Abstract
Historically, research has focused on the mean and often neglected the variance. However, variability in nature is observable at all scales: among cells within an individual, among individuals within a population and among populations within a species. A fundamental quest in biology now is to find the mechanisms that underlie variability. Here, we investigated behavioural variability in a unique unicellular organism, Physarum polycephalum. We combined experiments and models to show that variability in cell signalling contributes to major differences in behaviour underpinning some aspects of social interactions. First, following thousands of cells under various contexts, we identified distinct behavioural phenotypes: ‘slow–regular–social’, ‘fast–regular–social’ and ‘fast–irregular–asocial’. Second, coupling chemical analysis and behavioural assays we found that calcium signalling is responsible for these behavioural phenotypes. Finally, we show that differences in signalling and behaviour led to alternative social strategies. Our results have considerable implications for our understanding of the emergence of variability in living organisms.
Keywords: behavioural phenotype, calcium, social behaviour, slime mould, variability
1. Introduction
Differences in behaviour between individuals within a species lead to major consequences for the ecology and evolution of populations [1–7]. This is obvious when behavioural polymorphism is correlated with genetic polymorphism, but it is also true when behavioural differences can be traced to different environments or are based on underlying stochasticity [8]. Behavioural differences are observed at multiple levels of organization: among individuals within a group [2–6], among groups within a population [7] and among populations within a species [9]. Despite the pioneering studies of Jennings [10] and Gause [11], and the present-day interest in microbial social behaviour [12,13], studies on behavioural polymorphism tend to be focused on multicellular organisms, from insects to mammals [2–7]. We argue that an understanding of behavioural variability in unicellular organisms is crucial for understanding variability in more complex organisms. In this paper, we studied variability in an unusual unicellular organism, the ‘true’ slime mould Physarum polycephalum.
Physarum polycephalum is a unicellular multinucleate eukaryote that is often seen on damp vegetable matter [14]. It crawls around forming extended pseudopods in order to search for food. Slime moulds have been known for decades for their amazing abilities such as finding their way in a maze [15], solving nutritional challenge [16], avoiding traps [17] and anticipating periodic events [18]. These are just a few examples of our growing understanding of these unusual organisms. The vast majority of these studies focus on single strains (in fact genetically identical individuals, which are easy to obtain because a single cell of P. polycephalum can be cut into multiple viable cells). Hence, we are left wondering how well the behaviour observed in one strain will generalize to other strains.
In this study, we used the most studied strains of P. polycephalum in recent major studies: North American strain (e.g. [19,20]), Japanese strain (e.g. [21,22]) and Australian strain (e.g. [16,17]). We first quantified the degree of variability in their behavioural repertoire, focusing on exploration and foraging behaviour. Second, we identified the underlying processes yielding behavioural polymorphism. Third, we investigated how such variability among strains influence social interactions under foraging context. Finally, we proposed a mathematical model that explains our data qualitatively and quantitatively and made predictions beyond the particular strains used in this paper.
2. Material and methods
(a). Species
Physarum polycephalum is an acellular slime mould that inhabits shady, cool and humid organic substrates that have relatively uniform pattern and texture, such as tree bark or soil. Its vegetative morph, the plasmodium, a vast multinucleate cell, can grow to cover up to 900 cm2 and crawl at speeds from 0.1 to 5 cm h−1 [14]. The migrating cell extends a search front at the leading edge, which is followed by a system of intersecting tubules towards the trailing edge (electronic supplementary material, figure S1). In the presence of certain chemical stimulus in the environment, P. polycephalum shows directional movements (i.e. chemotaxis) [14].
We used three different strains of P. polycephalum: AUS (Southern Biological, Victoria, Australia), JPN (Hakodate University, Japan) and USA (Carolina Biological, South Carolina, USA). Experiments were initiated with a total of 30 sclerotia per strain, which are encysted resting stages. We cultivated cells on a 10% oat medium (rolled oat in a 1% agar solution). All experiments were carried out in the dark at 25°C temperature and 70% humidity, and run for 24 h. Pictures were taken every 5 min with a digital Canon 60D camera.
(b). Exploration behaviour
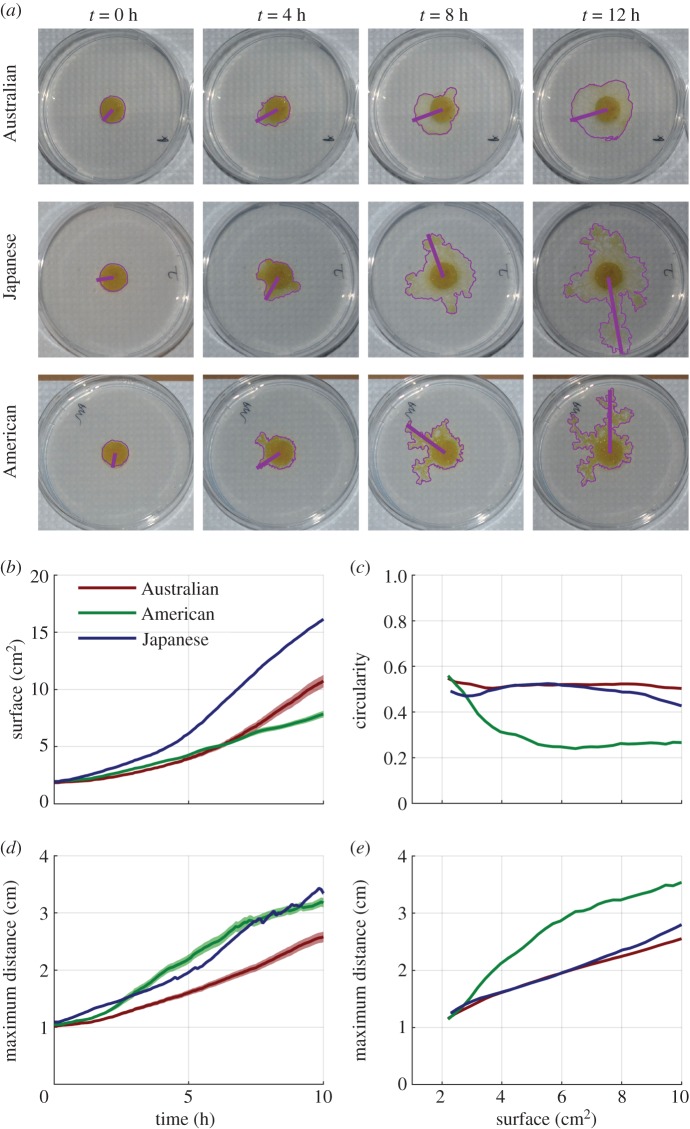
We began by monitoring the exploration movement evoked in cells in the absence of food in the environment. Each cell was introduced in the centre of a circular arena (Ø = 9 cm) containing a layer of agar (1% in distilled water). Once the agar had set, we punched one circular hole (Ø = 1.3 cm; figure 1). The hole was filled with a circular cell of diameter 1.3 cm sitting on a 10% oat medium (H = 0.3 cm) using a template. All cells were fed just before the experiment so we assumed that they were in the same physiological state. We monitored the cell until it reached the edge of the arena. Every 5 min, we recorded the area covered, the maximum distance to the centre and a measure of irregularity that we call circularity (figure 1a; for image processing, see electronic supplementary material, methods). We replicated the experiment 40 times for each strain.
Figure 1.
Exploration behaviour. (a) Photographs showing cells of Australian, Japanese and American strains at various times. (b) Surface covered by the cell as a function of time (average across all individuals). (c) Maximum distance from centre, as a function of time. (d) Circularity as a function of surface covered by the cell. We define circularity as s/(πd2), where s is the surface covered by the cell and d is the maximum distance from centre. Circularity is 1 for a circle and decreases as the cell creates branches. (e) Maximum distance from centre, as a function of surface covered by the cell. All pale patches are s.e.m.
(c) Foraging behaviour
We then monitored the directional movement response evoked in cells in the presence of food or food-related activities. Cells were allowed to choose between two localized stimuli placed at a distance of 2 cm from each other and from the cell within a circular test arena (Ø = 9 cm) containing a layer of agar (1% in distilled water; electronic supplementary material, figure S2). Once the agar had set, we punched three circular holes. The first hole was filled with a circular cell (Ø = 1.3 cm) sitting on a 10% oat medium (H = 0.3 cm) using a template. The two other holes, placed 2 cm away from the first, were filled with the stimuli presented (Ø = 1.3 cm, H = 0.3 cm). The stimuli were:
— AG, neutral stimulus: 1% plain agar substrate (stored for 24 h in sealed containers at 25°C and rinsed with distilled water just before the experiment). AG was taken as the background stimulus, namely the same agar as on the rest of the arena.
— FD, food stimulus: food 10% oatmeal–agar mixture (stored for 24 h in sealed containers at 25°C and rinsed with distilled water just before the experiment).
— FO, foraging stimulus: 1% plain agar substrate on which a cell was fed previously. We allowed a cell to cover the agar and fed it with oat flakes for 24 h, making sure that the food was in contact only with the cell and never in contact with the agar substrate. Then we removed the cell and food, and rinsed the agar substrate with distilled water just before the experiment.
The three stimuli were combined pairwise to offer cells the binary choices listed below.
(i) AG versus FD: to test whether cells were able to discriminate between food and a neutral stimulus.
(ii) AG versus FO: to test whether cells were able to discriminate between a foraging stimulus left by a congener while feeding and a neutral stimulus.
(iii) FD versus FO: to test whether cells were able to discriminate between food and a foraging stimulus left by a congener while feeding.
(iv) FO versus FO: to test whether cells were able to discriminate between foraging stimuli left by different strains, by responding differentially.
In the case of choices (ii) to (iv) cells were confronted in separate experiments by stimuli produced by either a cell of the same strain or a cell of a different strain.
Throughout a typical experiment, the cell explored its environment by expanding its network of tubules in all directions for a short distance and then building one or few search fronts (electronic supplementary material, figure S2). The patch that was reached first was taken to imply a positive response (i.e. a relative preference for the stimulus in that patch over the alternative). We recorded which stimulus was contacted first. We replicated each binary choice (30 in total) 100 times (3000 experiments in total).
For all binary choice experiments, we tested whether cells preferred one stimulus to the other using a binomial test. We also conducted generalized linear models using a logit-link function to test for the effect of the strain of the cell monitored (strain effect) and the strain of the cell that left the foraging stimulus (stimulus effect) on the probability of choosing FO for both binary choices (FO versus AG and FO versus FD).
(d). Foraging stimulus
Chemical products diffuse from the foraging stimulus into the agar, as evidenced by the apparition of concentric precipitate rings, also known as Liesegang's rings [23] (electronic supplementary material, figure S3). Calcium was considered as a likely candidate for the foraging stimulus because it is extruded together with indigestible food components when P. polycephalum cells are feeding [24] and it has been shown to be an attractant for P. polycephalum [25]. Hence, in the following experiment we investigated if calcium excretion could account for the positive chemotaxis response to FO.
(i) Presence of calcium. First, we verified that all strains were excreting calcium while feeding using complexometric titration of calcium. The FO was enclosed in agar gel and not in a solution. Agar gel can be turned into an aqueous solution but this necessitates continuous heating at 70°C. However, a temperature of 70°C promotes instability in the formation constant (Kf) and thus prevents any classic calcium titration using EDTA [26]. Therefore, we designed a different protocol to evaluate calcium excretion. We used Eriochrome Black T (EBT 0.05%, V = 100 ml), a complexometric indicator that turns red when it forms a complex with calcium and magnesium. The pHs of the solutions for titration were adjusted using 0.5 ml of sodium hydroxide solution (NaOH, 30%) to obtain a pH of 12 at which magnesium precipitated as the hydroxide and did not react with EBT. Stimuli embedded in agar patches (Ø= 1.3 cm, H = 0.3 cm) similar to the ones used in the ‘foraging experiment’ were added one by one every 30 s until the solution turned from blue to red, indicating the presence of calcium. We tested the following stimuli: FO produced by the three strains, FD, AG and calcium stimuli (Ca) of various concentrations (0.5, 0.05, 0.005 and 0.0005 M, CaHPO4 in 1% agar). For each stimulus, we recorded the number of patches added to the solution before it turned red and computed a ‘calcium index’ as the inverse function of the number of patches. We replicated this measure 10 to 14 times for each stimulus. As agar gel is not a natural substrate for P. polycephalum, we also checked if calcium could diffuse in paper, a substrate closer to natural conditions (electronic supplementary material, figure S4).
(ii) Attraction to calcium. To confirm that calcium was an attractant for all strains, we placed a cell in a test arena and confronted it with a binary choice between a calcium stimulus (CaHPO4 in 1% agar) varying in concentration (0.05, 0.005 and 0.0005 M) and a neutral stimulus (AG). Each combination was repeated at least 50 times for each strain. We monitored which stimulus was contacted first.
(iii) Sequestration of calcium. To prove that calcium excretion might be a good candidate to explain FO attraction, we sequestrated the calcium in FO using EDTA. We repeated the experiment FO versus AG. However, this time, previous to the experiment we soaked the patch FO with either EDTA (0.025 M) or distilled water for 1 h. Using the calcium titration described previously, we ensured that calcium was absent in patches rinsed with EDTA (calcium index < 0.05, n = 10) and still present in patches rinsed with distilled water (calcium index > 0.1, n = 10). The patches were then added to a test arena together with a neutral agar patch (AG). In a control experiment, we offered a choice between FD rinsed with EDTA and AG to demonstrate that EDTA is not repulsive. We monitored which stimulus was contacted first. We replicated each binary choice at least 50 times for each strain and each experiment.
We tested whether cells preferred one stimulus to the other using binomial tests. We also tested for stimulus attractiveness across strains tested and stimulus characteristics (calcium concentration or rinsing treatment) using a generalized linear model using a logit-link function.
(e). Intercellular interactions
After testing whether cells reacted to cues left by other cells, we looked at what happened when cells interacted in a foraging environment. Two cells belonging to the same strain were confronted with two identical food sources in a foraging environment consisting of a 9 cm arena lined with agar (1% in distilled water). Once the agar had set, we punched four holes (Ø = 1.3 cm) into the agar and filled them with either a 10% oat medium (Ø = 1.3 cm, H = 0.3 cm) or a cell sitting on a 10% oat medium (Ø = 1.3 cm, H = 0.3 cm). We considered two foraging environments (figure 4a). In the first one, cells were close (2 cm) and interactions were favoured. In the second one, cells were distant from one another (5 cm) and interactions were unlikely. Each cell was 2 cm away from the food sources in both environments. As a control, a single cell was confronted with two identical food sources in the absence of congeners in the same environments (figure 4a). We conducted more than 100 replicates for each strain and each environment. For each cell, we monitored the latency to reach the food and which food was reached first.
Figure 4.
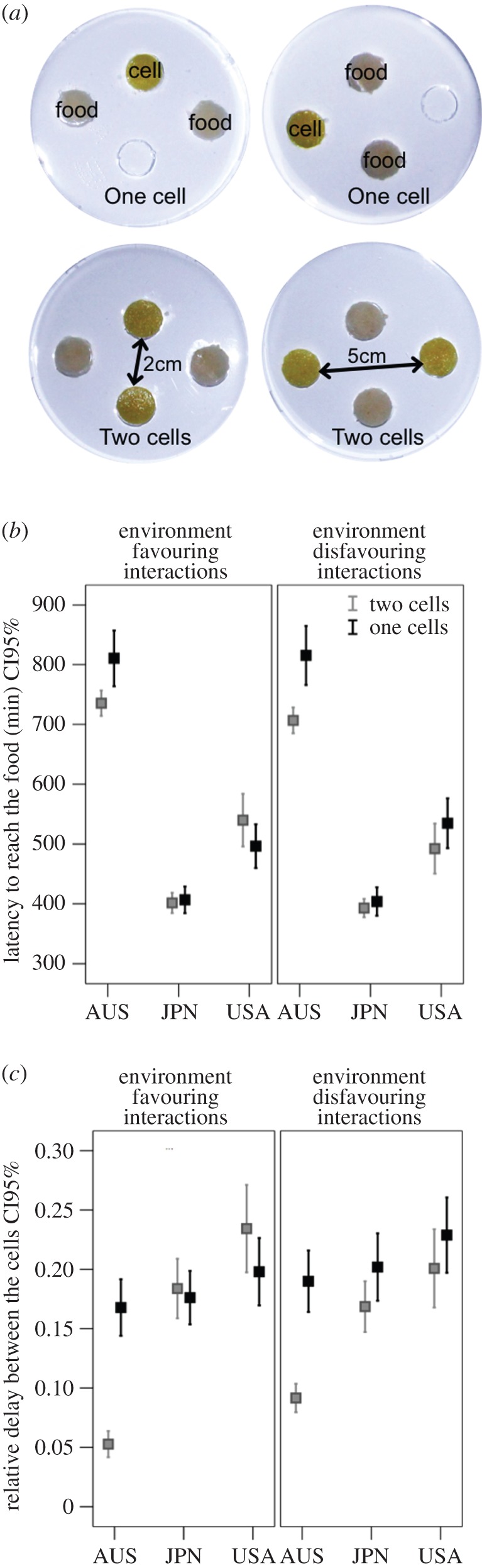
Intercellular interactions. (a) Pictures of the environment favouring interactions (short distance between the two cells) and the environment disfavouring interactions (long distance between the two cells). (b) Latency to reach the food for cells placed with a congener (two cells) and for cells on their own (one cell). (c) Expected and observed relative delay between cells. The relative delay is computed as (L1 – L2)/(L1 + L2), where L1 and L2 stand for the latencies to reach the food for the first and the second cell. The expected relative delay represents the value that we should obtain if both cells behaved independently (i.e. did not interact with each other). We simulated pairs of cells by randomly selecting two latency values from the experimental data obtained with cells of their own and computed the expected relative delay. For all experiments, we replicated each combination at least 100 times. (Online version in colour.)
The latencies were compared using a general linear model, with strain, presence of a congener and foraging environment as factors. To investigate if the cells' choices were independent from each other, the distribution of the two cells between the food sources were compared with a random distribution using a binomial test.
(f). Model
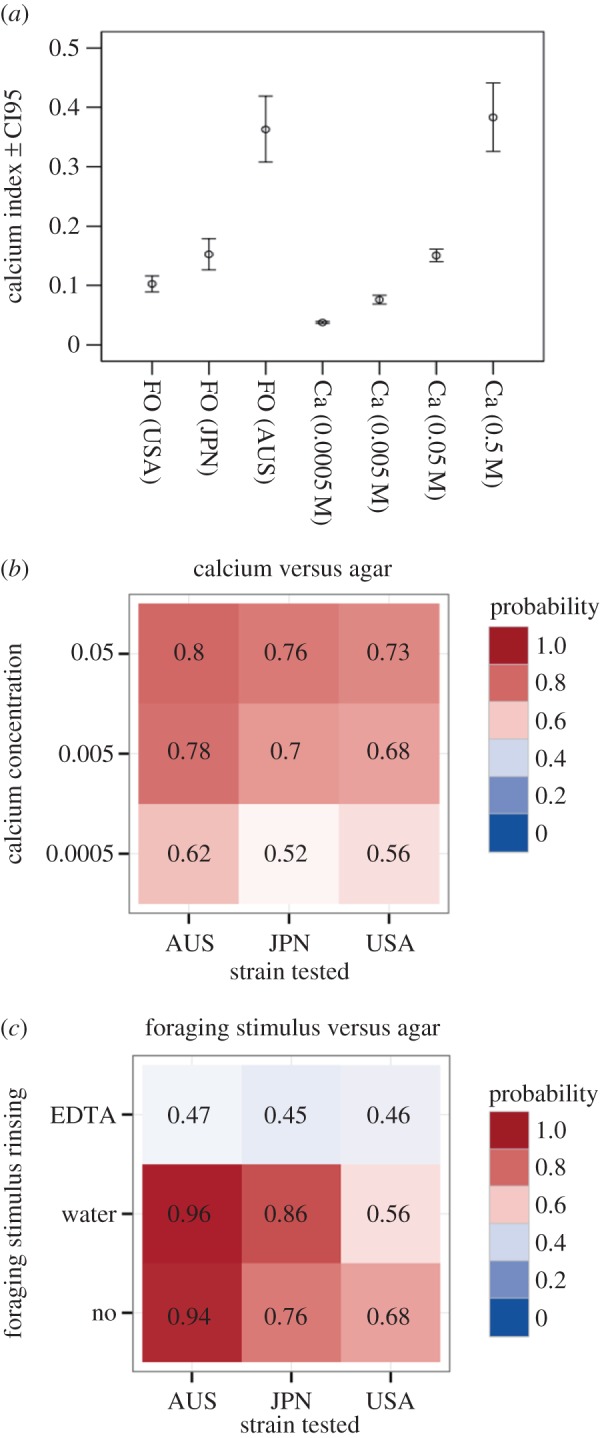
We proposed a dynamic model of cell–cell interactions in a foraging environment. The model explored the decision-making process when two cells X and Y were introduced in the same foraging environment with two identical food sources. As in the experiment, we designed two foraging environments, one favouring interactions and one disfavouring interactions. We extended previous models of how cells of P. polycephalum migrate [20,27] to simulate the cells growth rate towards the food sources. The cell fractions Xi and Yi on the source i (i = 1, 2) changed with time t according to the following set of ordinary differential equations:
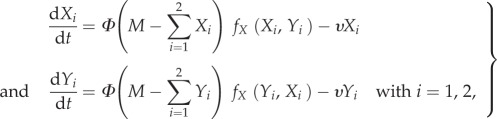 |
2.1a |
where M is the total cell size and  (or
(or 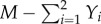 ) is the cell fraction left in the start position. ν accounts for the spontaneous departure from the food source i and Φ is the cell speed. As a cell moves towards the food source i, the rate of migration increases, resulting in further cell growth. This is accounted for by
) is the cell fraction left in the start position. ν accounts for the spontaneous departure from the food source i and Φ is the cell speed. As a cell moves towards the food source i, the rate of migration increases, resulting in further cell growth. This is accounted for by 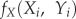 and
and  , which are translating positive feedbacks in the migration process: the cell fraction Xi (or Yi) on the food source i increases with the fractions of cell Xi and Yi already on the food source i. The functions describing these positive feedbacks are written as:
, which are translating positive feedbacks in the migration process: the cell fraction Xi (or Yi) on the food source i increases with the fractions of cell Xi and Yi already on the food source i. The functions describing these positive feedbacks are written as:
 |
2.1b |
The migration of a cell X (or Y) towards the food source i typically starts being appreciable once the cell fraction that has left the start position exceeds a threshold value K. The probability of spontaneously moving is Θ/K at t = 0. The Hill exponent n represents the strength of the positive feedback. Here, it is fixed to 2 but the properties of the model do not change qualitatively, as long as n > 1. Finally, β determines how X is attracted to Y (and conversely Y to X).
3. Results
(a). Exploration behaviour
JPN cells had the highest growth rate in terms of area covered, followed by the AUS cells and USA cells, which had similar growth rates (figure 1b). USA and JPN cells travelled very fast in terms of maximum distance from the centre (figure 1c). AUS and JPN cells grew uniformly in all directions (figure 1d), while the USA cells grew more directionally, reaching a larger distance for a given surface covered (figure 1e).
(b). Foraging behaviour
When given a choice between AG and FD, most AUS cells went directly to FD and began to exploit it (93 of 100 cases, binomial test p < 0.01; electronic supplementary material, figure S2). The seven cells that reached AG first soon moved to FD. Given a choice between AG and FO, AUS cells reached first the stimulus of a congener feeding (p < 0.01; figure 2a). Between FD and FO, AUS cells reached FO first and moved to FD afterwards (p < 0.01; figure 2b). JPN cells behaved comparably to AUS. They preferred FD to AG (in 88 of 100 cases, p < 0.01), FO to AG, and FO to FD (p < 0.01; figure 2a–b). USA cells too preferred FD to AG (92 of 100 cases, p < 0.01) and FO to AG (p < 0.01; figure 2a) but differed from the others in preferring clearly FD to FO (p < 0.01; figure 2b).
Figure 2.

Foraging behaviour. (a) Probability of choosing the foraging stimulus (FO) versus the neutral stimulus (AG). (b) Probability of choosing the foraging stimulus (FO) versus food (FD). (c) Probability of choosing one foraging stimulus (FO) over another one according to its strain source. The strain source of the foraging stimulus that significantly ‘wins’ (binomial test p < 0.05) is indicated in each box. AUS, Australian strain; USA, North American strain; JPN, Japanese strain. The x-axis indicates the strain of the cell whose response is being monitored (a–c). The y-axis indicates the strain of the cell that left the foraging stimulus (a–b) or the binary choice offered (c). Each probability was computed from 100 replicates (a–c).
Taken together, these results showed that AUS cells displayed the strongest attraction towards FO (GLM, strain: LRX = 22.49, p < 0.001 for FO versus AG and LRX = 231.73, p < 0.001 for FO versus FD; LRX stands for likelihood ratio χ2) and produced the most attractive FO (stimulus: LRX = 47.41, p < 0.001 for FO versus AG, LRX = 16.13, p < 0.001 for FO versus FD). Conversely, USA cells showed the weakest attraction and conveyed the least attractive FO. This was also confirmed by an experiment offering choices between two FO left by two different strains (figure 2c).
(c). Foraging stimulus
AUS secreted the highest quantity of calcium, followed by JPN cells and then USA cells (one-way ANOVA: F2,33 = 89.45, p < 0.001; figure 3a). Calcium was an attractant for all strains (binomial test: p < 0.01) and its attraction depended on its concentration (GLM, calcium: LRX = 14.76, p < 0.001; strain: LRX = 2.73, p = 0.255; figure 3b). FO was not attractive anymore when rinsed with EDTA while it remained attractive when rinsed with water (binomial test: p < 0.01; figure 3c). Hence, calcium excretion is a good candidate to explain FO attraction. AUS cells displayed again a stronger attraction to FO than USA cells (GLM, strain: LRX = 32.44, p < 0.001). In the control experiment, when offered a choice between FD rinsed with EDTA and AG, the cells reached FD first, showing that EDTA was not repulsive (in 42, 39 and 38 of 50 cases for AUS, JPN and USA, respectively; binomial test: p < 0.01).
Figure 3.
Foraging stimulus. (a) Calcium index quantified both in the foraging stimuli left by the three strains and in the calcium stimuli varying in concentration (CaHPO4 in 1% agar). (b) Probability of choosing the calcium stimulus (CaHPO4 in 1% agar) versus the neutral stimulus (AG). (c) Probability of choosing the foraging stimulus (FO) versus the neutral stimulus (AG). The foraging stimulus and the neutral stimulus were rinsed with distilled water or an EDTA solution (0.025 M), or not rinsed. The foraging stimulus was produced by the strain monitored. The x-axis indicates the strain of the cell whose response is being monitored. The y-axis indicates the calcium concentration (b) or the rinsing treatment (c). Each probability was computed from 50 to 65 replicates.
(d). Intercellular interactions
USA and JPN cells reached a food source faster than AUS cells (GLM, strain: F2,2033 = 508.66, p < 0.001; figure 4b). Interestingly, only AUS cells contacted a food source more rapidly when placed with a congener than when on their own in both environments (congener: F1,2033 = 11.12, p < 0.001; congener × strain F1,2033 = 9.42, p < 0.001; environment: F1,2033 = 0.56, p = 0.455; figure 4b). The time required for both cells to contact a food source (relative delay) was shorter than expected for AUS cells (GLM, strain: F2,1313 = 60.12, p < 0.001; observation: F1,1313 = 30.29, p < 0.001; figure 4c), regardless of the environment (environment: F1,1313 = 2.70, p = 0.101), indicating some sort of facilitation. Interestingly, the relative delay was somewhat longer than expected for USA cells, suggesting a slight conflict between cells when in an environment favouring interactions (strain × observation: F2,1313 = 26.78, p < 0.001; figure 4c).
The experiment could have two main outcomes: the two cells selected the same food source (SF) or different food sources (DF). When two AUS cells were placed in an environment favouring interactions the food exploitation was not random and they both often exploited the same food source (binomial test p < 0.001; figure 5). When they were placed in an environment disfavouring interaction, food exploitation was not different from a random distribution (i.e. the outcomes SF and DF happened with an equal probability; p > 0.05; figure 5). Regarding the USA and JPN cells, food exploitation was not different from a random distribution no matter the environment (p > 0.05; figure 5). This indicates that the presence of congeners influenced the decision-making process in terms of which food to exploit only for the AUS cells.
Figure 5.
Model. Theoretical and experimental frequency distributions of the proportion of cells selecting the same food source (SF) or different food sources (DF). The two cells are AUS cells (ΦAUS = 10) in an environment favouring interactions (βAUS = 0.2) and disfavouring interactions (βAUS = 0.08), JPN cells (ΦJPN = 15) in an environment favouring interactions (βJPN = 0.1) and disfavouring interactions (βJPN = 0.04), USA cells (ΦUSA = 15) in an environment favouring interactions (βUSA = 0.05) and disfavouring interactions (βUSA = 0.02), and one AUS cell and one USA cell (ΦAUS = 10 and ΦUSA = 15) in an environment favouring interactions (βAUS = 0.2 and βUSA = 0.05) and disfavouring interactions (βAUS = 0.08 and βUSA = 0.02). Each theoretical proportion is the result of 5000 realizations of stochastic integrations with an additive noise whose variance is 10−5 and initial conditions for all variables equal to zero (x1 = x2 = y1 = y2 = 0 at t = 0). We integrated the system for 5000 time units with a time step equal to 0.1. Each experimental proportion is the result of at least 100 replicates. (Online version in colour.)
(e). Model
Upon appropriate non-dimensionalization, the model defined by equations (2.1a) and (2.1b) could be reduced to
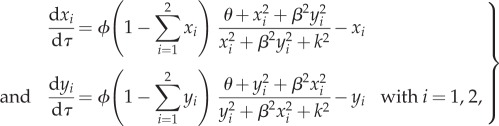 |
3.1 |
where 
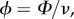

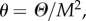
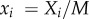 and
and 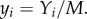 k is a threshold directly linked to the cell size M, thus K and M are expected to be of the same order. Here, we made the hypothesis that k is the same for all strains and is equal to 1. As for θ, we will take it to be small throughout the numerical solution of equations (3.1) (such that θ/K2 will be a small spontaneous probability of moving from the initial position at t = 0). To gain analytical insight, we will hereafter fix θ to 0, it being understood that the qualitative properties of the solutions, and in particular their stability, are not affected. Although all solutions were not analytically accessible even with θ = 0, it was straightforward to see that equation (3.1) accounted at the steady-state regime for the principal results of the experiments. Indeed, we found analytically (see the electronic supplementary material), in addition to the trivial solution x1 = x2 = y1 = y2 = 0, the following solutions:
k is a threshold directly linked to the cell size M, thus K and M are expected to be of the same order. Here, we made the hypothesis that k is the same for all strains and is equal to 1. As for θ, we will take it to be small throughout the numerical solution of equations (3.1) (such that θ/K2 will be a small spontaneous probability of moving from the initial position at t = 0). To gain analytical insight, we will hereafter fix θ to 0, it being understood that the qualitative properties of the solutions, and in particular their stability, are not affected. Although all solutions were not analytically accessible even with θ = 0, it was straightforward to see that equation (3.1) accounted at the steady-state regime for the principal results of the experiments. Indeed, we found analytically (see the electronic supplementary material), in addition to the trivial solution x1 = x2 = y1 = y2 = 0, the following solutions:
— x1 = y1 (or x2 = y2) and x2 = y2 = 0 (or x1 = y1 = 0) corresponding to the situation where both cells selected the same food source;
— x1 = y2 (or x2 = y1) and x2 = y1 = 0 (or x1 = y2 = 0) corresponding to the situation where the cells selected different food sources; and
— x1 = x2 = y1 = y2 corresponding to the situation where both cells selected both food sources (homogeneous state).
After checking numerically the stability of the solutions, it appeared that only these solutions could be stable (electronic supplementary material, figure S5). The non-dimensionalized model (with k = 1 and θ = 0) possesses now two key parameters that have specific values depending on the strains considered: the speed ϕ and the attraction level β. We showed that AUS cells were 1.5 times slower than JPN and USA cells (figure 1b). We therefore fixed 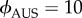 and
and 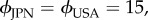 corresponding to regions where other solutions than the trivial one exist (electronic supplementary material, figure S5). We showed that the attraction towards a congener varied among strains as reflected by the probabilities of selecting FO when offered against FD (0.78, 0.64 and 0.12 for AUS, JPN and USA, respectively; figure 2b). In the model, β (and also the other parameters) acts on every time step until the final state is reached while the probabilities measured experimentally reflect the final state. Experiment provides thus a hint for choosing β-values rather than a direct access to the values. We therefore adopted β-values to secure compatibility with the experiments, and set βAUS = 0.2, βJPN = 0.1 and βUSA = 0.05 for the environment favouring interactions, and βAUS = 0.08, βJPN = 0.04 and βUSA = 0.02 for the environment disfavouring interactions (2.5 times less than the values for the environment favouring interactions, corresponding to a 2.5 times larger distance between the cells).
corresponding to regions where other solutions than the trivial one exist (electronic supplementary material, figure S5). We showed that the attraction towards a congener varied among strains as reflected by the probabilities of selecting FO when offered against FD (0.78, 0.64 and 0.12 for AUS, JPN and USA, respectively; figure 2b). In the model, β (and also the other parameters) acts on every time step until the final state is reached while the probabilities measured experimentally reflect the final state. Experiment provides thus a hint for choosing β-values rather than a direct access to the values. We therefore adopted β-values to secure compatibility with the experiments, and set βAUS = 0.2, βJPN = 0.1 and βUSA = 0.05 for the environment favouring interactions, and βAUS = 0.08, βJPN = 0.04 and βUSA = 0.02 for the environment disfavouring interactions (2.5 times less than the values for the environment favouring interactions, corresponding to a 2.5 times larger distance between the cells).
In order to see how often each solution was visited (basin of attraction) with the parameters chosen, we integrated numerically the full equation (3.1) with an additive white noise of variance equal to 10−5 and  As for the experiments, the simulations could have two main solutions: the two cells select the same food source (x1 = y1) or different food sources (x1 = y2). In the simulations, AUS cells often exploited the same food source in an environment favouring interactions while food selection was random in an environment disfavouring interactions (figure 5). Food selection was random for JPN and USA cells in both environments (figure 5). These theoretical predictions fully match the experimental results.
As for the experiments, the simulations could have two main solutions: the two cells select the same food source (x1 = y1) or different food sources (x1 = y2). In the simulations, AUS cells often exploited the same food source in an environment favouring interactions while food selection was random in an environment disfavouring interactions (figure 5). Food selection was random for JPN and USA cells in both environments (figure 5). These theoretical predictions fully match the experimental results.
Exploring the model, we predicted what would happen if the most different cells, namely AUS and USA, were introduced together with two food sources in both environments. According to the model, both cells would often select the same food source, showing that the AUS cell phenotype dominated the outcome. We then confirmed this prediction experimentally (figure 5). Often, the USA cell first found a food source and is followed by the AUS cell (electronic supplementary material, figure S6).
4. Discussion
Although many studies have focused on behavioural differences in numerous animals [2–7], few have taken the step of looking for behavioural differences among lower organisms [11,28,29]. This study extends our knowledge of the behavioural repertoire of a single-celled organism and shows the existence of various behavioural phenotypes among true slime moulds. The behavioural phenotypes that we observe form three clearly distinguishable categories: ‘slow, regular and social’ (AUS), ‘fast, irregular and asocial’ (USA) and ‘fast, regular and social’ (JPN).
We showed that the existence of behavioural type relies on a simple mechanism. While feeding, cells extrude calcium together with indigestible residuals in the environment—the so-called ‘foraging stimuli’ in our experiment. We demonstrated here that these excretions are attractive to other cells. We identified calcium as the main chemical signal that mediates this behaviour, confirming that chemotactic orientation can be induced by calcium [25]. We revealed clear differences in terms of stimulus attractiveness and stimulus responsiveness among strains. Foraging stimuli produced by AUS cells are richer in calcium and are the most attractive for all three strains. AUS cells are also the most responsive to the foraging stimuli. This might be due to difference in perception or response threshold to calcium among strains. AUS cells might respond to weaker concentration than USA cells. Our observations offer an intriguing parallel to what is seen in the (unrelated) cellular slime mould Dictyostelium discoideum: there, starved cells spontaneously develop differences in calcium content [30]. ‘High-calcium’ cells move faster than ‘low-calcium’ cells [29]; later, they go on to display other behavioural differences related to differentiation [31]
The response to calcium observed in our experiment might explain the difference in growth. We showed that cells exhibited differences in growth rate under exploration context. AUS cells grow tubes slowly in all directions while USA cells construct few tubes rapidly. In the exploration experiment no stimulus was present except the cell's own excretions. In that case, the excretions might have played a retention role and slowed down growth rate in AUS cells, which were shown to excrete a higher concentration of calcium than the two other strains. Conversely, in a foraging context, the observations on AUS cells demonstrate that the information supplied by these excretions is crucial. It allows them to coordinate their activities and detect the food faster, which is an advantage to monopolize a food resource. This could constitute a step towards cooperation [32–34]. Many microorganisms possess complex communication mechanisms that allow them to interact with one another [35]; now we can add the true slime moulds to the list. In a precedent study, we showed that cells leave an extracellular slime while exploring their environment, and use it to avoid areas previously explored and navigate in a complex environment [17]. Here we show that cells while feeding also leave chemicals in the environment, which are attractive to congeners. This means that P. polycephalum, an acellular slime mould, uses a system of communication similar to scent markings, with which they can signal to each other across time and space [36].
The model developed in this paper shares common features with previous models of decision-making in group-living organisms having different pheromones [37] or different types of individuals [38,39]. Exploring the model (electronic supplementary material, figure S5), we were allowed to make predictions for different values of speed and attraction level. These predictions would be straightforward to test either by using different strains or by changing environmental conditions. For example, cells of P. polycephalum can crawl at speeds ranging from 0.1 to 5 cm h−1 depending on the substrate or their nutritional state [14]. Moreover, thanks to the model's genericity, we can generate a number of predictions beyond the P. polycephalum case, applicable to group-living systems, in which by changing few parameters we go from a state of aggregation to one of segregation.
It is often observed that molecules initially serving one function in cells became co-opted as the signal molecule for another, thus playing numerous roles in the same organism. Such a mechanism has been proposed to explain major evolutionary transitions such as that of intercellular communication leading to multicellular organisms [40]. Following the same idea, our results show that the development of a response to molecules released passively while feeding is responsible for the emergence of communication-based cooperation between unicellular organisms and demonstrates how sociality may have emerged.
In conclusion, as proved by the model and the experiments, the existence of various phenotypes does not require highly complex neural machinery but relies on the interplay between speed and inter-attraction level. We demonstrated that responses to congeners are mediated by the use of calcium as a signal, and showed that conflict and cooperation among individuals might emerge due to slight differences in response to calcium. Differences in response threshold to stimuli might be amplified in the presence of congeners and lead to the emergence of behavioural phenotypes. This study proves once more that calcium plays a unique biological role in cell signalling and intercellular communication [41].
Supplementary Material
Acknowledgement
We thank Michael Travisano, Jacques Gautrais, Mathieu Lihoreau and two referees for their comments on the manuscript.
Data accessibility
Data are deposited in the Dryad repository: http://dx.doi.org/10.5061/dryad.s0qc1.
Authors' contributions
A.D. designed the study. A.D. and D.V. performed research. A.D. and A.P.-E. analysed the data. S.C.N. and D.J.T.S. performed modelling work. A.D. wrote the first draft of the manuscript, and all authors contributed to revisions. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
A.D. was supported by a grant from the ‘Agence Nationale de la Recherche’, reference no. JSV7-0009-01, D.J.T.S. and S.C.N. by the European Research Council Grant, reference no. IDCAB 220/104702003.
References
- 1.West-Eberhard MJ. 1989. Phenotypic plasticity and the origins of diversity. Ann. Rev. Ecol. Syst. 20, 249–278. ( 10.1146/annurev.es.20.110189.001341) [DOI] [Google Scholar]
- 2.Sih A, Bell A, Johnson JC. 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19, 372–378. ( 10.1016/j.tree.2004.04.009) [DOI] [PubMed] [Google Scholar]
- 3.Réale D, Reader SM, Sol D, McDougall PT, Dingemanse NJ. 2007. Integrating animal temperament within ecology and evolution. Biol. Rev. 82, 291–318. ( 10.1111/j.1469-185X.2007.00010.x) [DOI] [PubMed] [Google Scholar]
- 4.Sih A, Cote J, Evans M, Fogarty S, Pruitt J. 2012. Ecological implications of behavioural syndromes. Ecol. Lett. 15, 278–289. ( 10.1111/j.1461-0248.2011.01731.x) [DOI] [PubMed] [Google Scholar]
- 5.Wolf M, Weissing FJ. 2012. Animal personalities: consequences for ecology and evolution. Trends Ecol. Evol. 27, 452–461. ( 10.1016/j.tree.2012.05.001) [DOI] [PubMed] [Google Scholar]
- 6.Carere C, Maestripieri D. 2013. Animal personalities: behaviour, physiology, and evolution. Chicago, IL: University of Chicago Press. [Google Scholar]
- 7.Jandt JM, Bengston S, Pinter-Wollman N, Pruitt JN, Raine NE, Dornhaus A, Sih A. 2014. Behavioural syndromes and social insects: personality at multiple levels. Biol. Rev. 89, 48–67. ( 10.1111/brv.12042) [DOI] [PubMed] [Google Scholar]
- 8.Nanjundiah V. 2003. Phenotypic plasticity and evolution by genetic assimilation. In Origins of organismal form (eds Müller G, Newman SA), pp. 244–263. Cambridge, MA: MIT Press. [Google Scholar]
- 9.Foster SA, Endler JA. 1999. Geographic variation in behavior: perspectives on evolutionary mechanisms. New York, NY: Oxford University Press. [Google Scholar]
- 10.Jennings HS. 1906. The behaviour of the lower organisms. New York, NY: Columbia University Press. [Google Scholar]
- 11.Gause GF. 1934. The struggle for existence. Baltimore, MD: Williams and Wilkins. [Google Scholar]
- 12.Crespi BJ. 2001. The evolution of social behaviour in microorganisms. Trends Ecol. Evol. 16, 178–183. ( 10.1016/S0169-5347(01)02115-2) [DOI] [PubMed] [Google Scholar]
- 13.Bonner JT. 2009. The social amoebae: the biology of cellular slime moulds. Princeton, NJ: Princeton University Press. [Google Scholar]
- 14.Kessler D. 1982. Plasmodial structure and motility. In Cell biology of physarum and didymium (eds Aldrich HC, Daniel JW), pp. 145–207. New York, NY: Athlone Press. [Google Scholar]
- 15.Nakagaki T, Yamada H, Tóth Á. 2000. Intelligence: maze-solving by an amoeboid organism. Nature 407, 470 ( 10.1038/35035159) [DOI] [PubMed] [Google Scholar]
- 16.Dussutour A, Latty T, Beekman M, Simpson SJ. 2010. Amoeboid organism solves complex nutritional challenges. Proc. Natl Acad. Sci. USA 107, 4607–4611. ( 10.1073/pnas.0912198107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid CR, Latty T, Dussutour A, Beekman M. 2012. Slime mould uses an externalized spatial ‘memory’ to navigate in complex environments. Proc. Natl Acad. Sci. USA 109, 17 490–17 494. ( 10.1073/pnas.1215037109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saigusa T, Tero A, Nakagaki T, Kuramoto Y. 2008. Amoebae anticipate periodic events. Phys. Rev. Lett. 100, 018101 ( 10.1103/PhysRevLett.100.018101) [DOI] [PubMed] [Google Scholar]
- 19.Alim K, Amselem G, Peaudecerf F, Brenner MP, Pringle A. 2013. Random network peristalsis in Physarum polycephalum organizes fluid flows across an individual. Proc. Natl Acad. Sci. USA 110, 13 306–13 311. ( 10.1073/pnas.1305049110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabzina N, Dussutour A, Mann RP, Sumpter DJT, Nicolis SC. 2014. Symmetry restoring bifurcation in collective decision-making. PLoS Comput. Biol. 10, e1003960 ( 10.1371/journal.pcbi.1003960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tero A, Takagi S, Saigusa T, Ito K, Bebber DP, Fricker MD, Yumiki K, Kobayashi R, Nakagaki T. 2010. Rules for biologically inspired adaptive network design. Science 327, 439–442. ( 10.1126/science.1177894) [DOI] [PubMed] [Google Scholar]
- 22.Kunita I, Sato K, Tanaka Y, Takikawa Y, Orihara H, Nakagaki T. 2012. Shear banding in an F-actin solution. Phys. Rev. Lett. 109, 248303. [DOI] [PubMed] [Google Scholar]
- 23.Henisch HK. 2005. Crystals in gels and Liesegang rings. Cambridge, UK: Cambridge University Press. [DOI] [PubMed] [Google Scholar]
- 24.Wolf KV, Stockem W. 1979. Studies on microplasmodia of Physarum polycephalum: endocytotic activity, morphology of the vacuolar apparatus and defecation mechanism. Protoplasma 99, 125–138. ( 10.1007/BF01274074) [DOI] [Google Scholar]
- 25.Ueda T, Muratsugu M, Kurihara K, Kobatake Y. 1976. Chemotaxis in Physarum polycephalum: effects of chemicals on isometric tension of the plasmodial strand in relation to chemotactic movement. Exp. Cell Res. 100, 337–344. ( 10.1016/0014-4827(76)90157-9) [DOI] [PubMed] [Google Scholar]
- 26.Arena G, Musumeci S, Purrello R. 1983. Calcium-and magnesium-EDTA complexes. Stability constants and their dependence on temperature and ionic strength. Thermochim. Acta 61, 129–138. ( 10.1016/0040-6031(83)80309-8) [DOI] [Google Scholar]
- 27.Tero A, Kobayashi R, Nakagaki T. 2007. A mathematical model for adaptive transport network in path finding by true slime mould. J. Theor. Biol. 244, 553–564. ( 10.1016/j.jtbi.2006.07.015) [DOI] [PubMed] [Google Scholar]
- 28.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469, 393–396. ( 10.1038/nature09668) [DOI] [PubMed] [Google Scholar]
- 29.Goury-Sistla P, Nanjundiah V, Pande G. 2012. Bimodal distribution of motility and cell fate in Dictyostelium discoideum. Int. J. Dev. Biol. 56, 263–272. ( 10.1387/ijdb.113384ps) [DOI] [PubMed] [Google Scholar]
- 30.Azhar M, Manogaran PS, Kennady PK, Pande G, Nanjundiah V. 1996. A Ca2+-dependent early functional heterogeneity in amoebae of Dictyostelium discoideum, revealed by flow cytometry. Exp. Cell Res. 227, 344–351. ( 10.1006/excr.1996.0283) [DOI] [PubMed] [Google Scholar]
- 31.Azhar M, Saran S, Nanjundiah V. 1995. Spatial gradients of calcium in the slug of Dictyostelium discoideum. Curr. Sci. 68, 337–342. [Google Scholar]
- 32.Diggle SP, Griffin AS, Campbell GS, West SA. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414. ( 10.1038/nature06279) [DOI] [PubMed] [Google Scholar]
- 33.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607. ( 10.1038/nrmicro1461) [DOI] [PubMed] [Google Scholar]
- 34.West SA, Griffin AS, Gardner A. 2007. Evolutionary explanations for cooperation. Curr. Biol. 17, R661–R672. ( 10.1016/j.cub.2007.06.004) [DOI] [PubMed] [Google Scholar]
- 35.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. ( 10.1146/annurev.cellbio.21.012704.131001) [DOI] [PubMed] [Google Scholar]
- 36.Haldane JBS. 1955. Animal communication and the origin of human language. Sci. Prog. 43, 385–401. [Google Scholar]
- 37.Dussutour A, Nicolis SC, Shephard G, Beekman M, Sumpter DJ. 2009. The role of multiple pheromones in food recruitment by ants. J. Exp. Biol. 212, 2337–2348. ( 10.1242/jeb.029827) [DOI] [PubMed] [Google Scholar]
- 38.Ame JM, Rivault C, Deneubourg JL. 2004. Cockroach aggregation based on strain odour recognition. Anim. Behav. 68, 793–801. ( 10.1016/j.anbehav.2004.01.009) [DOI] [Google Scholar]
- 39.Nicolis SC, Despland E, Dussutour A. 2008. Collective decision-making and behavioural polymorphism in group living organisms. J. Theor. Biol. 254, 580–586. ( 10.1016/j.jtbi.2008.06.028) [DOI] [PubMed] [Google Scholar]
- 40.Bonner JT. 2000. First signals: the evolution of multicellular development. Princeton, NJ: Princeton University Press. [Google Scholar]
- 41.Campbell AK. 2014. Intracellular calcium, vol. 2 New York, NY: John Wiley & Sons. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are deposited in the Dryad repository: http://dx.doi.org/10.5061/dryad.s0qc1.