Abstract
During DNA replication, chromatin is reassembled by recycling of modified old histones and deposition of new ones. How histone dynamics integrates with DNA replication to maintain genome and epigenome information remains unclear. Here, we reveal how human MCM2, part of the replicative helicase, chaperones histones H3–H4. Our first structure shows an H3–H4 tetramer bound by two MCM2 histone-binding domains (HBDs), which hijack interaction sites used by nucleosomal DNA. Our second structure reveals MCM2 and ASF1 cochaperoning an H3–H4 dimer. Mutational analyses show that the MCM2 HBD is required for MCM2–7 histone-chaperone function and normal cell proliferation. Further, we show that MCM2 can chaperone both new and old canonical histones H3–H4 as well as H3.3 and CENPA variants. The unique histone-binding mode of MCM2 thus endows the replicative helicase with ideal properties for recycling histones genome wide during DNA replication.
To maintain genome integrity and function, both DNA sequence and DNA organization into chromatin must be faithfully duplicated during cell division. DNA replication and chromatin assembly are tightly linked to ensure that chromatin domains, such as heterochromatin and centromeres, are maintained in dividing cells1–4. A key factor linking these processes is proliferating cell nuclear antigen (PCNA), which recruits the DNA polymerases and chromatin assembly factor 1 (CAF-1)5, 6, the histone chaperone that deposits new canonical H3–H4. Another protein with a dual function is minichromosome maintenance 2 (MCM2), a component of the replicative helicase that binds H3–H4 in complex with the antisilencing function 1 (ASF1) histone chaperone7–9. The replicative helicase, consisting of CDC45, MCMs 2–7 (MCM2–7) and go-ichi-ni-san (GINS) (designated CMG), unwinds DNA at replication origins upon initiation and ahead of the DNA-synthesis machinery during elongation10, 11. The histone-binding property of MCM2 thus gives the MCM2–7 helicase the ability to orchestrate histone dynamics, which we explore in this study.
On the parental DNA strand, nucleosomes are disassembled immediately ahead of the replication fork12, 13, and the evicted histones are recycled onto the daughter strands14, 15. To maintain nucleosomal density, newly synthesized canonical histones are deposited in parallel via the ASF1–CAF-1 assembly pathway16–19. A fundamental question is how chromatin constituents, particularly modified histones and histone variants, are reassembled at the correct sites on daughter DNA strands. Although the mechanism underlying parental H3–H4 recycling remains unknown1, 20, 21, an attractive model is that MCM2, as part of the MCM2–7 helicase, binds evicted parental H3–H4 and collaborates with histone chaperones to mediate redeposition7, 9, 22. ASF1 forms a complex with MCM2 that is bridged together by H3–H4 in human cells7, and the facilitates chromatin transcription (FACT) histone chaperone forms a histone-containing complex with MCM2 in yeast22. However, an understanding of how MCM2–7 helicase may handle histones during DNA replication is limited by a lack of structural insights regarding the histone-chaperone function of MCM2.
We therefore undertook a structure-function analysis to elucidate how human MCM2, as part of the replicative helicase, can chaperone H3–H4. Here, we report two crystal structures that provide insight into how MCM2 chaperones an H3–H4 tetramer or, in collaboration with ASF1, an H3–H4 dimer. The former complex highlights how an H3–H4 tetramer can be chaperoned outside the nucleosomal context, whereas the latter represents the first structure, to our knowledge, of two chaperones cotargeting an H3–H4 dimer. MCM2 uses an unusual binding mode to capture histones, in which it interacts with the interface between H3–H4 and DNA observed in the nucleosome. Importantly, this binding mode allows MCM2 to chaperone canonical and variant H3, including centromere protein A (CENPA), at replication forks as part of the MCM2–7 helicase.
RESULTS
MCM2 hijacks the DNA-binding surface to chaperone H3–H4
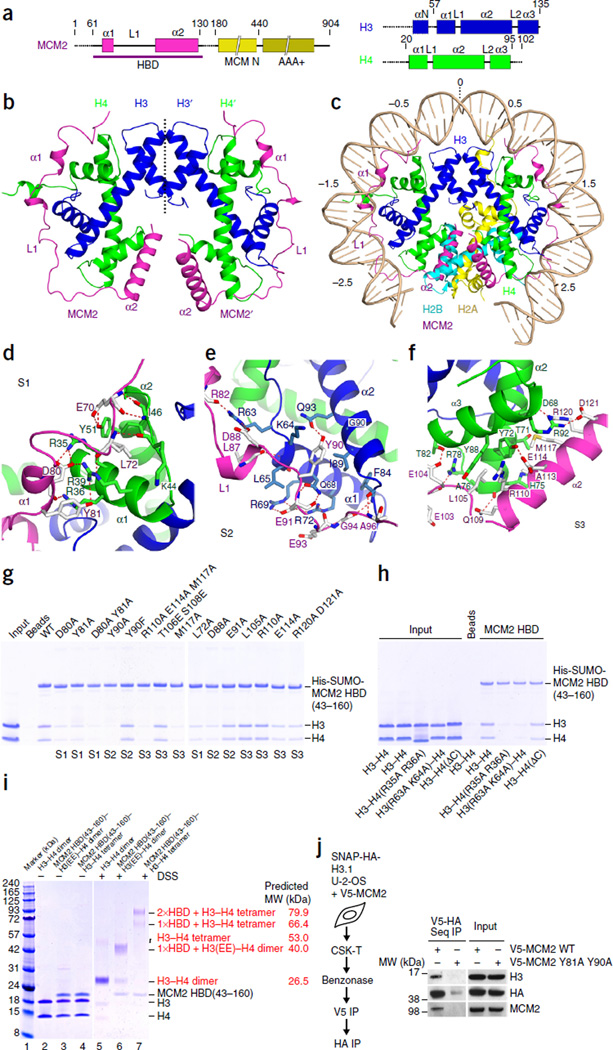
By using data from previous studies22, 23, we identified a minimal HBD of human MCM2 (amino acids 61–130; Fig. 1a) that preferentially binds histones H3–H4 rather than H2A–H2B (Supplementary Fig. 1a). The HBD formed stable complexes with histones H3.2–H4 and H3.3–H4 tetramers (Supplementary Fig. 1b–e), which, according to molecular weights estimated by size-exclusion chromatography with inline multiangle light scattering (SEC-MALS), comprised two molecules of MCM2 HBD bound to an H3–H4 tetramer (Supplementary Fig. 1f). We successfully crystallized a complex of the MCM2 HBD(61–130), H3.3(57–135) and H4, and we solved the structure to 2.85-Å resolution (Table 1). The structure shows a pair of MCM2 HBDs in an extended conformation wrapping around the lateral surface of an H3–H4 tetramer, with a pseudo–dyad axis running through the H3-H4 tetramerization interface (Fig. 1b). The two halves of the complex superposed quite well, with an r.m.s. deviation of 0.58 Å (Supplementary Fig. 1g). A related X-ray structure of an MCM2–H3–H4 tetramer complex was recently reported24, and we compared it with our structure in detail (Supplementary Fig. 1h). The conformational differences (Supplementary Fig. 1h) between these two structures may be due to different crystal packing or to the intrinsic dynamics of the complex.
Figure 1.
Structure of the MCM2 HBD–H3–H4 tetramer complex. (a) Schematics of domain architectures of MCM2, H3 and H4. (b) Ribbon representation of the structure of the MCM2 HBD–H3–H4 tetramer complex. (c) A model involving replacement of the H3–H4 tetramer in the nucleosome structure (PDB 1AOI51) by the structure of the MCM2 HBD–H3–H4 tetramer complex. (d–f) Details of the interactions between the MCM2 HBD and an H3–H4 tetramer spanning regions S1 (d), S2 (e) and S3 (f) of the complex. (g) Pulldowns of HBD mutants with H3–H4 tetramers. (h) Pulldowns of HBD WT with H3 and H4 mutants. (i) DSS cross-linking under physiological salt (150 mM). Lanes 2–4, controls without DSS; lane 5, H3–H4 cross-linked as a dimer; lane 6, cross-linked MCM2(43–160)–H3(EE)–H4 dimer, with H3(EE) denoting the L126E I130E mutant; lane 7, cross-linked MCM2(43–160)–H3–H4 tetramer. (j) Sequential (seq) IP of V5-MCM2 and SNAP-HA-H3.1 from solubilized chromatin. U-2-OS cells stably expressing SNAP-HA-H3.1 were transfected with V5-MCM2 WT or Y81A Y90A mutant as indicated. The V5-MCM2 complexes were isolated and then eluted with V5 peptide, which were subjected to a second IP with anti-HA before western blot analysis. Uncropped images of gels are shown in Supplementary Data Set 1. MW, molecular weight; His, histidine tag.
Table 1.
Data collection and refinement statistics
| MCM2 HBD–H3.3–H4 tetramer |
MCM2 HBD–H3.3–H4 dimer–ASF1 |
MCM2 HBD–H3.2–H4 dimer–ASF1 |
|
|---|---|---|---|
| Data collection | |||
| Space group | P41212 | P3121 | P3121 |
| Cell dimensions | |||
| a, b, c (Å) | 110.6, 110.6, 203.3 | 110.5, 110.5, 95.4 | 113.0, 113.0, 98.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50–2.85 (2.95–2.85)a | 50–2.30 (2.38–2.30) | 50–2.90 (3.00–2.90) |
| Rpim (%) | 4.8 (72.7) | 3.4 (34.6) | 2.6 (29.9) |
| I / σ I | 29.7 (2.0) | 29.4 (2.2) | 25.7 (1.8) |
| Completeness (%) | 100 (100) | 99.3 (98.4) | 100 (100) |
| Redundancy | 14.1 (13.1) | 11.9 (10.4) | 10.3 (10.8) |
| Refinement | |||
| Resolution (Å) | 50–2.85 | 50–2.30 | 50–2.90 |
| No. reflections (unique) | 30,366 | 29,872 | 16,336 |
| Rwork / Rfree | 19.6 / 22.0 | 18.4 / 21.4 | 19.0 / 23.8 |
| No. atoms | |||
| Protein | 3,267 | 2,960 | 2,964 |
| PO4 | 15 | – | – |
| Glycerol | – | 6 | 6 |
| Water | 31 | 124 | – |
| B factors | |||
| Protein | 77.9 | 52.6 | 64.4 |
| PO4 | 109.2 | – | – |
| Glycerol | – | 45.0 | 62.5 |
| Water | 55.6 | 48.6 | – |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.010 |
| Bond angles (°) | 1.212 | 1.254 | 1.398 |
Values in parentheses are for highest-resolution shell. One crystal was used for each data set.
A key feature in our structure is that a pair of MCM2 HBDs wraps around the lateral surface of an H3–H4 tetramer. Superposition of our structure with the H3–H4 tetramer from the nucleosome illustrates how MCM2 HBD hijacks the DNA-binding interface of the H3–H4 tetramer, with the segment before the α2 helix of the HBD following the trajectory of nucleosomal DNA (Fig. 1c). The α2 helix of the HBD also covers a relatively hydrophobic interface of H4, which forms the H4-H2B four-helix-bundle interaction in the nucleosome (Fig. 1c). Notably, the H3–H4 tetramer adopts the same topology either when bound by MCM2 HBDs or in the context of the nucleosome, as reflected in a small r.m.s. deviation of 0.66 Å (Supplementary Fig. 1i). In sum, the structure demonstrates that the pair of MCM2 HBDs chaperones the H3–H4 tetramer in a manner similar to that of DNA-histone binding interactions observed in the nucleosome and, moreover, that binding of MCM2 HBD and H2A–H2B is mutually exclusive.
Details of recognition in the MCM2 HBD–H3–H4 tetramer complex
To outline the intermolecular interactions, and for simplicity, we subdivide the MCM2 HBD into three segments (S1, S2 and S3; Fig. 1d–f). The S1 segment (68–81), including an N-terminal loop and the α1 helix, interacts primarily with the L1 loop and α1 helix of H4 (Fig. 1d and Supplementary Fig. 2a). Residues 70–72 form a β-strand–like structure with residues 44–46 in the L1 loop of H4, while the α1 helix of the HBD traverses the top of the α1 helix of H4, with strictly conserved Asp80 and Tyr81 covering Arg35, Arg36 and Arg39 of H4 (Fig. 1d). Concordantly, the HBD D80A Y81A mutant showed strongly reduced binding to H3–H4 (Fig. 1g). Furthermore, R35A R36A mutation of H4 reduced binding to the HBD, whereas deletion of the disordered C-terminal tail of H4 had no impact (Fig. 1h).
The S2 segment (82–96) of the HBD interacts with the α1 helix and the base of the α2 helix of H3 (Fig. 1e and Supplementary Fig. 2b), with primarily acidic residues directly covering primarily basic residues of the α1 helix of H3. The strictly conserved Tyr90 of the HBD penetrates into a relatively hydrophobic pocket in H3, and Tyr90 forms a strong hydrogen bond with Gln93 of H3 (Fig. 1e). Accordingly, mutation of HBD Tyr90 (Y90A) impaired binding to H3–H4 (Fig. 1g), and the H3 R63A K64A double mutant showed reduced interaction with the HBD (Fig. 1h).
The S3 segment (103–124) of the HBD, composed primarily of the α2 helix, traverses the L2 loop of H4 and packs against a relatively hydrophobic groove formed by the α2 and α3 helices of H4 (Fig. 1f and Supplementary Fig. 2c). The HBD α2 helix is anchored to H4 through several conserved residues (Fig. 1f). Met117 of the HBD appears to be very important, because the M117A mutant reduced the binding with H3–H4 (Fig. 1g).
In conclusion, all interacting residues along the extended MCM2 HBD contribute synergistically to binding H3–H4. Moreover, the structure explains the contribution of HBD Tyr81 and Tyr90, residues previously found, by mutational analysis, to mediate MCM2-histone binding22.
MCM2 binds H3–H4 tetramers under physiological conditions
Because histones H3 and H4 predominantly form dimers, not tetramers, under physiological conditions, we carried out cross-linking experiments to elucidate whether the MCM2 HBD chaperones H3–H4 tetramers under physiological conditions. SDS-PAGE revealed a major band corresponding to two HBDs bound per H3–H4 tetramer and a minor band corresponding to one HBD bound per H3–H4 tetramer (Fig. 1i). In contrast, H3 and H4 alone formed dimers and, in the presence of a tetramerization-disrupting H3 mutant (L126E I130E), the MCM2 HBD bound an H3–H4 dimer. These results establish that the MCM2 HBD can bind and stabilize H3–H4 tetramers. Free MCM2 HBD was in equilibrium with its complexes containing one or two equivalents bound to an H3–H4 tetramer, thus suggesting that two HBD are not strictly required for chaperoning an H3–H4 tetramer in vivo. To detect H3–H4 tetramers in complex with MCM2 from chromatin, we established a sequential immunoprecipitation (IP) assay and could identify endogenous H3 together with exogenous hemagglutinin (HA)- and SNAP-tagged H3.1 in complex with V5-tagged MCM2 wild type (WT) but not with an HBD mutant (Fig. 1j). This supports that MCM2 in chromatin can bind H3–H4 tetramers as a potential first step in histone recycling.
Structure of the MCM2 HBD–H3–H4 dimer–ASF1 complex
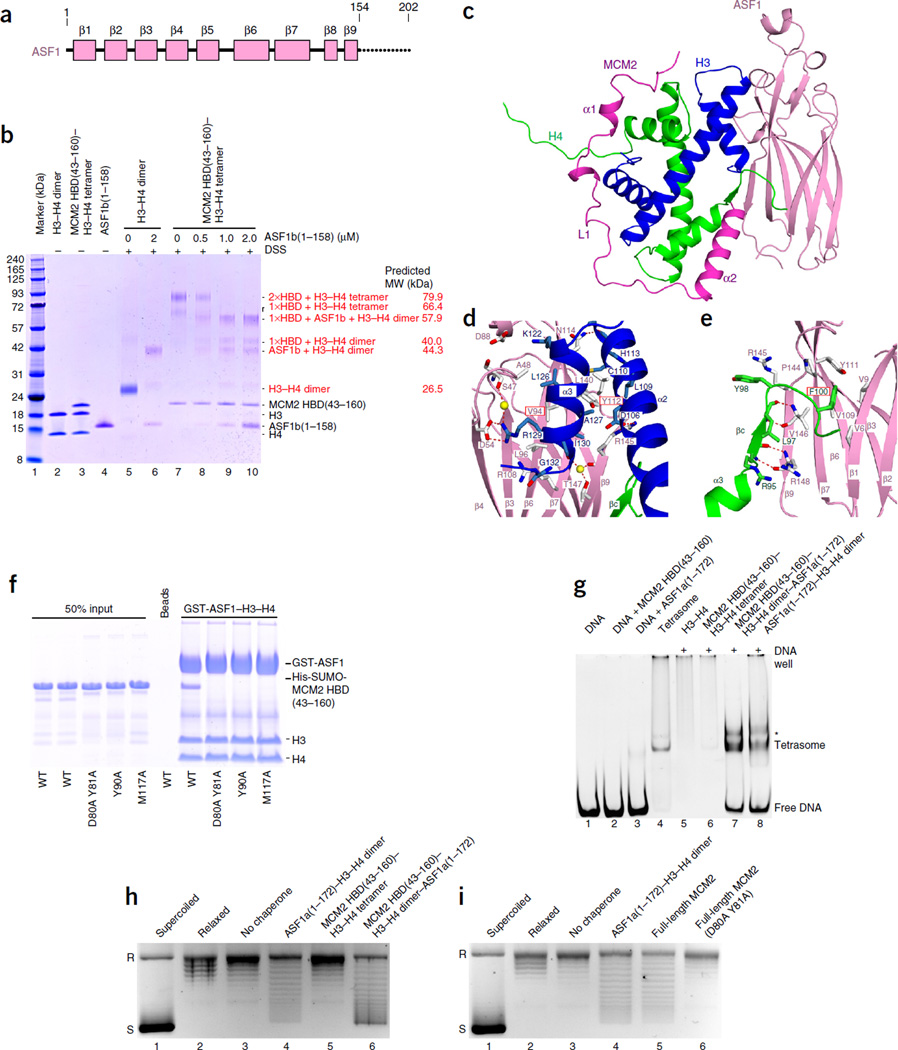
ASF1 (Fig. 2a) has been proposed to act together with MCM2 in H3–H4 recycling, as part of an MCM2–7–H3–H4–ASF1 complex7. Accordingly, ASF1 was capable of binding to the MCM2 HBD–H3–H4 tetramer complex (Supplementary Fig. 2d), thus resulting in formation of a new MCM2 HBD–H3–H4 dimer–ASF1 complex, as monitored by gel-filtration analysis (Supplementary Fig. 2e,f). We then monitored complex formation by using disuccinimidyl suberate (DSS) cross-linking under physiological conditions. Addition of ASF1 resulted in the formation of a MCM2 HBD–H3–H4 dimer–ASF1 complex as a major component in equilibrium with subcomplexes (Fig. 2b). This demonstrates that ASF1 can trigger a transition from an H3–H4 tetramer bound by MCM2 HBD to an H3–H4 dimer bound by both MCM2 HBD and ASF1.
Figure 2.
Structure of the MCM2 HBD–H3–H4 dimer–ASF1 complex. (a) Schematics of domain architecture of ASF1. (b) DSS cross-linking results showing the transition of MCM2-bound H3–H4 tetramer into dimer by addition of ASF1 under physiological salt (150 mM). Lanes 2–4, controls without DSS; lanes 5–10, titration of ASF1 into H3–H4 dimer or MCM2 HBD(43–160)–H3–H4 tetramer complex, with cross-linking after titrations. (c) Ribbon representation of the structure of the MCM2 HBD–H3–H4 dimer–ASF1 complex. (d,e) Details of the intermolecular interactions of ASF1 with H3 (d) and H4 (e) in the MCM2 HBD–H3–H4 dimer–ASF1 complex. (f) Pulldowns of immobilized GST-ASF1–H3–H4 dimer complex with key mutants of MCM2 HBD. (g) Tetrasome assembly on linear DNA, monitored by native PAGE. Lanes 1–4, controls; lanes 5–8, H3–H4 tetramer or different prepurified chaperone–H3–H4 complexes incubated with linear DNA. Asterisk, unassigned band. (h,i) Plasmid-supercoiling assays showing chaperone activities of the prepurified MCM2 HBD–H3–H4 dimer–ASF1 complex (h) and the full-length MCM2 (i). R, relaxed DNA; S, supercoiled DNA. Uncropped images of gels are shown in Supplementary Data Set 1.
To facilitate crystallization, we covalently fused the MCM2 HBD(61–130) with ASF1b(1–158) through a 12-mer GSG8SG peptide linker. We successfully purified and crystallized covalently linked MCM2 HBD–ASF1b in complex with H3.3(57–135) and H4 (Supplementary Fig. 2g,h) and solved the structure to 2.30-Å resolution (Table 1). The structure shows that one MCM2 HBD and one ASF1 together envelop an H3–H4 dimer (Fig. 2c). To our knowledge, this is the first structure of two chaperones cotargeting an H3–H4 dimer. Concordantly, the molecular weight of the complex, as estimated by SEC-MALS, corresponded to one molecule each of MCM2 HBD and ASF1 bound to an H3–H4 dimer (Supplementary Fig. 1f).
In the cochaperone structure, MCM2 HBD adopts a conformation similar to that observed in the structure of the HBD bound to an H3–H4 tetramer and buries approximately 2,000 Å2 of the H3-H4 dimer interface (Figs. 1b and 2c). Concomitantly, ASF1 binds the H3-H4 tetramerization interface and the C-terminal tail of H4, burying 1,400 Å2 of the H3-H4 dimer interface (Fig. 2c). The H3–H4 dimer bridges the MCM2 HBD and ASF1 so that there are no direct interactions between the two chaperones in the complex, a result consistent with previous biochemical analysis7. The MCM2 HBD and ASF1 also bind interfaces of H4 involved in binding H2B and H2A in the nucleosome (Figs. 1c and 2c and refs. 25,26). Thus, the two chaperones together occlude the DNA-binding, H3-H4 tetramerization and H2A- and H2B-binding interfaces of the H3–H4 dimer, efficiently averting spurious interactions before histone deposition.
Recognition details in the MCM2–H3–H4 dimer–ASF1 complex
In the structure of the MCM2 HBD– H3–H4 dimer–ASF1 complex, ASF1 uses its concave surface to envelop the tip of the α2 helix and the entire α3 helix of H3 (Fig. 2c). Val94 and Tyr112 of ASF1 are positioned in the center of the interacting surface (Fig. 2d), and the well-characterized V94R mutant of ASF1 disrupted the MCM2 HBD–H3–H4 dimer–ASF1 complex (Supplementary Fig. 2d). ASF1 also captures the C-terminal tail of H4, with Phe100 of H4 inserting into a hydrophobic pocket of ASF1 (Fig. 2e). These intermolecular contacts are similar to those reported previously25, 26 for the structure of the ASF1–H3–H4 dimer complex (Supplementary Fig. 3a,b; comparison listing small differences between the structures in Supplementary Figs. 3c–e and 4a).
The intermolecular contacts between the MCM2 HBD and the H3–H4 tetramer in our first structure (Supplementary Fig. 2a–c) are retained in the structure of the MCM2 HBD–H3–H4 dimer–ASF1 complex (Supplementary Fig. 2i–k). Accordingly, the same set of MCM2 HBD mutants also disrupted the integrity of the MCM2 HBD–H3–H4 dimer–ASF1 complex (Fig. 2f). However, we observed some changes when the H3–H4 tetramer in complex with two MCM2 HBDs transited into a dimer in complex with MCM2 HBD and ASF1 (Supplementary Figs. 3f–h and 4b,c).
Of note, the most C-terminal residues of MCM2 HBD(125–130) and the 12-mer linker were disordered in the cochaperone structure (Fig. 2c). Furthermore, cochaperone complexes showed similar profiles in gel-filtration analyses and exhibited tetrasome-assembly activities regardless of the presence of a linker (Supplementary Fig. 4d–f). Thus, despite the covalent nature of the MCM2 HBD–ASF1 cassette, our structure captured the native state of the complex.
MCM2 histone-chaperone activity
The MCM2 HBD–H3–H4 dimer–ASF1 complex assembled histones onto linear DNA and relaxed circular plasmid DNA with similar efficiency as that of the ASF1–H3–H4 dimer complex alone (Fig. 2g,h), but the MCM2 HBD did not have chaperone activity. However, full-length MCM2 did have chaperone activity (Fig. 2i), a result consistent with previous work8, and, importantly, this activity was lost upon mutation in the HBD (D80A Y81A). This result establishes that the chaperone function of full-length MCM2 relies on the histone binding mode described in our structures.
MCM2 binding stabilizes non-nucleosomal histones
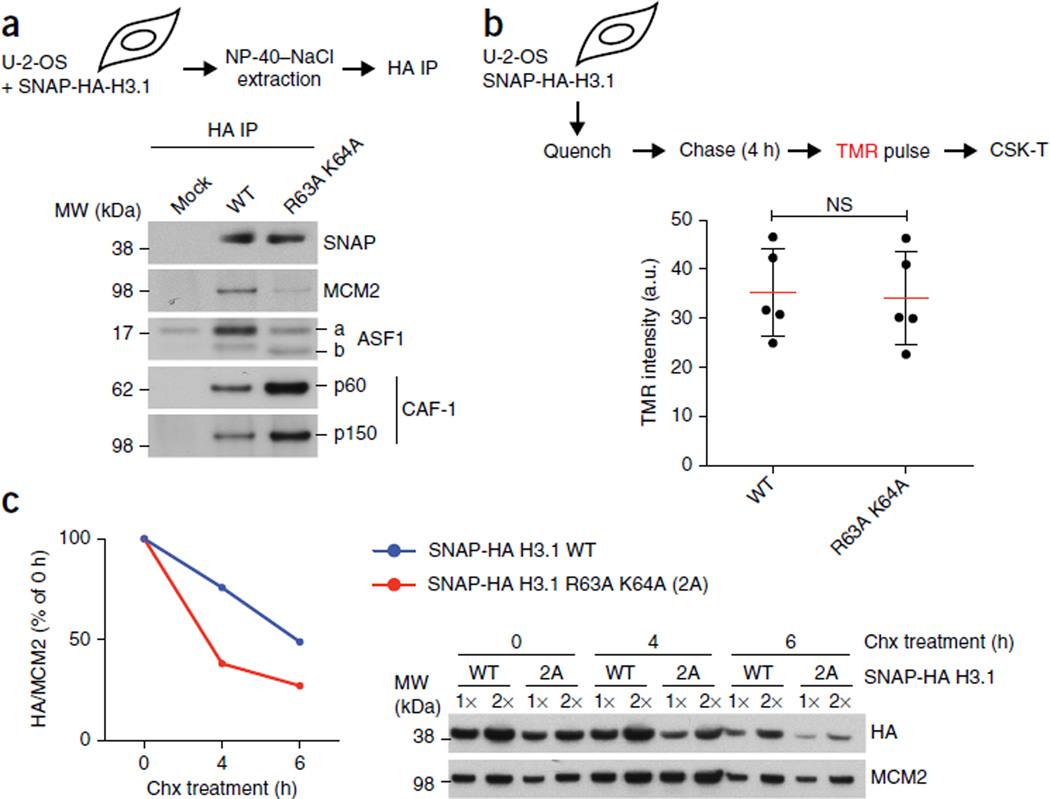
In human cells, MCM2 binds H3–H4 and ASF1, either as a soluble complex or as part of the chromatin-bound MCM2–7 helicase7, 9. We previously proposed that the histone-chaperone function of MCM2 could be important for histone recycling7, but no biological function has been suggested for MCM2 in chaperoning soluble newly synthesized histones H3 and H4 (refs. 9,27). To address this, we first determined how mutation of key MCM2-interacting residues (R63A K64A) affected the composition of soluble non-nucleosomal H3.1 complexes. The histone H3.1 R63A K64A mutant (Fig. 1e) showed a substantial reduction in MCM2 binding (Fig. 3a; input control in Supplementary Fig. 5a) as well as a moderate loss of binding of ASF1 (a and b forms). In contrast, CAF-1 binding was enhanced, thus suggesting that the assembly line from ASF1 to CAF-1 (refs. 19,28) was intact but that storage in the ASF1–MCM2 complex was lost9. Consistently with CAF-1 binding, newly synthesized histone H3.1 WT and the R63A K64A mutant were incorporated into chromatin with similar efficiency (Fig. 3b and Supplementary Fig. 6a), thus indicating that MCM2 binding is not required for replication-coupled de novo nucleosome assembly. However, we noted that the total level of H3.1 R63A K64A was reduced in comparison to that of H3.1 WT, despite the mRNA levels being identical (Supplementary Fig. 6b,c). This prompted us to test whether MCM2 binding could stabilize a pool of non-nucleosomal H3.1–H4. To assay stability, we blocked H3.1–H4 incorporation by inhibiting DNA synthesis; this inhibition leads to accumulation of histones in a soluble complex with ASF1 and MCM2 (refs. 7,9). In this setting, we found that the stability of soluble H3.1 R63A K64A was reduced in comparison with that of H3.1 WT (Fig. 3c). Collectively, this result supports the concept that cochaperoning by MCM2 and ASF1 maintains a pool of newly synthesized histones H3.1 and H4 in cells experiencing replication stress9, 29.
Figure 3.
MCM2 binding stabilizes non-nucleosomal H3.1–H4. (a) IP of SNAP-HA-H3.1 WT or R63A K64A mutant from transfected cells (input material for IP in Supplementary Fig. 5a). (b) Replication-coupled H3.1 incorporation, measured by SNAP-tag fluorescent tetramethylrhodamine (TMR) labeling in stable cells expressing SNAP-HA-H3.1 WT or R63A K64A. Top, schematic showing quenching of old SNAP-HA-H3.1 to block labeling; incorporation of newly synthesized histones for 4 h before TMR labeling; and preextraction with CSK 0.5% Triton X-100 (CSK-T). Bottom, dot plot showing mean (red lines) TMR intensities are from five independent cell cultures and experiments, each including five technical replicates including more than 5,000 TMR-positive cells (representative micrographs in Supplementary Fig. 6a). Error bars (black lines), s.d.; NS, not significant by two-tailed unpaired t test; a.u., arbitrary units. (c) Stability of non-nucleosomal H3.1 WT and R63A K64A. Left, quantification, with SNAP-HA-H3.1 levels shown relative to MCM2. Right, western blot. Samples are soluble histones extracted from stable SNAP-HA-H3.1 WT or mutant cell lines synchronized in S phase and treated with hydroxyurea (HU) and cycloheximide (chx) to inhibit replication-coupled histone deposition and protein synthesis, respectively. One representative experiment out of three is shown. Uncropped images of gels are shown in Supplementary Data Set 1.
MCM2 chaperones H3–H4 as part of the MCM2–7 helicase
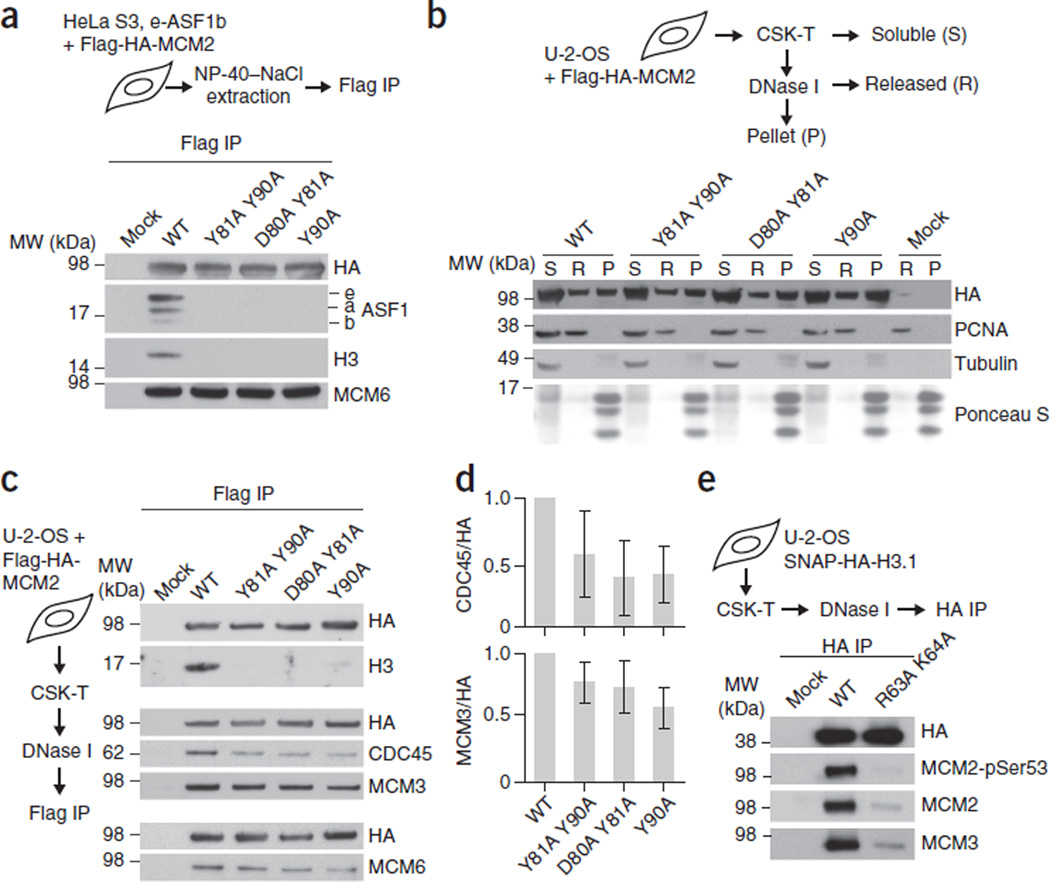
To test whether the intermolecular interactions identified in our structure are essential for the MCM2–H3–H4–ASF1 complex, we generated MCM2 mutants predicted to abrogate direct interactions with H3 (Y90A; Fig. 1e), H4 (D80A Y81A; Fig. 1d) or both (Y81A Y90A). All of the MCM2 mutants showed strongly reduced binding to non-nucleosomal H3–H4 and ASF1 (a and b forms) (Fig. 4a and Supplementary Fig. 5b). However, binding to MCM6 was not affected, a result corroborating that histone binding is not important for formation of the soluble MCM2–4–6–7 subcomplex (Fig. 4a and refs. 8,30). Histone binding was also not required for MCM2 loading onto chromatin, as shown by fractionating cells into soluble material, DNase I–sensitive chromatin and pellets containing undigested chromatin (Fig. 4b). Mild DNase I treatment solubilizes actively replicating regions, as indicated by the release of chromatin-bound PCNA (Fig. 4b). In this fraction, enriched for active replication forks, MCM2 bound H3–H4 in a manner dependent on the key residues identified in our structure (Fig. 4c and Supplementary Fig. 5c). Moreover, all the MCM2 HBD mutants showed moderately reduced binding to MCM3 and MCM6 as well as a substantial loss of CDC45 (Fig. 4c,d), which is loaded onto the MCM2–7 complex upon activation of the helicase. This suggests that histone binding stabilizes the CMG complex or alternatively has a role during its formation. The MCM2 Y81A Y90A mutant showed typical cell cycle–specific staining patterns31 (Supplementary Fig. 6d,e) consistent with normal loading and unloading dynamics. However, the proportion of cells with early S-phase patterns was increased in comparison to that of cells expressing MCM2 WT (Supplementary Fig. 6e), a result suggesting that impaired MCM2 histone binding hampers efficient S-phase progression. Importantly, histone H3.1 WT, but not the mutant deficient in MCM2 interaction coprecipitated MCM3 as well as MCM2 phosphorylated at Ser53 by the activating kinase Cdc7–DBF4 (ref. 32) (Fig. 4e and Supplementary Fig. 5d). Collectively, this suggests that the MCM2–7 helicase, via the MCM2 HBD, chaperones H3 and H4 at replication forks.
Figure 4.
MCM2 chaperones H3–H4 as part of the MCM2–7 helicase in chromatin. (a) IP of transiently expressed Flag-HA-MCM2 from extracts of HeLa S3 cells stably expressing e-ASF1b7. MCM2 mutations were designed to disrupt interactions with H3 (Y90A), H4 (D80A Y81A) and both H3 and H4 (Y81A Y90A). (b) Fractionation of cells expressing Flag-HA-MCM2 mutants. α-tubulin, fractionation control. (c) IP of Flag-HA-MCM2 mutants from solubilized chromatin. (d) Quantification of coimmunoprecipitated CDC45 and MCM3 relative to Flag-HA-MCM2. Error bars, s.d. from the mean for Y81A Y90A (n = 4 independent cell cultures and experiments), D80A Y81A (n = 4) and Y90A (n = 3). (e) IP of SNAP-HA-H3.1 WT or mutant from DNase I–solubilized chromatin of stable cell lines (input material for IPs in Supplementary Fig. 5b–d). pSer53, phosphorylated Ser53. Uncropped images of gels are shown in Supplementary Data Set 1.
Disruption of MCM2 chaperone function impairs cell growth
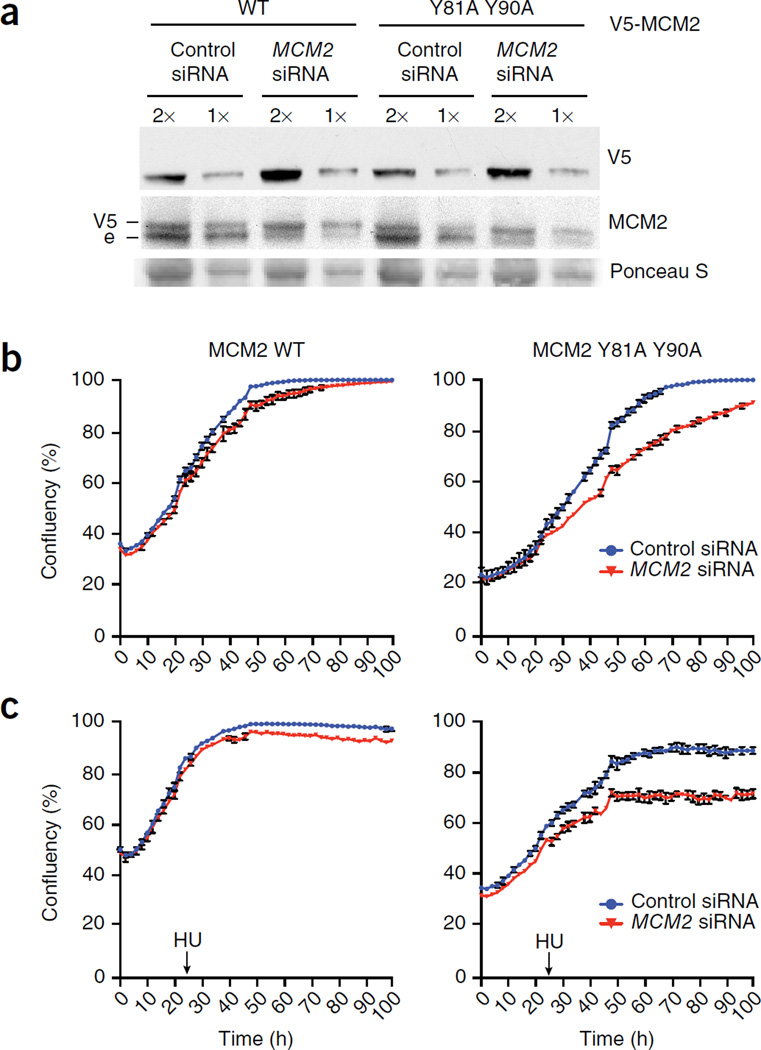
Like other components of the replicative helicase, MCM2 is essential for cell proliferation10. To determine whether the histone-chaperone function of MCM2 is required, we used a replacement strategy in which lentiviral transduction of short interfering RNA (siRNA)-resistant MCM2 WT or HBD mutant was combined with siRNA depletion of endogenous MCM2. High-resolution SDS-PAGE and western blotting confirmed expression of exogenous MCM2 at near-endogenous levels and depletion of endogenous MCM2 (Fig. 5a). Measurement of cell proliferation by high-content imaging in living cells showed that the MCM2 HBD mutant failed to rescue cell proliferation as efficiently as MCM2 WT (Fig. 5b). Moreover, cells expressing the MCM2 HBD mutant were also more sensitive to treatment with the replication inhibitor hydroxyurea (HU) (Fig. 5c), which, owing to activation of dormant origins, makes cells more dependent on a full complement of functional MCM2–7 (ref. 33). These cellular studies establish that the function of MCM2 as a histone chaperone, alone or as part of the MCM2–7 helicase, is required to sustain cell proliferation under unperturbed and replication-stress conditions.
Figure 5.
MCM2 histone-chaperone function is required for cell proliferation. (a–c) U-2-OS cells transduced with siRNA-resistant MCM2 WT or Y81A Y90A, treated with MCM2 siRNA and analyzed by western blotting (e, endogenous) (a) or high-content live-cell imaging to detect cell proliferation in the absence (b) or presence (c) of HU. Imaging was started 6 h after siRNA transfection; HU (0.5 mM) was added 24 h later as indicated. The graphs show the mean confluency (%) ± range from n = 2 technical replicates and are representative of three independent experiments and cell cultures. Uncropped images of gels are shown in Supplementary Data Set 1.
MCM2 chaperones new and old histones and all H3 variants
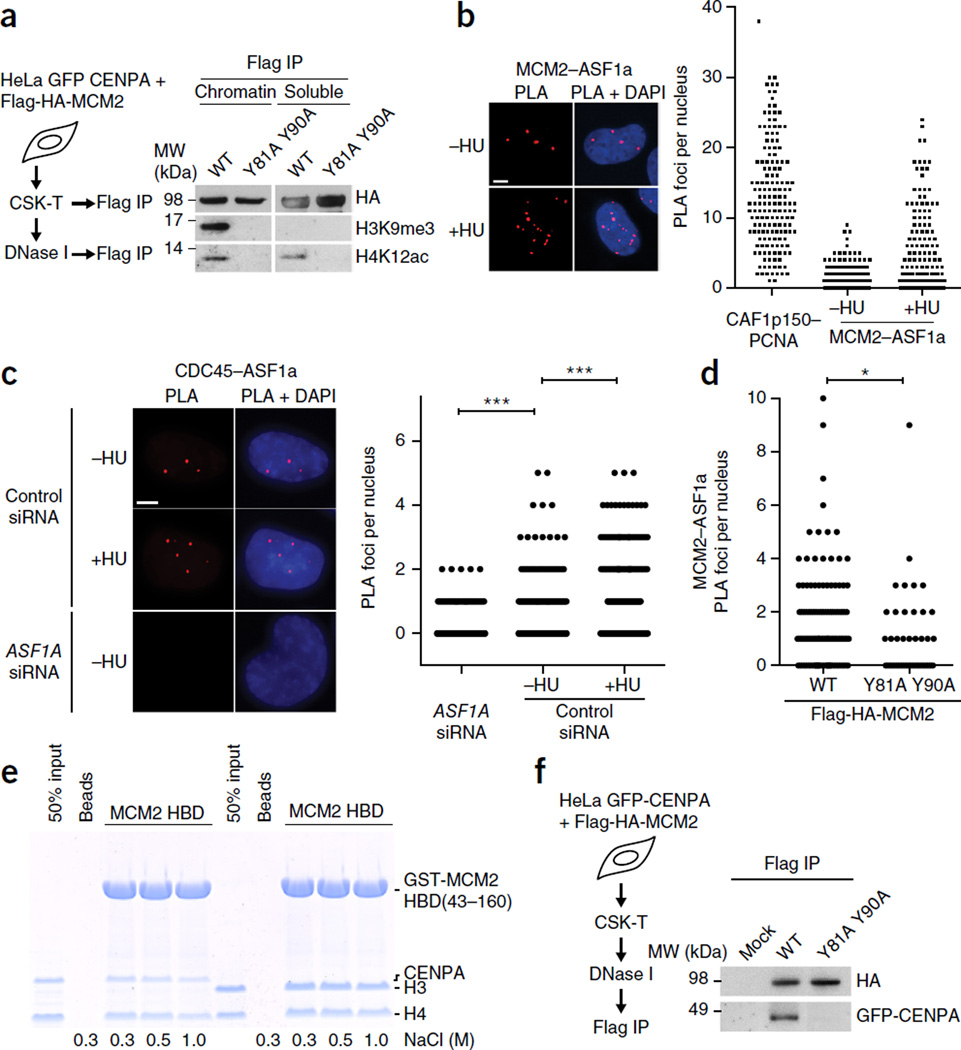
Research by us and others supports the notion that MCM2 could be handling both new histones and evicted old ones (this work and refs. 7,9,22). New histones carry diacetylated H4 K5 and K12 but lack trimethylated H3K9 (H3K9me3) which is established on nucleosomal histones9, 27. We found that H4 acetylated at K12 (H4K12ac) was present on histones associated with both soluble and chromatin-bound MCM2, whereas the H3K9me3 mark was detectable only on histones bound to MCM2 in chromatin (Fig. 6a and Supplementary Fig. 5e). This implies that MCM2 binds histones H3 and H4 that are available; i.e., soluble MCM2 associates with new histones, whereas chromatin-bound MCM2 can bind old histones as well as new ones. This is consistent with previous MS profiling data of marks on ASF1-bound histones and identification of H3K9me3 and trimethylated H3 K27 with ASF1 extracted by high salt from chromatin7, 9. Our findings that ASF1 can disrupt the H3–H4 tetramers bound by the MCM2 HBD (Fig. 2b and Supplementary Fig. 2d–f) and that MCM2 and ASF1 bind to distinct surfaces of the H3–H4 dimer (Fig. 2c) also support the possibility that these chaperones could collaborate in handling old H3–H4 during DNA replication. ASF1 colocalizes with MCM2–7 on chromatin9, but, because only a fraction of MCM2–7 complexes are engaged in replication34, immunofluorescence cannot place ASF1 at active replication forks. However, by proximity ligation assay (PLA) we found colocalization of ASF1 with MCM2 and CDC45 in chromatin (Fig. 6b,c and Supplementary Fig. 7a,b). The number of interaction foci increased significantly upon HU-induced replication stress (Fig. 6b,c), a condition under which ASF1 binding to MCM2 is also enhanced7, 9. Importantly, the interaction of ASF1 with MCM2 was disrupted in the HBD mutant (Fig. 6d and Supplementary Fig. 7c). This places the ASF1–H3–H4–MCM2–7 complex at active replication forks.
Figure 6.
MCM2 can handle new and old histones, including all H3 variants. (a) IP of Flag-HA-MCM2 WT and mutant from soluble fractions or DNase I–solubilized chromatin. (b,c) PLA of endogenous chromatin-bound ASF1a with MCM2 (b) or CDC45 (c). Soluble proteins were extracted by CSK-T. Positive control, PLA of CAF-1 p150 and PCNA; negative control, PLA in cells depleted for ASF1a. Dot plots show PLA interaction foci per nucleus and are representative of two independent cell cultures and stainings. In c, n = 270 cells (ASF1A siRNA), n = 260 cells (control siRNA, −HU) and n = 284 cells (control siRNA, +HU). ***P < 0.001 by unpaired two-tailed t test with Welch’s correction. (d) PLA of endogenous chromatin-bound ASF1a and MCM2 in U-2-OS cells transiently transfected with Flag-HA-MCM2 WT or Y81A Y90A mutant. Dot plot shows PLA interaction foci per nucleus and is representative of two independent cell cultures and stainings. n = 188 (WT); n = 44 (Y81A Y90A). *P < 0.05 by unpaired two-tailed t test with Welch’s correction. (Representative images are shown in Supplementary Fig. 7c.) (e) Pulldowns of GST-MCM2 HBD with H3–H4 or CENPA–H4 tetramers in different salt conditions. (f) IP of Flag-HA-MCM2 WT and mutant from DNase I–solubilized chromatin of cells stably expressing GFP-CENPA52. (Input material for IP is shown in Supplementary Fig. 5f. GFP-CENPA coprecipitation with MCM2 in CSK-T fractions is shown in Supplementary Fig. 8d.) Uncropped images of gels are shown in Supplementary Data Set 1.
Our data support the model whereby MCM2 captures parental H3–H4 tetramers during DNA replication, with ASF1 acting as a cochaperone that could mediate redeposition of H3–H4 dimers. Given that MCM2–7 is required for replication genome wide, and histone H3.3 and CENPA are both recycled during replication35, 36, this model raised the question of how histone variants are handled at the fork. MCM2 is found in H3.1, H3.2 and H3.3 complexes in vivo (ref. 37 and this work), and solving the structures of the MCM2 HBD–ASF1b cassette in complex with either H3.3–H4 or H3.2–H4 demonstrated that the interactions of MCM2 HBD with H3.3 or H3.2 are highly similar (Supplementary Fig. 7d). We then looked at centromeric histone H3 and noted that, although CENPA shares only about 50% amino acid identity with canonical H3, the residues mediating MCM2 binding are conserved (Supplementary Fig. 7e). Furthermore, the H3.3–H4 tetramer bound to the MCM2 HBD is highly similar to the nucleosomal CENPA–H4 tetramer (PDB 3AN2 (ref. 38)), with an r.m.s. deviation of 0.80 Å (Supplementary Fig. 7f). Concordantly, the MCM2 HBD bound a CENPA–H4 tetramer with high affinity in vitro (Fig. 6e) and formed a stable MCM2 HBD–CENPA–H4 tetramer complex in gel-filtration analyses (Supplementary Fig. 8a–c). Moreover, GFP-CENPA copurified with WT MCM2 but not the Y81A Y90A histone-binding mutant from stable GFP-CENPA cell lines (Fig. 6f and Supplementary Fig. 5f). This illustrates that MCM2 has the capability to handle all histone H3 subtypes, a predicted requirement for a factor handling evicted parental histones genome wide during DNA replication. Coimmunoprecipitation analysis showed that HJURP, a CENPA-specific chaperone that binds CENPA–H4 dimers39, 40, could associate with MCM2 in a histone-independent manner similar to that of FACT22 (Supplementary Fig. 8e), thus opening the possibility of a CENPA-specific cochaperone relationship.
DISCUSSION
Our structure-function study of MCM2 has identified an unusual mode of H3–H4 binding, which, because it hijacks histone sites interacting with nucleosomal DNA, allows MCM2 to bind a large variety of modified and variant histones H3 and H4. We show that MCM2 histone-chaperone function, alone or as part of the MCM2–7 helicase, relies on this binding mode and is required for normal rates of cell proliferation. In addition to storing and stabilizing new histones, MCM2 is also able to handle old histones. Histone binding could potentially fine-tune MCM2–7 helicase activity and thus coordinate DNA unwinding with nucleosome disassembly and assembly. Disruption of the MCM2 HBD did not affect cell-cycle progression in yeast22, thus suggesting that histone binding is not essential for CMG function. However, telomeric gene silencing was impaired22, as expected for a chromatin-assembly defect41. Given that yeast, unlike multicellular organisms19, 42–44, generally tolerate defects in chromatin assembly41, 44–46, the proliferation defect in human cells carrying the MCM2 HBD mutant could be linked to replication-coupled chromatin dynamics rather than to DNA replication.
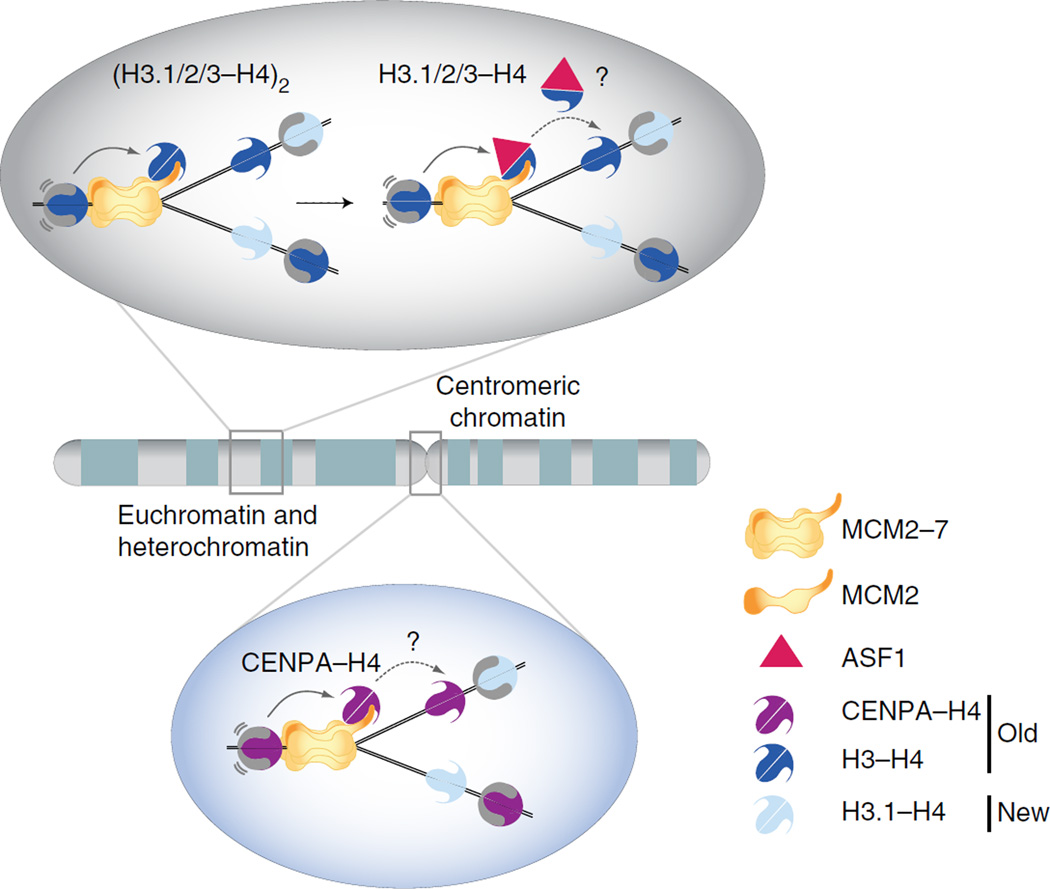
The recycling of parental H3–H4, whether as a tetramer or dimer, remains a matter of ongoing debate3, 4, 14, 18, 47, 48. Our data show that MCM2 can handle both tetramers and dimers, providing a molecular basis for understanding the transfer mechanisms that, as previously proposed48, 49, might be dependent on chromatin context (Fig. 7). We provide structural and functional evidence that ASF1 can act as a cochaperone for H3–H4 dimers together with MCM2 at replication forks, but we anticipate that other chaperones could also be involved in histone transfer (Fig. 7). On the basis of structural information and known interaction with MCM2, possible candidates include FACT22, which could bind to the N-terminal tail of H3, and HJURP (Supplementary Fig. 8e), which mainly recognizes the CATD region of CENPA50.
Figure 7.
Structure-based model for how MCM2 handles parental histones H3 and H4 genome wide during DNA replication. H3–H4 or CENPA–H4 tetramers, released from parental nucleosomes upon progression of the MCM2–7 helicase, are captured by MCM2 and held in proximity to the fork. MCM2-bound H3.1–H4, H3.2–H4 or H3.3–H4 tetramers can be disrupted by ASF1, which in turn could mediate redeposition of the dimers. Alternatively, other chaperones such as FACT might mediate histone transfer. Chaperone choice could be context dependent; for example, a CENPA–H4–specific chaperone should be brought in during replication of centromeric DNA. A key feature of this model is the ability of MCM2 to hijack the nucleosomal DNA-binding surface to chaperone H3–H4 tetramers, thus shielding the histones and ensuring their recycling regardless of their subtype and modifications. H3.1/2/3, histone H3 variants H3.1, H3.2 and H3.3.
In conclusion, our research provides structural and functional insight into the histone-chaperone properties of MCM2, a central component of the replicative DNA helicase. Further, it reveals how two chaperones, MCM2 and ASF1, can collaborate in shielding functional interfaces of histones, and it provides a paradigm for improving understanding of other cochaperone relationships. The MCM2 histone-binding domain is remarkably well suited to capture parental histones evicted ahead of the replicative helicase, and it provides a platform to ensure their subsequent reassembly on the daughter strands. Our research thus offers a framework for understanding how modified parental histones (canonical and variant) can be handled during replication, a long-standing open question with broad implications for biology.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Protein expression and purification
All the proteins used in this study were expressed in the BL21(DE3) RIL cell strain (Stratagene). The human MCM2 histone-binding domain (HBD), fragments 43–160 or 61–130, were cloned into a modified RSFDuet-1 vector (Novagen), with an N-terminal His6-SUMO tag. All the MCM2 HBD(43–160) mutants were introduced by standard PCR procedures. The expressed proteins were first purified on an Ni-NTA affinity column. After removal of the His6-SUMO tag by Ulp1 (SUMO protease), the proteins were further purified on a HiLoad 16/600 Superdex 200 column (GE Healthcare).
For preparation of GST-tagged proteins, human ASF1a(1–155), MCM2 HBD(43–160) and full-length MCM2 were cloned into the pGEX-6P-1 vector (GE Healthcare). The V94R mutation of ASF1a(1–155) and the D80A Y81A double mutation of full-length MCM2 were introduced by PCR. The proteins were first purified with glutathione–Sepharose 4B and were further purified by gel filtration. The GST tags of full-length MCM2 were removed with 3C protease before the gel-filtration step.
For coexpression of human H3.3–H4, H3.2–H4 and His6-CENPA–H4 complexes, the genes encoding H3.3, CENPA and H4 were cloned into the pETDuet-1 vector, whereas H3.2 and H4 were cloned into the RSFDuet-1 vector. The H3.3–H4 and H3.2–H4 complexes were first captured by a heparin column, and the His6-CENPA–H4 tetramer was captured by an Ni-NTA affinity column. Then all the complexes were further purified with a ceramic hydroxyapatite column (Bio-Rad). The complexes of histone mutants were purified in the same way.
The GST-tagged ASF1a(1–155)–H3.2–H4 complex was prepared by coexpression of plasmids of GST-tagged ASF1a(1–155) and H3.2–H4 and was purified sequentially with glutathione–Sepharose 4B and heparin columns. For preparation of the ASF1a(1–172)–H3.2–H4 complex, all three genes were cloned into one vector for coexpression. Then the complex was purified by heparin column.
For purification of MCM2 HBD(43–160)–H3.2–H4, MCM2 HBD(43–160)–H3.3(Δ56)–H4 and MCM2 HBD(61–130)–H3.3(Δ56)–H4 complexes, the plasmids of His6-SUMO–tagged MCM2 HBD and H3.2–H4 or H3.3(Δ56)–H4 were coexpressed. The complexes were purified sequentially by Ni-NTA affinity column, subsequent removal of the tag, loading onto heparin column and further purification to homogeneity by gel-filtration column.
For purification of MCM2 HBD(43–160)–H3.2–H4–ASF1a(1–172) and MCM2 HBD(43–160)–H3.2(Δ55)–H4–ASF1a(1–172) complexes, first the cells expressing His6-SUMO–tagged MCM2 HBD alone were mixed with the cells coexpressing ASF1a(1–172)–H3.2–H4 or ASF1a(1–172)–H3.2(Δ55)–H4. Then the mixed cells were lysed together and copurified with an Ni-NTA affinity column; this was followed by removal of the tag and purification with a gel-filtration column.
For preparation of MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.3(Δ56)–H4 and MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.2(Δ55)–H4 complexes, the MCM2 HBD(61–130) and ASF1b(1–158) were covalently linked into one expressed cassette, and the cassette was cloned into the RSFDuet-1 vector with an N-terminal His6-SUMO tag. The resulting plasmid was coexpressed with plasmid containing H3.3(Δ56)–H4 or H3.2(Δ55)–H4. The complexes were first purified with an Ni-NTA affinity column. After removal of the tag, the complex was further purified by heparin and gel-filtration columns.
Crystallization
The MCM2 HBD(61–130)–H3.3(Δ56)–H4 complex (herein denoted the MCM2 HBD–H3.3–H4 tetramer complex) at a concentration of 26 mg ml−1 was crystallized in 1.5 M Na-K phosphate, pH 8.0, with the hanging-drop vapor-diffusion method at 20 °C.
The MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.3(Δ56)–H4 complex (herein denoted the MCM2 HBD–H3.3–H4 dimer–ASF1 complex) and MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.2(Δ55)–H4 complex (herein denoted the MCM2 HBD–H3.2–H4 dimer–ASF1 complex) at concentrations of 19 mg ml−1 and 15 mg ml−1, respectively, were crystallized in 3.2 M Na formate, 0.1 M Tris, pH 8.5, with the hanging-drop vapor-diffusion method at 20 °C.
All the crystals were soaked in a cryoprotectant made from mother liquor supplemented with 20% glycerol, except for the crystals of the MCM2 HBD–H3.2–H4 dimer–ASF1 complex with 10% glycerol, before being flash frozen in liquid nitrogen.
Structure determination
The data sets for the MCM2 HBD–H3.3–H4 tetramer complex were collected at 1.075 Å on beamline X29 (Brookhaven NSLS), and the data sets for the MCM2 HBD–H3.3–H4 dimer–ASF1 and MCM2 HBD–H3.2–H4 dimer–ASF1 complexes were collected at 0.979 Å on beamline 24-ID-C/E NE-CAT (Advanced Photo Source, Argonne National Laboratory). All the data sets were processed with HKL 2000. The initial structure for the MCM2 HBD–H3.3–H4 tetramer complex was solved by molecular replacement in PHASER53 with a search model edited from PDB 1AOI51 and was manually refined and built with Coot54. The final structure of this complex was refined to 2.85-Å resolution with PHENIX55. (The Ramachandran plot showed 95.8% favored and 4.2% allowed.) The structure of the MCM2 HBD–H3.3–H4 dimer–ASF1 complex was solved by molecular replacement in PHASER with a search model edited from PDB 2IO5 (ref. 26), and the structure of the MCM2 HBD–H3.2-H4 dimer–ASF1 complex was solved with the structure of the MCM2 HBD–H3.3–H4 dimer–ASF1 complex as a search model. The structures of the MCM2 HBD–H3.3–H4 dimer–ASF1 (98.1% favored and 1.9% allowed) and MCM2 HBD–H3.2–H4 dimer–ASF1 (98.1 favored and 1.9% allowed) complexes were refined to 2.30-Å and 2.90-Å resolution, respectively, with PHENIX. Table 1 summarizes the statistics for data collection and structural refinement.
Histidine and GST pulldowns
For the pulldowns of His6-SUMO–tagged MCM2 HBD(43–160) and its mutants with H3.2–H4 tetramer and histone mutants, 40 µL of Ni-NTA beads was suspended with 200 µL of binding buffer (20 mM Tris, pH 7.5, and 1 M NaCl), and 1 nmol of His6-SUMO–tagged proteins was added and incubated at 4 °C for 10 min; then 0.5 nmol of H3.2–H4 tetramer and histone mutants was added and incubated for another 30 min; then the beads were washed quickly six times with 1 mL of washing buffer (binding buffer, 45 mM imidazole, 10% glycerol and 1% Triton X-100) before elution with 40 µL of elution buffer (20 mM Tris, pH 7.5, 0.5 M NaCl, and 500 mM imidazole).
For the pulldowns of GST-tagged ASF1a(1–155) and its mutant V94R with MCM2 HBD(43–160)–H3.2–H4 complex and H3.2–H4 tetramer, 20 µL of glutathione–Sepharose 4B beads was suspended with 200 µL of binding buffer, and 1 nmol of GST-tagged proteins was added and incubated at 23 °C for 20 min; then 0.5 nmol of prepurified MCM2 HBD(43–160)–H3.2–H4 complex or H3.2–H4 tetramer was added and incubated for another 3 h; then the beads were washed quickly four times with 1 mL of washing buffer (binding buffer, 1% Triton X-100) before addition of 50 µL of sample loading buffer. The pulldowns of GST-tagged ASF1a(1–155)–H3.2–H4 complex with His6-SUMO–tagged MCM2 HBD(43–160) and its mutants were done in the same way as above, whereas the pulldowns of GST-tagged MCM2 HBD(43–160) with His6-CENPA–H4 tetramer were performed similarly but at 4 °C. All the samples were analyzed with SDS-PAGE.
DSS cross-linking assay under physiological conditions
First, 2 µM of H3–H4 dimer or different chaperone–histone complexes (listed above) was incubated with 500 µM of DSS (Thermo) at 23 °C for 30 min in 20 mM HEPES, pH 7.5, and 150 mM NaCl. Then 50 mM Tris, pH 7.5, was added to quench the reactions. The samples were analyzed with SDS-PAGE.
To monitor the influence of ASF1 on the MCM2 HBD–H3–H4 tetramer complex, 2 µM of MCM2 HBD(43–160)–H3–H4 tetramer complex was titrated with 0, 0.5, 1 and 2 µM of ASF1b(1–158), and the resulting titrations were incubated for 30 min. Then the reaction products were cross-linked with DSS and analyzed as above.
Tetrasome assembly assay on linear DNA
For tetrasome assembly on linear DNA, 0.4 µM of 147-bp Widom 601 DNA was mixed with 1.2 µM of H3.2–H4 tetramer or different chaperone–histone complexes in assembly buffer (10 mM Tris, pH 7.5, 0.5 mM EDTA, 50 µg/mL BSA, 10% glycerol and 150 mM NaCl). The free DNA, DNA plus free chaperones and preformed tetrasome (generated through the dilution method) served as controls. The reactions were incubated at 23 °C for 1 h and analyzed on 6% PAGE in 0.2× TBE.
Plasmid supercoiling assay
H2A–H2B (150 ng) was mixed with H3–H4 (150 ng), prepurified ASF1a(1–172)–H3–H4 complex (270 ng), MCM2 HBD(43–160)–H3–H4 complex (230 ng) or MCM2 HBD(43–160)–H3–H4–ASF1a(1–172) complex (340 ng), in assembly buffer (10 mM Tris, pH 7.5, 125 mM NaCl, 2 mM MgCl2, 0.5 mM DTT and 0.1 mg/ml BSA) with a final volume of 25 µl. The reactions were incubated at 37 °C for 40 min. At the same time, phix174 RF1 DNA (100 ng, from NEB) was mixed with topoisomerase I (8 U, Promega) in assembly buffer and incubated for 25 min. Then the relaxation reaction was mixed into the chaperone-histone reaction and incubated for 80 min. After that, 50 µl of stop buffer (20 mM Tris, pH 8.0, 20 mM EDTA, 1% SDS and 0.5 mg/ml proteinase K) was added to stop the reaction and was incubated for 20 min. Phenol/chloroform DNA extraction and ethanol precipitation were then performed. The samples including supercoiled and relaxed DNA controls were analyzed on a 1% agarose gel with 1× TAE buffer. The gel was visualized by EB staining. For assay of full-length MCM2, H2A–H2B (150 ng), H3–H4 (150 ng) and MCM2 (2.3 µg) or D80A Y81A mutant (2.3 µg) were sequentially added into 25 µl assembly buffer. Then the same protocol was followed.
Gel-filtration assay of chaperone–histone complexes
0.5 mg of MCM2 HBD(61–130)–H3.3(Δ56)–H4 (complex 1); MCM2 HBD(43–160)–H3.3(Δ56)–H4 (complex 2); MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.3(Δ56)–H4 (complex 3); MCM2 HBD(43–160)–H3.2(Δ55)–H4–ASF1a(1–172) (complex 4); MCM2 HBD(43–160)–H3.2–H4–ASF1a(1–172) (complex 5); or H3.2–H4 tetramer complex and 0.8 mg of MCM2 HBD(43–160)–H3.2–H4 (complex 6); and 0.3 mg of MCM2 HBD(61–130)–H3.3(Δ56)–H4–ASF1b(1–158) (complex 3′) were analyzed with a Superdex 200 10/300 GL column in a buffer containing 20 mM Tris, pH 7.5, and 300 mM NaCl. The peak fractions were resolved with SDS-PAGE.
0.5 mg of MCM2 HBD(43–160)–CENPA–H4 tetramer (complex 7) and of CENPA–H4 tetramer was analyzed similarly as above in high-salt buffer (20 mM Tris, pH 7.5, and 1 M NaCl) and in low-salt buffer (20 mM Tris, pH 7.5, and 300 mM NaCl). The peak fractions were resolved by SDS-PAGE.
Details for the following are described in Supplementary Note: SEC-MALS; ASF1-mediated disruption of MCM2 HBD–bound H3–H4 tetramers into dimers in solution; cloning and mutagenesis; cell culture, transfection and drug treatment; immunofluorescence analysis; western blotting and antibodies; and quantitative real-time PCR.
Extract preparation
For DNase I extracts, soluble proteins were extracted by incubation for 10 min with CSK-T buffer (10 mM PIPES, pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% Triton X-100) supplemented with ATP (1 mM, Sigma) and protease and phosphatase inhibitors (5 mM Na fluoride, 10 mM β-glycerolphosphate, 0.2 mM sodium vanadate, 10 µg/ml leupeptin, 10 µg/ml pepstatin and 0.1 mM PMSF, Sigma). Insoluble material was collected by centrifugation at 1,500g for 5 min before incubation with DNase I (1,000 U/mL, Roche) in CSK buffer with 0.1% Triton X-100 and protease and phosphatase inhibitors for 30 min at 25 °C. DNase I–released material was isolated by centrifugation at 16,000g for 10 min, and the remaining pellet was resuspended in Laemmli sample buffer (50 mM Tris, pH 6.8, 100 mM DTT, 2% SDS, 7.5% glycerol, and bromophenol blue). For chromatin solubilization with benzonase, soluble protein was removed by CSK-T as described above. Insoluble material was incubated with benzonase (2,500 U/mL, Millipore) in CSK buffer with 0.1% Triton X-100 and protease and phosphatase inhibitors for 20 min at 30 °C. Benzonase-released material was isolated by centrifugation at 16,000g for 10 min. NP-40–NaCl extracts were made in buffer E (300 mM NaCl, 0.5% NP-40, 50 mM Tris, pH 7.8, 0.2 mM EDTA, 5% glycerol, and protease and phosphatase inhibitors). For measurement of histone stability, non-nucleosomal histones were extracted by NP-40–NaCl and cleared by centrifugation.
Complex purification and sequential immunoprecipitation
Coimmunoprecipitation experiments were performed on soluble fractions (NP-40–NaCl or CSK-T extracts) or DNase I–solubilized chromatin7, 56 with HA or Flag affinity resins. ATP was included in the reactions to preserve MCM2–7 complexes56, and DNase I digestion was limited to enrich for active helicase complexes in replicating chromatin. Extracts were precleared with Superflow 6 beads (IBA, 2-0412-000) for 1 h at 4 °C. This was followed by incubation with Flag magnetic beads (Sigma, M8823) or HA magnetic beads (Thermo Scientific, 88836) for 2–3 h at 4 °C. For the HA IPs, complexes were washed six times in wash buffer A (300 mM NaCl, 0.2% NP-40, 50 mM Tris, pH 7.8, 0.2 mM EDTA, 5% glycerol and protease and phosphatase inhibitors) and boiled in 1× LSB for 15 min. For Flag IP from soluble extracts, the beads were washed six times in wash buffer A. Beads were then equilibrated in elution buffer (150 mM NaCl, 0.2% NP-40, 50 mM Tris, pH 7.8, 0.2 mM EDTA, and protease and phosphatase inhibitors) and complexes were eluted with Flag peptide (Sigma) in three steps of 20 min each. For Flag IP and HA IP from DNase I extracts, complexes were washed six times in buffer B (CSK, 0.1% Triton X-100, 1 mM ATP, protease and phosphatase inhibitors, and 0.5 mM DTT) and eluted by Flag peptide in buffer B or by glycine (100 mM, pH 2.5). Samples for CENPA coprecipitation were washed in buffer B with 300 mM NaCl.
Sequential coimmunoprecipitation experiments were performed with benzonase-solubilized chromatin. Extracts were precleared with Superflow 6 beads (IBA, 2-0412-000) for 30 min at 4 °C. This was followed by incubation with V5 beads (Sigma, 121M4843) for 2.5 h. Beads were washed six times for 2 min each in buffer C (CSK, 0.1% Triton X-100, 200 mM NaCl, 1 mM ATP, and inhibitors). Complexes were eluted three times for 20 min each with V5 peptide (6 mg/ml, Sigma V7754), diluted to a final volume of 300 µl in buffer C and incubated with HA magnetic beads (Thermo Scientific, 88836) for 2.5 h. Beads were washed six times for 2 min each in buffer C and finally boiled in 1× LSB.
Proximity ligation assay
Coverslips were incubated with primary antibodies anti-mouse MCM2 (Bethyl, A300-122A), anti-rabbit ASF1a (Cell Signaling Technology, 2990S), and/or anti-mouse CDC45 (Santa Cruz, sc-55569) overnight at 4 °C. Validation of the primary antibodies is provided on the manufacturers’ websites. Secondary antibodies (Duolink In situ PLA probes mouse plus (DUO92001) and rabbit minus (DUO92005)) and detection reagents (Duolink In situ Detection Reagents Red (DUO92008)) were from Sigma, and the assay was performed according to the manufacturer’s instructions. All fluorescence images were collected on a DeltaVision system and analyzed by Volocity 6.2.1. Brightness and contrast were adjusted with Adobe Photoshop CS6. For the ASF1a–CDC45 PLA, images were taken as Z-stack projections, and Quick Projections of the Z-stacks were subsequently analyzed.
SNAP-HA-H3.1 labeling
U-2-OS cells stably expressing SNAP-HA-H3.1 WT or R63A K64A were seeded onto 96-well Screenstar microplates (Greiner Bio-One) 48 h before analysis. SNAP labeling was performed as described previously57. In brief, cells were quenched with SNAP-Cell Block (10 µM, NEB) for 30 min, washed twice in PBS, recovered for 30 min in complete medium, washed with PBS twice, chased for 4 h in complete medium, washed with PBS twice, and finally pulse labeled with SNAP-Cell TMR (2 µM, NEB) for 20 min. After labeling, cells were washed in PBS twice, allowed to recover for 30 min in complete medium, washed in PBS twice and preextracted in CSK-T. Microscopic analysis was performed with a 20× dry objective on an automated IX-83 fluorescence imaging system combined with ScanR image acquisition and analysis software (Olympus). Median TMR signal intensity was determined for TMR-positive nuclei. On average, we scored >1,000 TMR-positive cells per technical replicate. Each biological replicate represents the average of five technical replicates.
Analysis of cell proliferation by live-cell imaging
Cell proliferation was monitored by live-cell high-content imaging in an IncuCyte (Essen Bioscience) for 100 h. U-2-OS cells stably expressing siRNA-resistant V5-MCM2 were plated in 12-well plates and transfected with MCM2-targeting siRNAs or control siRNAs. 6 h after transfection, the medium was changed, and the plates were placed in the IncuCyte for cell-proliferation analysis. 30 h after transfection, cells were treated with 500 µM HU where indicated.
Supplementary Material
ACKNOWLEDGMENTS
We thank the beamline staff at the synchrotrons at the Argonne National Laboratory (NE-CAT) and the Brookhaven National Laboratory (XL-29) for technical assistance. We thank H. Wu for access to SEC-MALS equipment for molecular-weight measurements. We thank Y. Feng and C. Alabert for help with immunofluorescence and PLA analysis. We thank C. Alabert for comments on the manuscript and Z. Jasencakova for help with experiments and for input in the manuscript and the model. We also thank P. Meraldi (University of Geneva) and L. Jansen (Gulbenkian Institute) for reagents and K. Labib (University of Dundee) for sharing information before publication. D.J.P. was supported in part by grants from the Leukemia and Lymphoma Society (LLS 7006-13) and the Starr foundation (I5-A554). A.G. is supported as a European Molecular Biology Organization Young Investigator, and her research is supported by the Danish National Research Foundation to the Center for Epigenetics (DNRF82), the European Commission ITN FP7 ‘aDDRess’, a European Research Council Starting Grant (ERC2011StG, no. 281,765), the Danish Cancer Society, the Danish Medical Research Foundation and the Lundbeck Foundation.
Footnotes
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes PDB 5BNV (MCM2 HBD–H3.3–H4 tetramer complex), PDB 5BNX (MCM2 HBD–H3.3–H4 dimer–ASF1 complex) and PDB 5BO0 (MCM2 HBD–H3.2–H4 dimer–ASF1 complex).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
H.H. conceived and led the generation of cassettes for crystallization of the complexes, and H.H. solved the structures under the supervision of D.J.P. C.B.S. and A.G. conceived and led the functional studies. G.S. performed immunoprecipitation of HA–H3.1 from chromatin. M.H. analyzed histone incorporation by SNAP-tag assay. A.S. carried out lentiviral transduction and cell characterization. C.G.-A. helped with cloning and data analysis. S.C. performed SEC-MALS molecular-weight measurements. H.H., C.B.S., A.G. and D.J.P. wrote the manuscript, and all authors commented on the manuscript.
COMPETING FINANCIALINTERESTS
The authors declare no competing financial interests.
References
- 1.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 2.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 5.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 7.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 8.Ishimi Y, Komamura-Kohno Y, Arai K, Masai H. Biochemical activities associated with mouse Mcm2 protein. J. Biol. Chem. 2001;276:42744–42752. doi: 10.1074/jbc.M106861200. [DOI] [PubMed] [Google Scholar]
- 9.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 10.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: breaking up is hard to do. Curr. Opin. Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 12.McKnight SL, Miller OL. Jr. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12:795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 13.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes: the replication fork, core histone segregation and terminal structures. J. Mol. Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 14.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 15.Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- 16.Annunziato AT. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta. 2013;1819:196–210. doi: 10.1016/j.bbagrm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 18.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 19.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 20.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foltman M, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Reports. 2013;3:892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Ishimi Y, Komamura Y, You Z, Kimura H. Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 24.Richet N, et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 27.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Mello JA, Almouzni G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 29.Groth A, et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montagnoli A, et al. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- 33.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillespie PJ, Blow JJ. Clusters, factories and domains: the complex structure of S-phase comes into focus. Cell Cycle. 2010;9:3218–3226. doi: 10.4161/cc.9.16.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latreille D, Bluy L, Benkirane M, Kiernan RE. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Res. 2014;42:3542–3550. doi: 10.1093/nar/gkt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 39.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 40.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 42.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klapholz B, et al. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118:235–248. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- 44.Mejlvang J, et al. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houlard M, et al. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, et al. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev. Biol. 2007;311:213–222. doi: 10.1016/j.ydbio.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 47.Groth A. Replicating chromatin: a tale of histones. Biochem. Cell Biol. 2009;87:51–63. doi: 10.1139/O08-102. [DOI] [PubMed] [Google Scholar]
- 48.Xu M, et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 49.Huang C, et al. H3.3–H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 2013;9:e1003558. doi: 10.1371/journal.pgen.1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 52.Amaro AC, et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 56.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. In vivo interaction of human MCM heterohexameric complexes with chromatin: possible involvement of ATP. J. Biol. Chem. 1997;272:10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- 57.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.