Abstract
Symbioses include some of the clearest cases of coevolution, but their origin, loss or reassembly with different partners can rarely be inferred. Here we use ant/plant symbioses involving three plant clades to investigate the evolution of symbioses. We generated phylogenies for the big-eyed arboreal ants (Pseudomyrmecinae), including 72% of their 286 species, as well as for five of their plant host groups, in each case sampling more than 61% of the species. We show that the ant-housing Vachellia (Mimosoideae) clade and its ants co-diversified for the past 5 Ma, with some species additionally colonized by younger plant-nesting ant species, some parasitic. An apparent co-radiation of ants and Tachigali (Caesalpinioideae) was followed by waves of colonization by the same ant clade, and subsequent occupation by a younger ant group. Wide crown and stem age differences between the ant-housing genus Triplaris (Polygonaceae) and its obligate ant inhabitants, and stochastic trait mapping, indicate that its domatium evolved earlier than the ants now occupying it, suggesting previous symbioses that dissolved. Parasitic ant species evolved from generalists, not from mutualists, and are younger than the mutualistic systems they parasitize. Our study illuminates the macroevolutionary assembly of ant/plant symbioses, which has been highly dynamic, even in very specialized systems.
Keywords: ant/plant coevolution, co-radiation, secondary colonization, molecular clocks, Pseudomyrmex, symbiosis
1. Introduction
The origin, maintenance, and breakdown of mutualisms are key questions in ecology and evolutionary biology [1–3]. Mapping traits of the mutualists and non-mutualist relatives on time-calibrated phylogenies has proved a powerful approach to unveil the temporal and geographical origin of mutualisms. A finding of co-phylogenetic studies of mutualisms is that co-speciation is rare (reviewed in [4]) and restricted to a few symbioses, especially those with vertical transmission, such as Buchnera bacterial endosymbionts and aphids [5–7]. Co-speciation in mutualistic partnerships that do not involve vertical transmission may exist in some obligate systems—for instance figs and their wasp pollinators as suggested by matching divergence times, although occasional wasp switches to other figs have been documented [8]. Other obligate mutualisms, such as the Yucca/yucca moth pollination mutualism, were found to have evolved multiple times [9,10]. Non-specialized mutualisms can exist between partners of highly discordant ages, for example, between introduced plants and native insect or bird pollinators [11]. Only species-dense molecular clock-dated phylogenies of both partner lineages therefore can elucidate the evolution of mutualistic systems. Such analyses over the past few years have revealed that cheaters rarely evolve from mutualists, contrary to theory [3].
Ant/plant symbioses involve plants with modified structures (domatia) that house ants, in return for protection or extra nutrients and sometimes also the physical or chemical removal of competing plant species [12–14]. Ant/plant symbioses appear to be younger than seed dispersal by ants or extrafloral nectary-mediated plant defence by ants, with no extant domatium-bearing clade older than 20 Myr [14]. Few co-phylogenetic studies of ant/plant systems have been conducted. In the African Leonardoxa africana, two of four subspecies have specialized domatia that were colonized in parallel by pre-adapted ant species [15,16]. Species of the Southeast Asian Crematogaster borneensis-group (former subgenus Decacrema) independently colonized three species groups of Macaranga, with an apparent matching of plant stem morphology and associated ant behaviour [17]. Co-radiation has been inferred in Pseudomyrmex and Mesoamerican Vachellia [18].
Pseudomyrmecinae comprise 230 described species in three genera [19–22], with 32 of the species living in plant domatia [14,19], making Pseudomyrmecinae the most diverse plant-occupying ant group worldwide [14]. Of the three genera, Myrcidris includes two species (one undescribed) from northern South America, Pseudomyrmex has 134 species, also confined to the New World, and Tetraponera comprises 95 species in Africa and Australasia [23]. Most species nest in dead hollow twigs of living plants, others nest only in the domatia of particular species that they protect against herbivores (figure 1), and some are parasites of other ant/plant symbioses [19,21,24,25]. Obligate domatium-nesting big-eyed ants have entered into more or less tight symbioses with species of the Fabaceae genera Vachellia, Tachigali and Platymiscium, and the Polygonaceae genera Triplaris and Ruprechtia [18,20,26,27]. This system is therefore ideal to study the evolution of ant/plant symbioses.
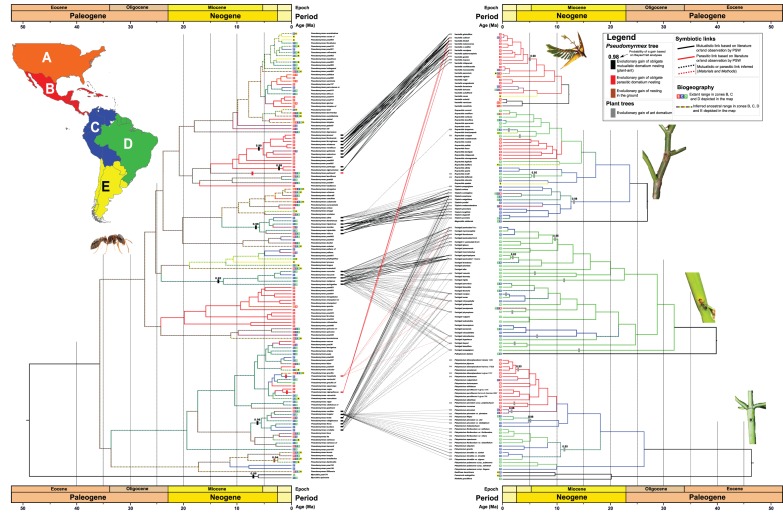
Figure 1.
Examples of Pseudomyrmex/plant symbioses. (a–c) Vachellia/Pseudomyrmex peperi symbiosis. (a) Vachellia habit with stipular thorn domatia. (b) Pseudomyrmex peperi worker feeding on the large Vachellia extrafloral nectaries. (c) Pseudomyrmex peperi collecting a protein-rich Beltian body from the Vachellia leaflet tips. (d) Triplaris americana domatium inhabited by Pseudomyrmex triplarinus. (e) Tachigali myrmecophila/Pseudomyrmex concolor-group symbiosis. (e, inset) P. concolor entering in a Tachigali myrmecophila leaf domatium. (e) Pseudomyrmex penetrator entering the leaf rachis domatium, where an entrance hole has been chewed. Photo credits: (a–c) Alexander Wild, (d) Fabian Michelangeli, (e, inset), Heraldo Vasconcelos, (e) Ricardo Solar. (Online version in colour.)
We had three expectations concerning the evolution of big-eyed ant/plant symbioses: (i) co-radiation (co-diversification) would be seen only in relatively young clades because of the increasing probability of partner loss over time, (ii) non-mutualistic domatium-nesting big-eyed ant species (i.e. parasites of existing symbioses) would be younger than mutualistic species, and (iii) highly age-discrepant partners would be rare in specialized symbioses. To evaluate geographical range shifts in both partners, we rely on a statistical biogeographic approach that allows comparing models with and without the assumption of speciation-with-dispersal [28,29]. With respect to geographical evolution, we expected that for specialized symbioses, ancestral areas of plant–ant clades should match those of their plant hosts.
2. Material and methods
(a). Taxon sampling, DNA isolation and amplification
The most important myrmecophyte genera associated with Pseudomyrmex ants are: Vachellia (Fabaceae: Mimosoideae), Platymisicum (Fabaceae: Faboideae), Tachigali (Fabaceae: Caesalpinoideae), Triplaris (Polygonaceae: Eriogonoideae) and Ruprechtia, the latter two being sister groups [30]. Our plant sampling ranged from 61 to 75% (see the electronic supplementary material, Material and Methods for details).
We sampled 64% of Pseudomyrmecinae including 78% of Pseudomyrmex species. Ten non-pseudomyrmecine ant species, including representatives of the sister-group (Myrmeciinae), were used as outgroups. Building on previous studies [22], we compiled or newly generated sequences from 10 nuclear markers, namely 28S rRNA, Wg, AbdA, LW Rh, EF1αF2, ArgK, Enolase, CAD, Top1 and Ubx. Out of 2150 sequences in the Pseudomyrmecinae matrix, 1990 are new (GenBank accession no. KR828817–KR830806). Taxon names, permanent voucher numbers with linked geographical information, and GenBank accession numbers are listed in the electronic supplementary material, table S1. The aligned data matrix for Pseudomyrmecinae has been deposited in TreeBase (study accession S17550). Primer sequences are given in the electronic supplementary material, table S2.
For Vachellia, Platymiscium and the Triplaris/Ruprechtia clades, we used sequences from published studies [18,30,31]; markers and alignment length are described in the electronic supplementary material, Material and Methods. For Tachigali, we sequenced ITS1 (nuclear) and matK, trnL intron, trnL–trnF and trnH-PsbA spacers (plastid) for 36 specimens. DNA isolation, purification and amplification followed standard methods [32]. Taxon names, vouchers, geographical information and GenBank accession numbers are listed in the electronic supplementary material, tables S3 (Vachellia), S4 (Platymiscium), S5 (Triplaris/Ruprechtia) and S6 (Tachigali). For more details see the electronic supplementary material, Materials and methods.
(b). DNA sequence alignment and phylogenetic analyses
Sequence alignments were performed in MAFFT v. 7 [33] (plants) or Clustal X v. 2.1 [34] (Pseudomyrmecinae), manually edited and concatenated in Mesquite v. 2.75 [35] (plants) or MacClade v. 4.08 [36] (Pseudomyrmecinae). Maximum-Likelihood tree inference relied on RAxML v. 8.1 [37] (plants) or GARLI v. 2.0 [38] (Pseudomyrmecinae), with 100 ML bootstrap replicates. For all plant analyses, we used the GTR + Γ substitution model in RAxML, while Pseudomyrmecinae were analysed using the partition scheme identified by PartitionFinder [39] (electronic supplementary material, table S7). For Tachigali and the Pseudomyrmecinae, we also conducted Bayesian analyses in MrBayes v. 3.2 [40], with partitioning by gene region for Tachigali, using the best-fitting models identified by jModelTest2 [41], and using the scheme identified by PartitionFinder for the Pseudomyrmecinae (electronic supplementary material, table S7). Further details are provided in the electronic supplementary material, Materials and Methods.
(c). Molecular clock dating
Molecular clock dating relied on BEAST v. 2 [42] and the GTR + Γ substitution model with empirical nucleotide frequencies and six rate categories. In all cases, we used the uncorrelated lognormal relaxed clock model, since its standard deviation was always more than 0.5. We used Yule tree priors, with Markov chain Monte Carlo (MCMC) chain lengths between 20 and 60 million generations, sampling every 10 000th generation with chain length depending on convergence as determined by examining the log files in Tracer v. 1.5 [43] after removal of an initial burn-in proportion of 10% of the trees. Fossil and secondary calibrations for all five DNA matrices are explained in detail in the electronic supplementary material, Material and methods.
(d). Ancestral state reconstructions
We coded Pseudomyrmecinae as (0) ‘ground nesting’, for species nesting in the ground; (1) ‘arboreal generalist’, for unspecialized arboreal species nesting in dead twigs or branches of various plants, but not usually in domatia; (2) ‘domatium mutualist’, for plant-ants nesting obligately in domatia and presenting aggressive behaviour, and (3) ‘domatium parasite’, for species obligately living in domatia but with a timid behaviour that results in the absence of defense payback to their host. Species assignments to these categories are based on published studies [19,20,27,44–47] and personal observations by P.S.W. over the past 30 years. To infer the ancestral states of nesting habits, we used the ML approach implemented in Mesquite v. 2.75 [35] with the MK1 model and the R package Ape (Ace function) [48], using as input trees both the maximum clade credibility tree from BEAST and the best ML tree from GARLI. To take into account topological uncertainties, we used two approaches: we ran MK1 reconstructions on a sample of 1000 Bayesian trees from the BEAST MCMC runs, and we used the Bayesian reversible jump MCMC approach in BayesTraits [49], which allows transition rates between character states to vary. The chain was run for 50 × 106 generations, and rate coefficients and ancestral states were sampled every 1000th generation. We ensured that the acceptance rate was between 20 and 40%, as recommended in the manual, and reconstructed the nodes of interest using the command ‘addnode’.
To reconstruct the evolutionary gains and losses of domatia in Vachellia, Tachigali, Triplaris/Ruprechtia and Platymiscium, we coded each tip for domatium absence (0) or domatium presence (1), using the world list of domatium-bearing plants [14]. We performed ancestral state reconstructions using the same approaches as for the Pseudomyrmecinae. These reconstructions, including the assumptions of the model used, are described further in the electronic supplementary material, Material and methods.
(e). Historical biogeography and range sizes
We coded the geographical ranges of all Pseudomyrmecinae and of all plant species in the phylogenies as: A = USA, B = Mexico to Panama including the Caribbean, C = Northern and Central Andes (Venezuela, Colombia, Ecuador, Peru, Bolivia), D = Brazil and the Guianas, E = Chile, Argentina, Paraguay and Uruguay, F = Afrotropics and G = India, southeast Asia and Australia. The coded Neotropical regions are shown in figure 2. To infer whether (i) ancestral areas of Pseudomyrmecinae clades match those of their plant hosts and whether (ii) our focal symbioses coincide with geographical range shifts, we used ancestral range reconstruction (back to 33.7 Ma) using the multimodel approach implemented in the R package BioGeoBEARS [28,29] on the BEAST chronograms.
Figure 2.
Dated phylogenies of Pseudomyrmex and its five main plant host groups: Vachellia, Triplaris/Ruprechtia, Tachigali and Platymiscium. Colour coding (map) shows the ancestral range resulting from the best-fit model (§2). Links between ant and plant species are solid black for documented mutualistic interactions, red for documented parasitic interactions, and dotted for inferred interactions. Black rectangles mark the evolutionary gain of mutualistic obligate plant nesting; red rectangles mark parasitic obligate plant nesting; and brown rectangles indicate ground nesting. The remaining Pseudomyrmex species are generalist arboreal ants. Grey rectangles mark the evolutionary gain of ant domatia. Numbers above rectangles refer to the probability of an inferred gain based on BayesTraits analyses (this cannot be inferred for single species; see §2). A rectangle positioned next to a crown group means that the trait originated at that node, while the rectangle position for branches leading to single species is arbitrary. Ancestral state reconstructions are shown in the electronic supplementary material, figures S5–S10 and S12. (Online version in colour.)
To determine whether increased Pseudomyrmex specialization (here obligate nesting in a particular plant species) coincides with range narrowing or broadening, we evaluated the range size of each plant-ant species and compared it to that of its sister group based on occurrence data from a database of vouchered material compiled by P.S.W. (electronic supplementary material, table S8). We calculated range sizes as the extent of occurrence using the software DIVA-GIS [50], following an approximate minimum convex polygon. Given the dense geographical sampling of Pseudomyrmecinae (electronic supplementary material, table S8), this approach reduces the risk of overestimating range sizes. Range size calculation and sister group taxonomic composition are described in detail in the electronic supplementary material, Material and methods.
(f). Interactions
We searched the literature to obtain information about the types of interactions between the plant and ant species sampled in our study. Data for Triplaris and the Pseudomyrmex triplarinus group come mainly from [51], those for Vachellia and the P. ferrugineus group from [20] (summarized in fig. 73) and [18]. Apart from these two groups, species-level information is scarce since botanists at best note the ant genus and entomologists the plant genus. We thus included indirect data from morphological traits and notes on genera (without species names), as long as there was a geographical overlap. All inferred links are depicted as dotted lines in the respective figures.
3. Results
(a). Phylogenetics of Neotropical Pseudomyrmecinae and their plant hosts
Both ML and Bayesian phylogenetic inference showed a well-supported Neotropical Myrcidris + Pseudomyrmex clade and four maximally supported Pseudomyrmex plant-ant groups (P. ferrugineus group, P. concolor group, P. triplarinus group and P. sericeus group; electronic supplementary material, figures S1 and S2). An unexpected result is that the Vachellia (‘ant-Acacia’) ants are not monophyletic, but instead form two clades separated by two species of arboreal generalists from Central America (figure 2), extending a previous finding [47].
Phylogenetic relationships in Triplaris/Ruprechtia, Platymiscium and Vachellia are as found in previous studies [18,30,31]. The monophyly of the newly investigated genus Tachigali is maximally supported in ML and Bayesian analyses (electronic supplementary material, figure S3), and the sister species relationships involving the position of myrmecophytes relevant to this study are well to moderately supported (electronic supplementary material, figure S3).
(b). Times of origin of Pseudomyrmecinae and their plant hosts
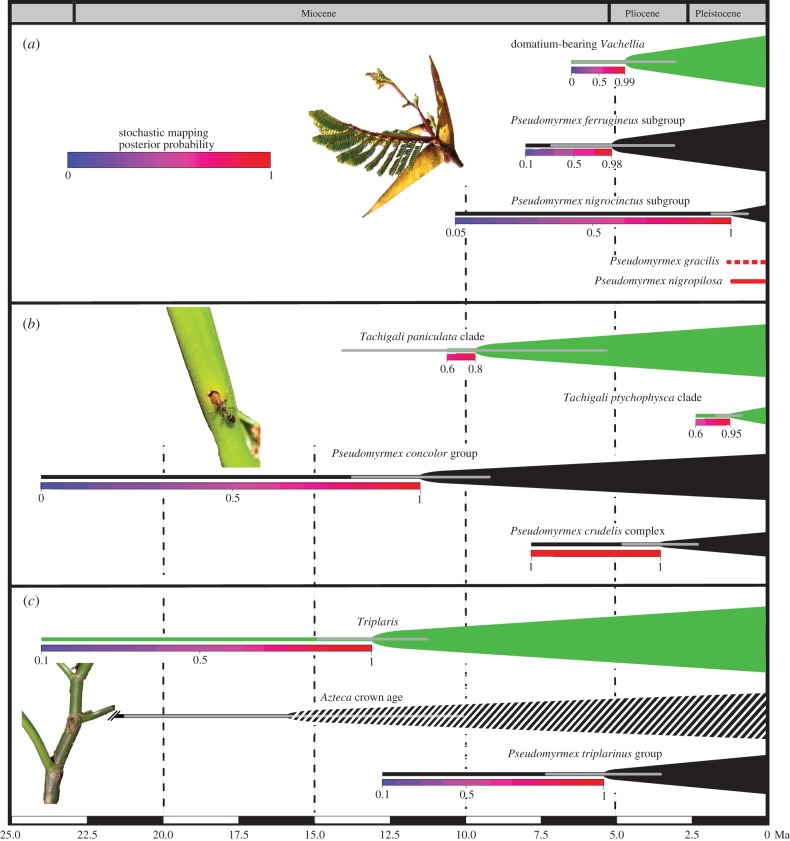
The most recent common ancestor (MRCA) of Pseudomyrmecinae dates to 71.7 ± 7 Ma, significantly older than found in chronograms that focused on all ants and therefore included only a few Pseudomyrmecinae [52,53]. The stem age of Pseudomyrmex is 49.0 ± 4 Ma, its crown age 35.8 ± 4 Ma (figure 2; electronic supplementary material, figure S4). The main clade of Vachellia-inhabiting species in the P. ferrugineus species group—here referred to as the P. ferrugineus subgroup—dates to 5.1 ± 1.5 Ma, matching the age of the MRCA of the Mesoamerican Vachellia clade, 4.7 ± 2 Ma. Two related Vachellia-inhabiting Pseudomyrmex species, P. nigrocinctus and P. particeps, forming the P. nigrocinctus subgroup, however, evolved 1.5 ± 1 Ma, after the radiation of the Vachellia species that they currently inhabit (figures 2 and 3a). Similarly, the P. triplarinus group evolved 5.7 ± 2 Ma, after the radiation of its obligate host clade, Triplaris (18 species, 61% sampled), here dated to 13 ± 2 Ma (figures 2 and 3c). The P. concolor species group dates to 12.2 ± 3 Ma, overlapping the age range inferred for the Tachigali clade that it inhabits (the T. paniculata group, 9.3 ± 5 Ma; figures 2 and 3b). Other ant-housing Tachigali species originated between 9.3 and 1.5 Ma, but the origin of domatia in single species cannot be dated (figure 2). Lastly, the P. fortis subgroup, a clade of myrmecophyte-inhabiting species within the P. sericeus species group, whose species nest in Tachigali, Triplaris, Platymiscium and other ant-plants, dates to 5.5 ± 1.5 Ma, and does not show any obvious crown matching with any of its hosts. Within this clade is a subgroup of strict Tachigali specialists, the P. crudelis complex, originating 3.7 ± 1 Ma, well after the P. concolor group.
Figure 3.
Macroevolutionary patterns of age and trait matching of interacting Pseudomyrmex ants and domatium-bearing plant lineages and hypothetical-associated evolutionary processes. (a) Co-radiation of Vachellia and the P. ferrugineus subgroup, followed by secondary colonization by mutualistic species of the P. nigrocinctus species complex, parasitic P. nigropilosus and the generalist P. gracilis. (b) Potential initial co-radiation of Tachigali and the P. concolor species group, followed by host broadening to other Tachigali lineages and secondary colonization of Tachigali by members of the P. crudelis species complex. (c) In domatium-bearing Triplaris, crown and stem ages and ancestral state reconstruction suggest that the ant mutualists (the P. triplarinus group) that currently nest in Triplaris domatia are younger by approximately 8 Myr than is domatium-presence in Triplaris, suggesting possible symbioses with other (earlier) ant species, such as Azteca whose crown age (banded) matches Triplaris and which sometimes forms symbioses with the latter (see §4). Grey error bars show the 95% CI from BEAST. Black (ants) or green (plant) bars depict stem branches. Colour gradient along the stem branch shows the posterior probability of a density plot summarizing 1000 stochastic simulations of trait evolution. See also the associated electronic supplementary material, figure S12. Below the arbitrary threshold of 0.5, the traits (domatium or domatium-nesting) are unlikely to have been present. (Online version in colour.)
(c). Biogeography of plant-nesting Pseudomyrmex and their plant hosts
BioGeoBEARS model comparison yielded the BAYAREA + J model as best fitting the ant data, significantly better than DEC + J (Lnl = −451.87 versus −608.91; electronic supplementary material, table S9a shows all statistics of BioGeoBEARS runs). DEC + J was the best-fit model for Triplaris/Ruprechtia and Platymiscium, while for Vachellia and Tachigali DEC had the same likelihood as DEC + J (electronic supplementary material, table S9b-e). Because many Pseudomyrmecinae species are widespread, the inferred ancestral ranges are also wide (figure 2). The ancestral area of Pseudomyrmex includes Central and Northern South America (ML probability = 0.8), and the ancestral ranges of the P. ferrugineus and the P. nigrocinctus subgroups are Central America (ML prob. = 1 and = 0.95, respectively), matching the inferred ancestral range of their Vachellia host plants (ML prob. = 0.99). The P. triplarinus group originated in Northern South America (ML prob. = 0.8), matching the ancestral range of its host, Triplaris (ML prob. = 0.8), and the same holds for the P. concolor group (ML prob. = 0.9) and its host Tachigali (ML prob. = 0.75–1 depending on lineage). The P. fortis species group within the P. sericeus group evolved in Northern South America (ML prob. = 0.9), where some of its hosts also arose (Triplaris and some myrmecophytic lineages of Platymiscium). In Pseudomyrmex, the evolution of obligate plant nesting correlates with a reduction in the number of ancestral areas (figure 2), which is partially confirmed by range size analysis (electronic supplementary material, figure S11).
(d). Gains or losses of plant nesting in Pseudomyrmecinae and of domatia in their major plant hosts
Our ML and Bayesian reconstructions (electronic supplementary material, Material and Methods; and figures S5 and S6) of plant nesting in Pseudomyrmecinae strongly support 10 origins of obligate domatium living in this subfamily, including five in the genus Pseudomyrmex alone. This result was highly supported across methodological approaches (figure 2; electronic supplementary material, figures S5, S6 and S12; and also electronic supplementary material). Two independent origins of domatium living are supported within the P. ferrugineus group, one in the P. ferrugineus subgroup and one in the P. nigrocinctus subgroup (Bayesian prob. 0.98, ML prob. 0.97–0.99; electronic supplementary material, figures S5, S6 and S12). No loss of obligate plant nesting was detected. In the plants, we inferred single gains of domatia in Vachellia (electronic supplementary material, figures S7 and S12) and Triplaris (electronic supplementary material, figures S8 and S12), confirming previous results [14], and three gains of domatia in Ruprechtia (electronic supplementary material, figures S8 and S12), at least nine in Tachigali (electronic supplementary material, figures S9 and S12) and five in Platymiscium (electronic supplementary material, figure S10 and S12). No domatium loss was inferred. Stochastic trait mapping (electronic supplementary material, Materials and Methods; and electronic supplementary material, figure S12) confirmed the results obtained with other methods.
4. Discussion
(a). Macroevolutionary assembly of ant/plant symbioses
The expectation that highly age-discrepant partners would be rare turned out to be wrong, while our expectations that co-diversification would be seen only in relatively young clades and that parasitic species would be younger than mutualistic species were both met. Temporally matched radiation (co-radiation) of interacting clades has occurred in the P. ferrugineus group and its Vachellia host species in Central America (figure 3a). Most ant species in this group can nest in several Vachellia species, with the exception of P. satanicus, which seems restricted to V. melanoceras ([20,24]; figure 2). We found no obvious matching of the DNA tree topologies, suggesting the absence of co-speciation, and the branching times of interacting species are not always temporally matched (figure 2), further pointing to host broadening and host switching. The limited dispersal ability of symbiotic ants and plants and their typically low specificity probably hamper co-speciation in ant/plant symbioses [12,14]. Reciprocally matching traits in Vachellia and their big-eyed ant symbionts include protein-rich food bodies (Beltian bodies) that are more effectively digested by P. ferrugineus ants than by generalist species [54], enlarged extrafloral nectaries (EFNs) with post-secretory nectar sucrose hydrolysis and the ants' ability to feed on sucrose-poor nectar [55]. A novel finding of this study is that P. nigrocinctus and P. particeps form a distinct lineage much younger than the remaining Vachellia ants, which apparently colonized already domatium-possessing Vachellia species (figure 3a; electronic supplementary material, figures S5–S7). Pseudomyrmex nigrocinctus is widespread, occupying several Vachellia species, while P. particeps is known only from V. allenii, a species that can also be inhabited by P. spinicola, a member of the P. ferrugineus subgroup [20]. Vachellia allenii thus represents a clear case of symbiont broadening, with the younger ant species P. particeps now competing with P. spinicola for domatia to live in. Both P. nigrocinctus and P. particeps patrol their host plants aggressively and gather Beltian bodies and extrafloral nectar ([56]; P.S.W., personal observation), but the extent to which they have adapted nutritionally to Vachellia, perhaps with traits similar to those found in the P. ferrugineus subgroup [54,55], remains to be investigated.
Tachigali domatia, which evolved at least nine times (figure 2), arise from an enlargement of the leaf rachis (and in some cases also the inflorescence stem), which may be developmentally ‘easy’ and happen readily under selection pressure from domatium-nesting ants, in this case ants of the P. concolor group (figure 2), as long as founder queens can cover the distance between domatium-bearing and non-domatium-bearing species occurring sympatrically [57]. The repeated evolution of domatia in related Tachigali species provides a striking example of parallel evolution that results from recurrent colonization by P. concolor group. The significantly younger age of the P. crudelis species group (3.7 Ma versus 12.2 for the P. concolor group and 8 Ma for the main domatium-bearing Tachigali lineage) strongly suggests that it secondarily colonized Tachigali (figures 2, 3b; electronic supplementary material, figures S5, S6 and S9). Secondary colonization, such as that of Vachellia by the P. nigrocinctus species complex and of Tachigali by the P. crudelis species complex, results in symbiont broadening for the plants and enables entry into ‘new adaptive zones’ represented by the myrmecophytes. The P. triplarinus group is 5–8 Myr younger than its obligate host Triplaris (figures 2 and 3; electronic supplementary material, figures S5, S6, S8 and S12). Wide crown and stem age differences between the ant-housing genus Triplaris (Polygonaceae) and its obligate ant inhabitants, and stochastic trait mapping (figures 2 and 3; electronic supplementary material, figure S12), indicate that its domatium evolved earlier than the ants now occupying it, suggesting previous symbioses that dissolved. Triplaris might thus represent a later stage in the evolution of coevolution as envisioned by Ehrlich & Raven [58], namely the complete switching to a new partner. Partner replacement could come about through colonization of domatia by generalist plant-ants [59,60]. A potential candidate for an earlier symbiosis with Triplaris is Azteca, a clade whose crown age matches that of Triplaris (figure 3c, [61]) and which contains both Triplaris specialists [46,62] and infrequent occupants of Triplaris domatia [51]. Alternatively, the inferred domatium trait might be an exaptation that would only have been converted later into a domatium, or Triplaris might have been associated with (now extinct) stem lineages species of the P. triplarinus group.
(b). Recent colonization of mutualistic symbioses by parasitic ant species
Our time-calibrated phylogenetic framework for the evolution of big-eyed ants and their plant host groups reveals that specialized mutualist species form well-defined clades, while parasite species consist of singletons (figure 2). Although the time of origin of a parasitic lifestyle in single species cannot be inferred, the relevant sister species divergence times imply that parasites evolved later than mutualists: P. nigropilosus, a specialist ant species that obligately nests in Vachellia and feeds on its food bodies and extrafloral nectar but does not protect it against herbivores or encroaching vegetation [27], split from its sister species P. major only 1.5 Ma. Similarly, P. gracilis, an arboreal generalist that occasionally occupies Vachellia and prevents queens of mutualistic ants from founding a new colony [63], split from the related species, P. hospitalis, only 1.7 Ma. Younger ages of ant parasites compared to mutualists are expected since mutualistic selection pressure must first have led to the evolution of domatia before parasitic ants could exploit these nesting structures. In all cases, we found that parasites evolved from generalists and not from mutualists, contrary to a common prediction in mutualism models [3], but consistent with previous phylogenetic analyses with less dense species sampling [47].
5. Conclusion
Our study reveals macroevolutionary patterns that may represent different stages in the evolution of ant/plant symbioses. Based on crown ages, we inferred co-diversification in the Vachellia/Pseudomyrmex ferrugineus-group system over a few million years and secondary and parallel colonizations of Vachellia, Tachigali and Platymiscium by other ant groups that entered new ‘adaptive zones' (mutualistic or parasitic). In Triplaris, we present evidence that the current Pseudomyrmex partners are secondary colonists that displaced earlier symbiont species, possibly as a consequence of locally reduced abundances and competition among plant-ants for nesting sites. The repeated evolution of domatia in Tachigali (26 of its 54 species have domatia; figure 2) may provide an example of a guild, namely the Pseudomyrmex concolor species group, imposing selection pressures on related plant species. Altogether, our study reveals that assemblage of ant/plant symbioses has been highly dynamic, even in very specialized systems, such as the iconic Central American Vachellia and its thorn-domatium-nesting P. ferrugineus group.
Acknowledgements
We thank Alex Wild, Heraldo Vasconcelos, Fabian Michelangeli and Ricardo Solar for photos, and curators of the herbaria of the Missouri Botanical Garden and the Catholic University of Quito for permission to isolate DNA from specimens under their care. We thank Rumsais Blatrix for a Tachigali leaf sample, Rosa Elena Andrade and Christian Feregrino for help in the lab, and Jeremy Aroles for proofreading the manuscript. P.S.W. extends gratitude to numerous collectors and curators who provided access to ant specimens, especially Gary Alpert, Woody Benson, Roberto Brandão, Stefan Cover, Fernando Fernández, Brian Fisher, John Lattke, Jack Longino, Bill MacKay, Roy Snelling and Jim Wetterer. We are grateful to the associate editor Megan Frederickson and three anonymous reviewers for critical comments that improved the manuscript.
Data accessibility
Data are available in TreeBase accessions S17550 (Pseudomymecinae) and Dryad accession http://dx.doi.org/10.5061/dryad.5p51r (plants).
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a grant from the German Research Foundation (DFG), RE 603/20 and grants from the Society of Systematic Biologists and the American Association of Plant Taxonomy to G.C. Research on Pseudomyrmecinae ants was supported by a series of grants to P.S.W. from the US National Science Foundation, most recently DEB-1354996.
References
- 1.Bronstein JL, Alarcón R, Geber M. 2006. The evolution of plant–insect mutualisms. New Phytol. 172, 412–428. ( 10.1111/j.1469-8137.2006.01864.x) [DOI] [PubMed] [Google Scholar]
- 2.Frederickson ME. 2013. Rethinking mutualism stability: cheaters and the evolution of sanctions. Q. Rev. Biol. 88, 269–295. ( 10.1086/673757) [DOI] [PubMed] [Google Scholar]
- 3.Sachs JL, Simms EL. 2006. Pathways to mutualism breakdown. Trends Ecol. Evol. 21, 585–592. ( 10.1016/j.tree.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 4.de Vienne DM, Refrégier G, López-Villavicencio M, Tellier A, Hood ME, Giraud T. 2013. Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198, 347–385. ( 10.1111/nph.12150) [DOI] [PubMed] [Google Scholar]
- 5.Clark MA, Moran NA, Baumann P, Wernegreen JJ. 2000. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution 54, 517–525. ( 10.1111/j.0014-3820.2000.tb00054.x) [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4, e337 ( 10.1371/journal.pbio.0040337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jousselin E, Desdevises Y, Coeur d'Acier A. 2009. Fine-scale cospeciation between Brachycaudus and Buchnera aphidicola: bacterial genome helps define species and evolutionary relationships in aphids. Proc. R. Soc. B 276, 187–196. ( 10.1098/rspb.2008.0679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruaud A, et al. 2012. An extreme case of plant–insect codiversification: figs and fig-pollinating wasps. Syst. Biol. 61, 1029–1047. ( 10.1093/sysbio/sys068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogler DJ, Neff JL, Simpson BB. 1995. Multiple origins of the yucca-yucca moth association. Proc. Natl Acad. Sci. USA 92, 6864–6867. ( 10.1073/pnas.92.15.6864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CI, Pellmyr O, Althoff DM, Balcazar-Lara M, Leebens-Mack J, Segraves KA. 2008. Pattern and timing of diversification in Yucca (Agavaceae): specialized pollination does not escalate rates of diversification. Proc. R. Soc. B 275, 249–258. ( 10.1098/rspb.2007.1405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu L, Luo Z, Zhang D, Renner SS. 2010. Pollination of Rhodoleia championii (Hamamelidaceae) in subtropical China. Biotropica 42, 336–341. ( 10.1111/j.1744-7429.2009.00585.x) [DOI] [Google Scholar]
- 12.Davidson DW, McKey D. 1993. The evolutionary ecology of symbiotic ant/plant relationships. J. Hymenopt. Res. 2, 13–83. [Google Scholar]
- 13.Frederickson ME, Greene MJ, Gordon DM. 2005. Ecology: ‘Devil's gardens’ bedevilled by ants. Nature 437, 495–496. ( 10.1038/437495a) [DOI] [PubMed] [Google Scholar]
- 14.Chomicki G, Renner SS. 2015. Phylogenetics and molecular clocks reveal the repeated evolution of ant-plants after the late Miocene in Africa and the early Miocene in Australasia and the Neotropics. New Phytol. 207, 411–424. ( 10.1111/nph.13271) [DOI] [PubMed] [Google Scholar]
- 15.Chenuil A, McKey D. 1996. Molecular phylogenetic study of a myrmecophyte symbiosis: did Leonardoxa/ant associations diversify via cospeciation? Mol. Phylogenet. Evol. 6, 270–286. ( 10.1006/mpev.1996.0076) [DOI] [PubMed] [Google Scholar]
- 16.Brouat C, McKey D, Douzery EJP. 2004. Differentiation in a geographical mosaic of plants coevolving with ants: phylogeny of the Leonardoxa africana complex (Fabaceae: Caesalpinioideae) using amplified fragment length polymorphism markers. Mol. Ecol. 13, 1157–1171. ( 10.1111/j.1365-294X.2004.02113.x) [DOI] [PubMed] [Google Scholar]
- 17.Quek SP, Davies SJ, Itino T, Pierce NE. 2004. Codiversification in an ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 58, 554–570. ( 10.1111/j.0014-3820.2004.tb01678.x) [DOI] [PubMed] [Google Scholar]
- 18.Gómez-Acevedo S, Rico-Arce L, Delgado-Salinas A, Magallón S, Eguiarte LE. 2010. Neotropical mutualism between Acacia and Pseudomyrmex: phylogeny and divergence times. Mol. Phylogenet. Evol. 561, 393–408. ( 10.1016/j.ympev.2010.03.018) [DOI] [PubMed] [Google Scholar]
- 19.Ward PS. 1991. Phylogenetic analysis of pseudomyrmecine ants associated with domatia-bearing plants. In Ant-plant interactions (eds Huxley CR, Cutler DF), pp. 335–352. New York, NY: Oxford University Press. [Google Scholar]
- 20.Ward PS. 1993. Systematic studies on Pseudomyrmex acacia-ants Hymenoptera: Formicidae: Pseudomyrmecinae. J. Hym. Res. 2, 117–168. [Google Scholar]
- 21.Ward PS. 1999. Systematics, biogeography and host plant associations of the Pseudomyrmex viduus group Hymenoptera: Formicidae, Triplaris- and Tachigali-inhabiting ants. Zool. J. Linn. Soc. 126, 451–540. ( 10.1111/j.1096-3642.1999.tb00157.x) [DOI] [Google Scholar]
- 22.Ward PS, Downie DA. 2005. The ant subfamily Pseudomyrmecinae Hymenoptera: Formicidae: phylogeny and evolution of big-eyed arboreal ants. Syst. Entomol. 30, 310–335. ( 10.1111/j.1365-3113.2004.00281.x) [DOI] [Google Scholar]
- 23.AntCat. 2015. AntCat. An online catalog of the ants of the world. See http://antcat.org (accessed 24 April 2015).
- 24.Janzen DH. 1974. Swollen-thorn Acacias of Central America. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 25.Heil M, González-Teuber M, Clement LW, Kautz S, Verhaagh M, Silva Bueno JC. 2009. Divergent investment strategies of Acacia myrmecophytes and the coexistence of mutualists and exploiters. Proc. Natl Acad. Sci. USA 106, 18 091–18 096. ( 10.1073/pnas.0904304106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hocking B. 1970. Insect associations with the swollen thorn acacias. Trans. R. Entomol. Soc. Lond. 122, 211–255. ( 10.1111/j.1365-2311.1970.tb00532.x) [DOI] [Google Scholar]
- 27.Janzen DH. 1975. Pseudomyrmex nigropilosa: a parasite of a mutualism. Science 188, 936–937. ( 10.1126/science.188.4191.936) [DOI] [PubMed] [Google Scholar]
- 28.Matzke NJ. 2012. Founder-event speciation in BioGeoBEARS package dramatically improves likelihoods and alters parameter inference in dispersal–extinction–cladogenesis DEC analyses. Front. Biogeogr. 4, 210. [Google Scholar]
- 29.Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. ( 10.1093/sysbio/syu056) [DOI] [PubMed] [Google Scholar]
- 30.Sanchez A, Kron KA. 2008. Phylogenetics of Polygonaceae with an emphasis on the evolution of Eriogonoideae. Syst. Bot. 33, 87–96. ( 10.1600/036364408783887456) [DOI] [Google Scholar]
- 31.Saslis-Lagoudakis C, Chase MW, Robinson DN, Russell SJ, Klitgaard BB. 2008. Phylogenetics of neotropical Platymiscium Leguminosae: Dalbergieae: systematics, divergence times, and biogeography inferred from nuclear ribosomal and plastid DNA sequence data. Am. J. Bot. 95, 1270–1286. ( 10.3732/ajb.0800101) [DOI] [PubMed] [Google Scholar]
- 32.Chomicki G, Renner SS. 2015. Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytol. 205, 526–532. ( 10.1111/nph.13163) [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 24, 4876–4882. ( 10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddison WP, Maddison DR. 2011. Mesquite 2.75: a modular system for evolutionary analysis. See http://mesquiteproject.org (accessed 1 April 2015).
- 36.Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. Version 4.08 Sunderland, MA: Sinauer Press. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bazinet AL, Zwickl DJ, Cummings MP. 2014. A gateway for phylogenetic analysis powered by grid computing featuring GARLI 2.0. Syst. Biol. 63, 812–818. ( 10.1093/sysbio/syu031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 296, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 40.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 613, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 104, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rambaut A, Drummond AJ. 2007. Tracer—MCMC trace analysis tool version v1.5. See http://beast.bio.ed.ac.uk.
- 44.Janzen DH. 1966. Coevolution of mutualism between ants and acacias in Central America. Evolution 20, 249–275. ( 10.2307/2406628) [DOI] [PubMed] [Google Scholar]
- 45.Wheeler WM, Bailey IW. 1920. The feeding habits of pseudomyrmine and other ants. Trans. Am. Philos. Soc. 22, 235–279. ( 10.2307/1005485) [DOI] [Google Scholar]
- 46.Wheeler WM. 1942. Studies of Neotropical ant-plants and their ants. Bull. Mus. Comp. Zool. 90, 1–262. [Google Scholar]
- 47.Kautz S, Lumbsch HT, Ward PS, Heil M. 2009. How to prevent cheating: a digestive specialization ties mutualistic plant-ants to their ant-plant partners. Evolution 63, 839–853. ( 10.1111/j.1558-5646.2008.00594.x) [DOI] [PubMed] [Google Scholar]
- 48.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 49.Pagel M, Meade A. 2013. BayesTraits v. 2.0. Reading, MA: University of Reading; See http://www.evolution.rdg.ac.uk/BayesTraits.html. [Google Scholar]
- 50.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. ( 10.1002/joc.1276) [DOI] [Google Scholar]
- 51.Sanchez A.2011. Evolutionary relationships in Polygonaceae with an emphasis on Triplaris. PhD thesis, Wake Forest University, NC, USA.
- 52.Moreau CS, Bell CD, Villa R, Archibald SB, Pierce N. 2006. Phylogeny of ants: diversification in the age of angiosperms. Science 312, 101–104. ( 10.1126/science.1124891) [DOI] [PubMed] [Google Scholar]
- 53.Moreau CS, Bell CD. 2013. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution 67, 2240–2257. ( 10.1111/evo.12105) [DOI] [PubMed] [Google Scholar]
- 54.Orona-Tamayo D, Wielsch N, Blanco-Labra A, Svatos A, Farías-Rodríguez R, Heil M. 2013. Exclusive rewards in mutualisms: ant proteases and plant protease inhibitors create a lock–key system to protect Acacia food bodies from exploitation. Mol. Ecol. 2215, 4087–4100. ( 10.1111/mec.12320) [DOI] [PubMed] [Google Scholar]
- 55.Heil M, Rattke J, Boland W. 2005. Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science 308, 560–563. ( 10.1126/science.1107536) [DOI] [PubMed] [Google Scholar]
- 56.Beulig ML, Janzen DH. 1969. Variation in behavior among obligate acacia-ants from the same colony (Pseudomyrmex nigrocincta). J. Kansas Entomol. Soc. 42, 58–67. [Google Scholar]
- 57.van der Werff H. 2008. A synopsis of the genus Tachigali (Leguminosae: Caesalpinioideae) in Northern South America. Ann. Miss. Bot Gard. 95, 618–661. ( 10.3417/2007159) [DOI] [Google Scholar]
- 58.Ehrlich PR, Raven PH. 1964. Butterflies and plants: a study in coevolution. Evolution 18, 586–608. ( 10.2307/2406212) [DOI] [Google Scholar]
- 59.McKey D. 1984. Interaction of the ant-plant Leonardoxa africana Caesalpiniaceae with its obligate inhabitants in a rainforest in Cameroon. Biotropica 16, 81–99. ( 10.2307/2387840) [DOI] [Google Scholar]
- 60.Maschwitz U, Fiala B, Davies SJ, Linsenmair KE. 1996. A South-East Asian myrmecophyte with two alternative inhabitants: Camponotus or Crematogaster as partners of Macaranga lamellata. Ecotropica 2, 29–40. [Google Scholar]
- 61.Pringle EG, Ramirez SR, Bonebrake TC, Gordon DM, Dirzo R. 2012. Diversification and phylogeographic structure in widespread Azteca plant-ants from the northern Neotropics. Mol. Ecol. 21, 3576–3592. ( 10.1111/j.1365-294X.2012.05618.x) [DOI] [PubMed] [Google Scholar]
- 62.Longino JT. 2007. A taxonomic review of the genus Azteca (Hymenoptera: Formicidae) in Costa Rica and a global revision of the aurita group. Zootaxa 1491, 1–63. [Google Scholar]
- 63.Clement LW, Köppen S, Brand WA, Heil M. 2008. Strategies of a parasite of the ant–Acacia mutualism. Behav. Ecol. Sociobiol. 26, 953–962. ( 10.1007/s00265-007-0520-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available in TreeBase accessions S17550 (Pseudomymecinae) and Dryad accession http://dx.doi.org/10.5061/dryad.5p51r (plants).