more than four reads. In total, 19.65 million K. marmoratus reads of genomic DNA reads mapped to the tilapia genome of a total of 29.04 million (i.e. 67.65%). For the K. marmoratus methyl captured DNA, 4.39 million reads (male) mapped of a total of 7.24 million (60.75%), and 4.04 million reads (hermaphrodite) mapped from a total of 6.67 million (60.66%). Read numbers showed a simple linear correlation between both captured genomes (electronic supplementary material, figure S5).
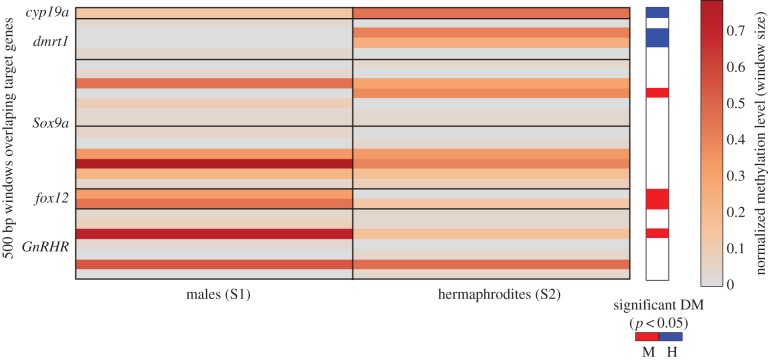
A total of 18 225 windows across the compared genomes presented a significantly different normalized number of reads (p < 0.001) and were considered DMRs. We then analysed genome locations where 11 K. marmoratus sequences related to sex determination or sex differentiation [35] aligned to tilapia with a minimum BLAST bit score of 192 (electronic supplementary material, figure S3). Six of these sequences were present among the methylated-enriched sequences and overlapped with DMRs. These included homologues of sex determination genes, namely cyp19a (GL831420.1 on the tilapia genome), Sox9a (two different loci on the tilapia genome GL831217.1 and GL831136.1), dmrt1 (locus GL831221 on the tilapia genome), foxl2 (locus GL831570 on the tilapia genome) and gonadotropin-releasing hormone receptor (GnRHR) (locus GL831133 on the tilapia genome) (figure 2). A further 35 894 windows were significant DMRs at p < 0.01. Using the MACS peak-calling algorithm [34], 1837 peaks with a statistically significant different number of reads were called between the two samples (827 hypermethylated in males and 1010 hypermethylated in hermaphrodites). Of these, 1023 were located in gene regions, while 55 overlapped with promoter regions (defined as 1.5 kb before the transcription start site) according to tilapia annotation Oreni1.0 (ensembl.org/Oreochromis_niloticus/Info/Annotation) (electronic supplementary material, table S4). Observed methylation level differences in these DMRs ranged from twofold to almost 300-fold (electronic supplementary material, figure S4).
Figure 2.
Heatmap of normalized (per window) DNA methylation levels of target genes in male and hermaphrodite samples. The sex showing significantly higher level of methylation (p < 0.05) on 500 bp windows overlapping with a DMR is indicated on the left of the heatmap (red indicates higher levels of methylation on males and blue on hermaphrodites). (Online version in colour.)
(d). Validation of putative DMRs using high-resolution melting analysis
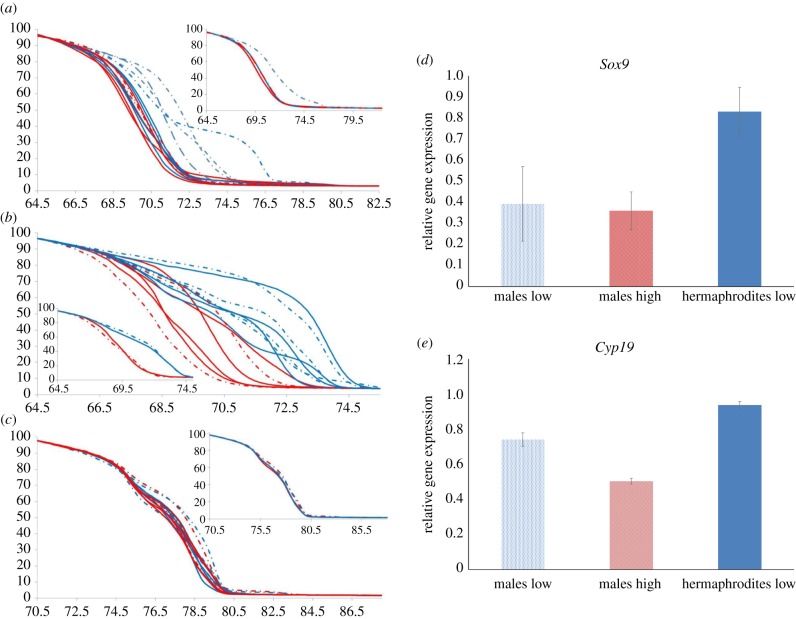
We then used methylation-sensitive MS-HRM analysis to validate the methylated status of three of the genes (cyp19a, Sox9a and dmrt1) in 17 individuals from the R strain from all experimental groups. These genes had been previously identified as being involved in sex reversal regulated by temperature and controlled by DNA methylation [36,40]. We found significantly higher levels of methylation in males incubated at low temperature compared with the other three groups (hermaphrodites at low temperature and males and hermaphrodites at high temperature for Sox9a; figure 3a). Conversely, differences in methylation related to incubation temperature during embryo development but not sex in cyp19a (figure 3b). Finally, HRM analysis for dmrt1 did not display methylation differences between sexes or temperatures (figure 3c).
Figure 3.
(a–c) Determination of gene methylation level using MS-HRM analysis. Normalized melting curves for (a) Sox9a, (b) cyp19a and (c) dmrt1 promoters in hermaphrodite (solid line) and male (dotted line) fish incubated at 25°C (red) or 18°C (blue) from a single selfing line (R). Small squares represent the average melting curves for each sex/temperature combination. (d,e) Determination of expression using RT-qPCR. Relative expression of (d) Sox9a and (e) cyp19a genes in males (dotted bar) and hermaphrodites (filled bar) incubated at 25°C (red) or 18°C (blue) using 18S rRNA as endogenous control and normalized against the expression value of the most common phenotype in nature (hermaphrodites incubated at 25°C). (Online version in colour.)
(e). Analysis of temperature-induced differential expression of differentially methylated sex determination genes
Relative expression of differentially methylated genes sox9a and cyp19a determined by quantitative RT-qPCR indicated differences in relative expression for sox9a with males at both temperatures showing lower expression in relation to all hermaphrodites (figure 3d). Based on the ΔCt values, the differences among the four groups were significant (KW = 11.7, p = 0.008), and paired tests indicated that there were significant differences between males and hermaphrodites incubated at low temperatures (Mann–Whitney U = 6.8, p = 0.009), hermaphrodites incubated at high temperature, and males at low (U = 6.0, p = 0.014) and high temperatures (U = 3.4, p = 0.000), but not between hermaphrodites incubated at different temperatures (U = 1.5, p = 0.221) or males incubated at different temperatures (U = 0.6, p = 0.439). By contrast, the differences in expression of cyp19a were less clear, males incubated at both temperatures displayed lower relative expression with males at high temperature presenting the lowest levels of expression (figure 3e), but the ΔCt values indicated no significant differences among the four groups (KW = 3.1, p = 0.376).
4. Discussion
Taken together, our results show that temperature-related variation in the sex ratio of the mixed-mating fish K. marmoratus related to changes in methylation patterns. Outcrossing between males and hermaphrodites in this species results in offspring with increased genetic diversity and lower parasite loads compared with selfed offspring [23,24]. Therefore, changes in the proportion of males are likely to influence outcrossing rates, offspring genetic diversity and ultimately population fitness. Differences in the proportion of males found in natural populations of K. marmoratus are attributed to environment-dependent sex determination [41], with population sex ratios controlled by local environmental conditions [42], but a regulatory mechanism has thus far not been described. However, the proportion of males varies significantly between selfing lines even under constant rearing conditions, suggesting a genetic component of sex determination in this species [43]. The combination of environmental and genetic factors in sex regulation, a mechanism more common to fish than previously recognized [44], seems the most plausible explanation for our results. Epigenetic regulation of gene expression during gonad differentiation is involved in the adaptive sex reversal observed in some fish and other organisms with genetic sex determination [45]. For example, methylation has been related to sexual reversal in relation to incubation temperature in the half-smooth tongue sole [36]. This is because, although also linked to genomic imprinting [46], transposon immobilization [47] and suppression of transcriptional noise [48], the major biological consequence of DNA methylation throughout plants, fungi and animals is gene silencing [49]. Thus, transcription of sex-related genes can be directly inhibited by methylated cytosine bases that preclude the association of DNA transcription factors [50] or indirectly mediated by methyl-CpG binding proteins [51]. DNA methylation states are affected by the environment, providing a potential link between phenotypic variation and genotype–environment interaction [52]. Epigenetic mechanisms, especially DNA methylation, may be critical factors in sex determination and reproductive development of certain plants and animals [53], and K. marmoratus, because of its life history and genetic architecture, represents a particularly good model for epigenetic studies. We have identified a number of genes involved in temperature sex regulation potentially modulated by DNA methylation in the mangrove killifish. We tested three of them (cyp19a, sox9a and dmrt1) using three methods (MethylCap-seq, well suited to assess relative differences between samples [54] particularly when used in combination with next-generation sequencing [55], MS-HRM and RT-qPCR of total RNA), and found evidence suggesting that two of them (cyp19a, sox9a) could be involved in modulating K. marmoratus sex ratios in response to environmental change during embryo development. However, although methylation changed significantly at a local level, these changes were not always in the same direction (i.e. all DMRs showing higher methylation levels in males when compared with hermaphrodites, or vice versa). Therefore, no significant global changes in methylation levels were observed across the studied genomes. Differential DNA methylation of promoters can suppress the transcription of either male or female specific loci, thus determining an individual's sex [56]. The inactivation of one X chromosome in female mammals [46] is a well-known example of epigenetic control, and there are indications that there is a similar mechanism in some dioecious plants [57]; experimental modification of DNA methylation in plants has also been shown to induce sex reversal [58]. For some time DNA methylation was hypothesized to be involved in sex determination of fish [59], and more recently methylation of the gonadal aromatase promoter (cyp 19a) has been related to the regulation of temperature-mediated sex ratios in two fish with environmental and genetic sex determination, the European sea bass (Dicentrarchus labrax) [40] and the half-smooth tongue sole (Cynoglossus semilaevis) [36], suggesting that DNA methylation is indeed a crucial mechanism linking environmental temperature and sex determination in some fish species.
The duplicated sox9a and sox9b are critical for testis differentiation in fish mammals, but also seem to be highly expressed in the brain [60], particularly sox9a. We found that sox9a was hypermethylated in K. marmoratus males incubated at low temperature and the expression of the gene was lower in males than in hermaphrodites, providing for the first time an explanation for the increase of males at low incubation temperatures. We also found that the expression of cyp19a was downregulated in the brain of males incubated at 25°C, with lower levels of methylation at higher temperatures in both sexes. This contradicts previous studies in this species, where cyp19a was downregulated in fish incubated at 20°C [35], but coincides with those in sea bass where high incubation temperatures result in higher methylation of the promoter and decreased expression of this gene in males [40]. Expression levels of cyp19a vary temporally and between tissues during fish embryogenesis [40], and possibly in relation to the genotype in K. marmoratus [35]. In contrast with mammals, adult teleost fish display a very intense expression of aromatase genes (cyp19a1a and cypa19a1b) in the brain, but less is known regarding differences in brain aromatase activities between sexes [61]. A more detailed temporal study including different tissues may be needed to fully understand the role of methylation in the regulation of this gene in K. marmoratus under different environmental conditions. Finally, we found that dmrt1 was differentially methylated between male and hermaphrodite in the MethylCap-seq analysis, but the MS-HRM results did not show any differences between any of the sex/temperature groups, possibly reflecting individual differences that could not be taken into account with the initial sequencing.
Sex ratios also vary with environmental conditions in other mixed-mating species; for example, the proportion of males increases in Caenorhabditis elegans under stressful conditions [62], and populations that strictly outcross with males show a clear fitness advantage with respect to obligate selfed populations, displaying better ability to adapt to the environmental change [62,63]. Thus, males may play an important adaptive role in mixed-mating species where hermaphrodites can self-fertilize but not outcross with other hermaphrodites. The environmental instability of the K. marmoratus habitat [64], widely different availability of males in different areas (less than 1% to greater than 20%) and the uniform distribution of isogenic lineages, without evidence of high local frequencies [65], suggest that reproductive assurance [66] may be the main adaptive advantage of selfing for K. marmoratus, as for other selfing species [65].
Our results serve to illustrate the role of an epigenetic mechanism (methylation) in modulating sex ratios in a selfing species, and provide a potential mechanistic explanation for the influence of the environment in maintaining variable selfing rates by modulating mate availability. Methylation has also been associated with environment-dependent inbreeding depression [18], which in turn can result in variable selfing rates. We suggest that this form of epigenetic modification could provide a mechanism of control of sexual identity in species exhibiting mixed-mating systems, and thereby a means by which environmental change may directly influence the balance between selfing and outcrossing, optimizing population resilience in the light of inbreeding depression, mate availability and environmental instability.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr Tina Bianco-Miotto of the University of Adelaide for support and reagents for HRM analysis. Carlos Garcia de Leaniz, Karl Hoffmann, Stuart Campbell, Kelly Zamudio and Zamudio laboratory members provided very valuable discussion and comments.
Ethics
All fish experimentation was carried out following the Association for the Study of Animal Behaviour/Animal Behavior Society Guidelines for the Use of Animals in Research [67], in consultation with the NACWO officer of Aberystwyth University.
Data accessibility
Sequences were archived under SRA project accession no. SRP033207.
Competing interests
We have no competing interests.
Funding
Funding was provided by a Waitt grant from the National Geographic Society (W76-09) to S.C. and an IBERS PhD scholarship to A.E.
References
- 1.Darwin CR. 1876. The effects of cross and self-fertilization in the vegetable kingdom. London, UK: John Murray. [Google Scholar]
- 2.Fisher RA. 1941. Average excess and average effect of a gene substitution. Ann. Eugenet. 11, 53–63. ( 10.1111/j.1469-1809.1941.tb02272.x) [DOI] [Google Scholar]
- 3.Herlihy CR, Eckert CG. 2002. Genetic cost of reproductive assurance in a self-fertilizing plant. Nature 416, 320–323. ( 10.1038/416320a) [DOI] [PubMed] [Google Scholar]
- 4.Jarne P, Auld JR. 2006. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 60, 1816–1824. ( 10.1554/06-246.1) [DOI] [PubMed] [Google Scholar]
- 5.Prahlad V, Pilgrim D, Goodwin EB. 2003. Roles for mating and environment in C. elegans sex determination. Science 302, 1046–1049. ( 10.1126/science.1087946) [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. ( 10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 7.Eckert CG, et al. 2010. Plant mating systems in a changing world. Trends Ecol. Evol. 25, 35–43. ( 10.1016/j.tree.2009.06.013) [DOI] [PubMed] [Google Scholar]
- 8.Busch JW, Delph LF. 2012. The relative importance of reproductive assurance and automatic selection as hypotheses for the evolution of self-fertilization. Ann. Bot. 109, 553–562. ( 10.1093/aob/mcr219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheptou PO, Donohue K. 2012. Epigenetics as a new avenue for the role of inbreeding depression in evolutionary ecology. Heredity 110, 205–206. ( 10.1038/hdy.2012.66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsitrone A, Jarne P, David P. 2003. Delayed selfing and resource reallocations in relation to mate availability in the freshwater snail Physa acuta. Am Nat 162, 474–488. ( 10.1086/376889) [DOI] [PubMed] [Google Scholar]
- 11.Schjørring S. 2009. Delayed selfing in relation to the availability of a mating partner in the cestode schistocephalus solidus. Evolution 58, 2591–2596. ( 10.1111/j.0014-3820.2004.tb00887.x) [DOI] [PubMed] [Google Scholar]
- 12.Morran LT, Cappy BJ, Anderson JL, Phillips PC, Schoen DJ. 2009. Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution 63, 1473–1482. ( 10.1111/j.1558-5646.2009.00652.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver B, Christina LR, Massimo P. 2008. Epigenetics for ecologists. Ecol. Lett. 11, 106–115. [DOI] [PubMed] [Google Scholar]
- 14.Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H. 2002. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 277, 37 741–37 746. ( 10.1074/jbc.M204050200) [DOI] [PubMed] [Google Scholar]
- 15.Cooney CA, Dave AA, Wolff GL. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132, 2393S–2400S. [DOI] [PubMed] [Google Scholar]
- 16.Castonguay E, Angers B. 2012. The key role of epigenetics in the persistence of asexual lineages. Genet. Res. Int. 2012, 1–9. ( 10.1155/2012/534289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox CW, Reed DH. 2011. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution 65, 246–258. ( 10.1111/j.1558-5646.2010.01108.x) [DOI] [PubMed] [Google Scholar]
- 18.Vergeer P, Wagemaker N, Ouborg NJ. 2012. Evidence for an epigenetic role in inbreeding depression. Biol. Lett. 8, 798–801. ( 10.1098/rsbl.2012.0494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheptou PO, Mathias A. 2001. Can varying inbreeding depression select for intermediary selfing rates? Am. Nat. 157, 361–373. ( 10.1086/319320) [DOI] [PubMed] [Google Scholar]
- 20.Taylor DS. 2012. Twenty-four years in the mud: what have we learned about the natural history and ecology of the mangrove rivulus, Kryptolebias marmoratus? Integr. Comp. Biol. 52, 724–736. ( 10.1093/icb/ics062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackiewicz M, Tatarenkov A, Taylor DS, Turner BJ, Avise JC. 2006. Extensive outcrossing and androdioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc. Natl Acad. Sci. USA 103, 9924–9928. ( 10.1073/pnas.0603847103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington RWJ. 1967. Environmentally controlled induction of primary male gonochorists from eggs of the selffertilizing hermaphroditic fish, Rivulus marmoratus Poey. Biol. Bull. 132, 174–199. ( 10.2307/1539887) [DOI] [PubMed] [Google Scholar]
- 23.Ellison A, Cable J, Consuegra S. 2011. Best of both worlds? Association between outcrossing and parasite loads in a selfing fish. Evolution 65, 3021–3026. ( 10.1111/j.1558-5646.2011.01354.x) [DOI] [PubMed] [Google Scholar]
- 24.Ellison A, Allainguillaume J, Girdwood S, Pachebat J, Peat KM, Wright P, Consuegra S. 2012. Maintaining functional major histocompatibility complex diversity under inbreeding: the case of a selfing vertebrate. Proc. R. Soc. Lond. B 279, 5004–5013. ( 10.1098/rspb.2012.1929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellison A, Jones J, Inchley C, Consuegra S. 2013. Choosy males could help explaining androdioecy in a selfing fish. Am. Nat. 181, 855–862. ( 10.1086/670304) [DOI] [PubMed] [Google Scholar]
- 26.Tatarenkov A, Ring BC, Elder JF, Bechler DL, Avise JC. 2010. Genetic composition of laboratory stocks of the self-fertilizing fish Kryptolebias marmoratus: a valuable resource for experimental research. PLoS ONE 5, e12863 ( 10.1371/journal.pone.0012863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez López CM, Morán P, Lago F, Espiñeira M, Beckmann M, Consuegra S. 2011. Detection and quantification of tissue of origin in salmon and veal products using methylation sensitive AFLPs. Food Chem. 4, 1493–1498. [Google Scholar]
- 28.Schjørring S, Jäger I. 2007. Incestuous mate preference by a simultaneous hermaphrodite with strong inbreeding depression. Evolution 61, 423–430. ( 10.1111/j.1558-5646.2007.00028.x) [DOI] [PubMed] [Google Scholar]
- 29.Caballero A, Quesada H, Rolan-Alvarez E. 2008. Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics 179, 539–554. ( 10.1534/genetics.107.083246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peakall ROD, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel: population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. ( 10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. 2010. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185, 1108–1118. ( 10.1111/j.1469-8137.2009.03121.x) [DOI] [PubMed] [Google Scholar]
- 32.Motulsky H, Christopoulos A. 2004. Fitting models to biological data using linear and nonlinear regression: a practical guide to curve fitting. New York, NY: Oxford University Press. [Google Scholar]
- 33.Barton K. 2012. MuMIn: multi-model inference. R package version 2.15. See http://cran.r-project.org/web/packages/MUmIn/MUmIn.pdf. [Google Scholar]
- 34.Zhang Y, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 ( 10.1186/gb-2008-9-9-r137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strykowski J. 2011. Effects of temperature on gene expression and sex determination in the mangrove rivulus, Kryptolebias marmoratus. Master's thesis, University of Maryland, College Park, MD. [Google Scholar]
- 36.Shao C, et al. 2014. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604–615. ( 10.1101/gr.162172.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parrott BB, Kohno S, Cloy-McCoy JA, Guillette LJ. 2013. Differential incubation temperatures result in dimorphic DNA methylation patterning of the SOX9 and aromatase promoters in gonads of alligator (Alligator mississippiensis) embryos. Biol. Reprod. 90, 2 ( 10.1095/biolreprod.113.111468) [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan JS, Stewart CN Jr. 2005. Real-time PCR statistics. PCR Encyclopedia 1, 101 127–101 149. [Google Scholar]
- 40.Navarro-Martin L, Vinas J, Ribas L, Diaz N, Gutierrez A, Di Croce L, Piferrer F. 2011. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7, e1002447 ( 10.1371/journal.pgen.1002447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington RW. 1968. Delimitation of the thermolabile phenocritical period of sex determination and differentiation in the ontogeny of the normally hermaphroditic fish Rivulus marmoratus Poey. Physiol. Zool. 41, 447–460. [Google Scholar]
- 42.Weibel AC, Dowling TE, Turner BJ. 1999. Evidence that an outcrossing population is a derived lineage in a hermaphroditic fish (Rivulus marmoratus). Evolution 53, 1217–1225. ( 10.2307/2640824) [DOI] [PubMed] [Google Scholar]
- 43.Turner BJ, Fisher MT, Taylor DS, Davis WP, Jarrett BL. 2006. Evolution of ‘maleness’ and outcrossing in a population of the self-fertilizing killifish, Kryptolebias marmoratus. Evol. Ecol. Res. 8, 1475–1486. [Google Scholar]
- 44.Ospina-Alvarez N, Piferrer F. 2008. Temperature-dependent sex determination in fish revisited: prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 3, e2837 ( 10.1371/journal.pone.0002837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manolakou P, Lavranos G, Angelopoulou R. 2006. Molecular patterns of sex determination in the animal kingdom: a comparative study of the biology of reproduction. Reprod. Biol. Endocrinol. 4, 59 ( 10.1186/1477-7827-4-59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaenisch R. 1997. DNA methylation and imprinting: why bother? Trends Genet. 13, 323–329. ( 10.1016/S0168-9525(97)01180-3) [DOI] [PubMed] [Google Scholar]
- 47.Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340. ( 10.1016/S0168-9525(97)01181-5) [DOI] [PubMed] [Google Scholar]
- 48.Bird AP. 1995. Gene number, noise reduction and biological complexity. Trends Genet 11, 94–100. ( 10.1016/S0168-9525(00)89009-5) [DOI] [PubMed] [Google Scholar]
- 49.Colot V, Rossignol J. 1999. Eukaryotic DNA methylation as an evolutionary device. Bioessays 21, 402–411. () [DOI] [PubMed] [Google Scholar]
- 50.Watt F, Molloy PL. 1988. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2, 1136–1143. ( 10.1101/gad.2.9.1136) [DOI] [PubMed] [Google Scholar]
- 51.Boyes J, Bird A. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134. ( 10.1016/0092-8674(91)90267-3) [DOI] [PubMed] [Google Scholar]
- 52.Richards EJ. 2006. Inherited epigenetic variation—revisiting soft inheritance. Nat. Rev. Genet. 7, 395–401. ( 10.1038/nrg1834) [DOI] [PubMed] [Google Scholar]
- 53.Spielman M, Vinkenoog R, Dickinson HG, Scott RJ. 2001. The epigenetic basis of gender in flowering plants and mammals. Trends Genet. 17, 705–711. ( 10.1016/S0168-9525(01)02519-7) [DOI] [PubMed] [Google Scholar]
- 54.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Wang J. 2010. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 52, 203–212. ( 10.1016/j.ymeth.2010.04.009) [DOI] [PubMed] [Google Scholar]
- 55.Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ. 2011. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6, 34–44. ( 10.4161/epi.6.1.13313) [DOI] [PubMed] [Google Scholar]
- 56.Gorelick R. 2003. Evolution of dioecy and sex chromosomes via methylation driving Muller's ratchet. Biol. J. Linn. Soc. 80, 353–368. ( 10.1046/j.1095-8312.2003.00244.x) [DOI] [Google Scholar]
- 57.Vyskot B, Araya A, Veuskens J, Negrutiu I, Mouras A. 1993. DNA methylation of sex chromosomes in a dioecious plant, Melandrium album. Mol. Gen. Genet. 239, 219–224. ( 10.1007/bf00281621) [DOI] [PubMed] [Google Scholar]
- 58.Janoušek B, Široký J, Vyskot B. 1996. Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Mol. Gen. Genet. 250, 483–490. ( 10.1007/bf02174037) [DOI] [PubMed] [Google Scholar]
- 59.Martin CC, McGowan R. 1995. Genotype-specific modifiers of transgene methylation and expression in the zebrafish, Danio rerio. Genet. Res. 65, 21–28. ( 10.1017/S0016672300032973) [DOI] [PubMed] [Google Scholar]
- 60.Böhne A, Heule C, Boileau N, Salzburger W. 2013. Expression and sequence evolution of aromatase cyp19a1 and other sexual development genes in East African cichlid fishes. Mol. Biol. Evol. 30, 2268–2285. ( 10.1093/molbev/mst124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diotel N, et al. 2010. Aromatase in the brain of teleost fish: expression, regulation and putative functions. Front. Neuroendocrinol. 31, 172–192. ( 10.1016/j.yfrne.2010.01.003) [DOI] [PubMed] [Google Scholar]
- 62.Morran LT, Parmenter MD, Phillips PC. 2009. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462, 350–352. ( 10.1038/nature08496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morran LT, Schmidt OG, Gelarden IA, Parrish RC, Lively CM. 2011. Running with the red queen: host–parasite coevolution selects for biparental sex. Science 333, 216–218. ( 10.1126/science.1206360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellison A, Wright P, Taylor DS, Cooper C, Regan K, Currie S, Consuegra S. 2012. Environmental diel variation, parasite loads, and local population structuring of a mixed-mating mangrove fish. Ecol. Evol. 2, 1682–1695. ( 10.1002/ece3.289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avise JC, Tatarenkov A. 2012. Allard's argument versus Baker's contention for the adaptive significance of selfing in a hermaphroditic fish. Proc. Natl Acad. Sci. USA 109, 18 862–18 867. ( 10.1073/pnas.1217202109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker HG. 1955. Self-compatibility and establishment after 'long-distance' dispersal. Evolution 9, 347–349. ( 10.2307/2405656) [DOI] [Google Scholar]
- 67.Association for the Study of Animal Behaviour; Animal Behavior Society. 2006. Guidelines for the treatment of animals in behavioural research and teaching. Anim. Behav. 71, 245–253. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences were archived under SRA project accession no. SRP033207.