Abstract
The genus Phasmarhabditis is an economically important group of rhabditid nematodes, to which the well-known slug-parasite P. hermaphrodita belongs. Despite the commercial use of Phasmarhabditis species as an attractive and promising approach for pest control, the taxonomy and systematics of this group of rhabditids are poorly understood, largely because of the lack of diagnostic morphological features and DNA sequences for distinguishing species or inferring phylogenetic relationship. During a nematode sampling effort for identifying free-living relatives of Caenorhabditis elegans in Huizhou City, Guangdong, China, a novel species belonging to the genus Phasmarhabditis was isolated from rotting leaves. Detailed morphology of the gonochoristic P. huizhouensis sp. nov. was described and illustrated. The adult female has a robust body, a relatively short and wide buccal capsule conjoined by a rhabditiform pharynx. Females are characterized by a short cupola-shaped tail end bearing a slender pointed tip, with the junction flanked by a pair of ‘rod-like’ phasmids. Males have an open peloderan bursa that is supported by 9 pairs of genital papillae and 1 terminal pair of phasmids. P. huizhouensis sp. nov. is morphologically very similar to the type species Phasmarhabditis papillosa but is distinguishable by its male caudal traits. The new species is readily differentiated from other taxa in the genus by its female tail shape. Molecular phylogenetic inferences based on small subunit (SSU) and the D2-D3 domain of large subunit (LSU) ribosomal DNA genes reveal that P. huizhouensis sp. nov. forms a unique branch in both phylogenies which is genetically related to P. hermaphrodita and other parasites such as Angiostoma spp. The host associations of P. huizhouensis sp. nov. and its ability to parasitize slugs are unknown.
Introduction
During our sampling of free-living nematodes in south China’s Guangdong Province, a novel rhabditid nematode was recovered and reared on standard C. elegans culture media seeded with Escherichia coli strain OP50. The rhabditid has a relatively short and wide buccal capsule with simple metastegostom ridges, female ‘rod-like’ prominent phasmids and male open peloderan bursa with 10 pair of bursal papillae, morphologically belonging to the ‘Papillosa’ group in the genus Pellioditis Dougherty, 1953 sensu Sudhaus [1–3]. However, the revision of Pellioditis by Andrássy was disparate, with the generic name assigned to the revised ‘Papillosa’ group being completely different to that assigned by Sudhaus. Andrássy established a new genus of Phasmarhabditis including Pelodera papillosa (syn. Pellioditis papillosa) as the type species, P. hermaphrodita and P. neopapillosa from the ‘Papillosa’ group, and two other species: Rhabditis (Cephaloboides) nidrosiensis with a marine habitat and R. (C.) valida with a littoral habitat [4–5]. Nevertheless, in a recent revision to the family Rhabditidae by Sudhaus, both species of nidrosiensis and valida were classified into another genus (Buetschlinema Sudhaus, 2011) and the remaining Phasmarhabditis taxa were replaced in Pellioditis [3]. In Sudhaus’ revision of the original Pellioditis, all species were moved into other genera except the ‘Papillosa’ group [3], whereas only this group was transferred in the revision by Andrássy [5]. In other words, the revised Pellioditis sensu Sudhaus [1–3] is a taxon basically equivalent to Phasmarhabditis sensu Andrássy [4–5] containing stem species of the ‘Papillosa’ group, i.e., P. papillosa, P. phasmarhabditis and P. neopapillosa. Though the taxon Pelodytes Schneider, 1895 with type species hermaphrodites (= hermaphrodita) preceded both taxa of Pellioditis and Phasmarhabditis, it was preoccupied by an amphibian genus [6–8]. The nomenclature ‘Pellioditis’ has priority over ‘Phasmarhabditis’, however, both generic names assigned to a synonymous group of rhabditids were simply indicative of diverse perspectives of revisions by Sudhaus and Andrássy respectively. To avoid taxonomic confusion, the new species is herein named as Phasmarhabditis huizhouensis sp. nov., with its specific epithet derived from its locality (Huizhou City).
Phasmarhabditis hermaphrodita is a well-known deadly parasite of slug pests of agricultural and horticultural crops. It is known to be capable of killing many species of slugs from several families. This nematode has been studied intensively and formulated into an effective biological control agent called Nemaslug® which was sold in Europe [9–13]. Recently, P. phasmarhabditis was reported for the first time in North America from the invasive slugs Deroceras reticulatum, D. laeve and Lehmannia valentiana [8]. P. neopapillosa is a close relative of P. hermaphrodita that is morphologically indistinguishable by females or hermaphrodites. The species can be differentiated by the frequency of males. Males are common in P. neopapillosa, but rare or absent in P. hermaphrodita [14]. Although P. papillosa was isolated from decaying organic materials, this species is also associated with terrestrial snails and slugs [15–16]. Later, P. tawfiki was described from Greater Cairo, Egypt, where it was isolated from the snail Eobania vermiculata Müller and the slug Limax flavus Linneaus, with males rare [17]. In addition, two undescribed species were found associated with the slug D. reticulatum and its relative Ariostralis nebulosa in South Africa [18], and another unidentified isolate was reported to infect the earthworm Lumbricus terrestris in Champaign, Illinois, USA [19]. All these Phasmarhabditis species have similar host associations. They are either slug-parasites or associated with slugs, snails or earthworms. Recent molecular studies revealed that all Phasmarhabditis species are in a monophyletic group [8].
Materials and Methods
Nematode isolation, culture and microscopic observation
Phasmarhabditis huizhouensis sp. nov. was described and illustrated using a wild isolate ZZY0412 that was recovered from rotting leaves in Kowloon Peak of Huizhou City, Guangdong Province, China (our sampling activity was conducted using rotting plant leaves, so no specific permissions were required, and the field studies did not involve endangered or protected species). Samples were placed onto a clean agar plate. Nematode individuals were picked out then cultured at room temperature using the C. elegans food (E. coli OP50). Measurements [20], drawings and descriptions were conducted from quiescent adult worms incubated at 4°C for 12h after recovery from 3-day culture plates. Differential interference contrast (DIC) images were taken with an Eclipse Ti Inverted Microscope (Nikon). Scanning electron microscopy (SEM) of P. huizhouensis sp. nov. was performed either using the Hitachi_S-450 scanning electron microscope at Hong Kong Baptist University or the Hitachi_S-3400N at China Agricultural University by the methods described previously [21]. Permanent slides were made using the specimens fixed in 3% formaldehyde, which were subsequently mounted in anhydrous glycerin under a paraffin seal after gradual dehydration of nematodes on a hot plate at 70°C[22].
DNA sequencing and phylogenetic analysis
For molecular analysis, genomic template DNA of Phasmarhabditis huizhouensis sp. nov. strain ZZY0412 was extracted using PureLinkTM Genomic DNA Mini Kit (Invitrogen, USA). SSU rDNA gene fragment of 1633 base pairs (bp) was amplified by PCR using three sets of universal primers shown below: (1) SSU18A (5’-AAAGATTAAGCCATGCATG-3’) and SSU26R (5’-CATTCTTGGCAAATGCTTTCG-3’) [23–24]; (2) RHAB1350F (5’-TACAATGGAAGGCAGCAGGC-3’) and RHAB1868R (5’-CCTCTGACTTTCGTTCTTGATTAA-3’) [23, 25]; and (3): SSU24F (5’-AGRGGTGAAATYCGTGGACC-3’) and SSU18P (5’-TGATCCWMCRGCAGGTTCAC-3’) [26–27]. Forward and reverse primer sequences for amplification of 902-bp D2-D3 LSU rDNA sequence were No. 391 (5’-AGCGGAGGAAAAGAAACTAA-3’) [28–29] and No. 501 (5’-TCGGAAGGAACCAGCTACTA-3’) [28, 30], respectively. PCR amplification and sequencing were performed as previously described [21]. The SSU and the D2-D3 domain of LSU rDNA sequences were deposited in GenBank database with accession numbers of KP017252 and KP017253 respectively, and were aligned against the sequences from GenBank to identify closely related species.
The model of base substitution was evaluated using MODELTEST [31–32]. The estimates of base frequencies, the proportion of invariable sites, the gamma distribution shape parameters and substitution rates suggested by the Akaike Information Criterion were used in phylogenetic analyses. Bayesian analysis was performed to confirm the tree topology for each gene separately using MrBayes 3.1.0 [32] running the chain for 1 × 107 generations and setting the “burnin” at 2,5000 which is 25% of the run. The sampling and printing frequencies are 100 and 1000 respectively. We used the Markov Chain Monte Carlo (MCMC) method within a Bayesian framework to estimate the posterior probabilities of the phylogenetic trees using 50% majority rule [33]. Tracer was used for assessing and summarizing the posterior estimates of the various parameters sampled by the Markov Chain and confirming that the burnin was approaching stationarity (http://tree.bio.ed.ac.uk/software/tracer).
Nomenclatural acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature, and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to the prefix "http://zoobank.org/". The LSID for this publication is: urn:lsid:zoobank.org:pub:47BE7062-4ECC-41B3-85CE-C3A53BE6C399. The electronic edition of this work was published in a journal with an ISSN, and has been archived and is available from the following digital repositories: PubMed Central, LOCKSS.
Results
Phasmarhabditis huizhouensis Huang, Ye, Ren & Zhao sp. nov.
urn:lsid:zoobank.org:act:B24611B5-9A23-4B2D-BAC4-D6EA2647E57B (Figs 1–3)
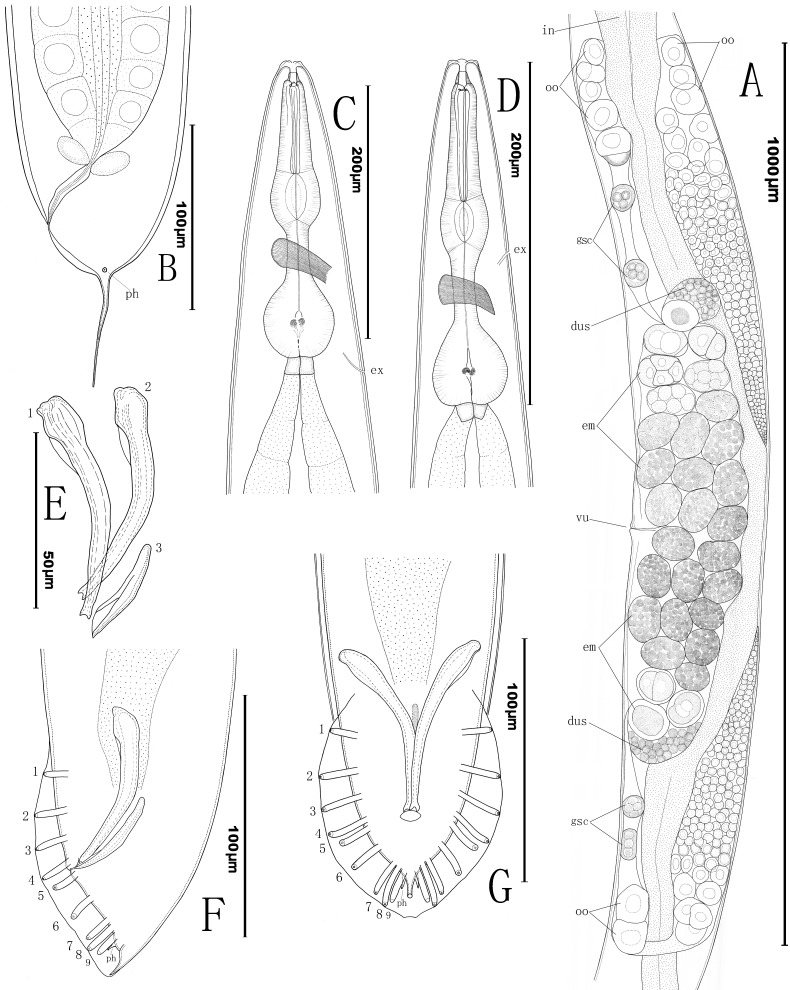
Fig 1. Line drawings of Phasmarhabditis huizhouensis sp. nov.
(A) Female reproductive system (in: intestine; oo: oocytes; gsc: aggregated sperm cells; dus: distal part of uteri filled with sperms; em: embryos; vu: vulva). (B) Female tail (ph: phasmid). (C) Anterior region of female (ex: excretory pore). (D) Anterior region of male. (E) Spicules and gubernaculum (1: spicule in dorsal view; 2: spicule in ventral view; 3: gubernaculum from sublateral view). (F) Lateral view of male tail (1–9: nine pairs of genital papillae; ph: phasmid). (G) Ventral view of male tail with an open peloderan bursa supported by 9 pairs of genital papillae and 1 terminal pair of phasmids.
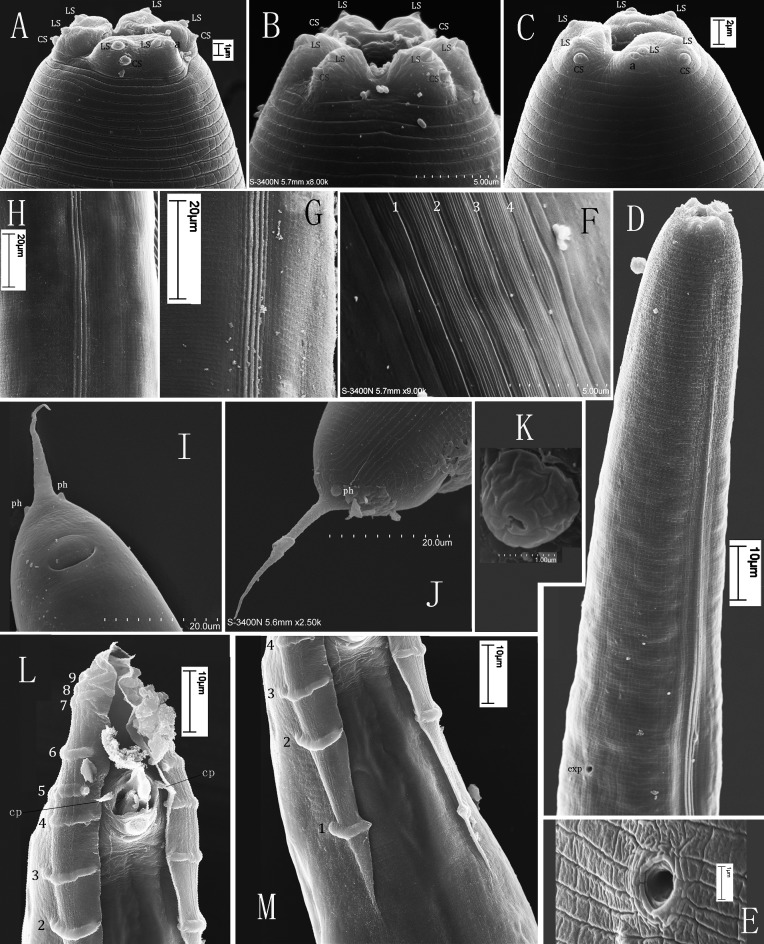
Fig 3. Scanning electron micrographs of Phasmarhabditis huizhouensis sp. nov.
(A) and (B) Head of females (LS: labial sensilla; a: amphid; CS: cephalic sensilla). (C) Head of male. (D) Anterior region of male showing excretory pore (exp). (E) Magnification of the excretory pore. (F) and (G) Lateral fields of females (1–4: 4 incisures compose the three middle ridges/bands). (H) Lateral field of male. (I) and (J) Ventral and lateral views of female tails (ph: phasmid). (K) Magnification of female phasmid. (L) and (M) Male caudal region with an open bursa as well as its 9 pairs of genital papillae (1–9) (cp: cloacal papillae). [(B), (F), (I), (J) and (K) were captured by the Hitachi_S-3400N scanning electron microscope, others by the Hitachi_S-450].
Description
Adults
Cuticle surface smooth or finely annulated. Lateral field longitudinally striated, often bearing 7–11 lines (ridges or incisures) with 3 prominent ridges or bands in the middle (Fig 3F–3H). Lip region flattened anteriorly, slightly higher in females than males. Six lips bearing two circles of sensilla arranged in three pairs: one dorsal and the others sub-ventral, encircling a triangular mouth opening (Fig 3A–3C). Each lip with one apically-inserted, papilliform sensillum. Lateral lips present a pair of small, pore-like amphidial openings, located near the labial sensilla. The other four sub-lateral lips each with 1 papilliform cephalic sensillum posterior to the labial one (Fig 3A–3C). Stoma capsule relatively wide and short (15–24 μm × 5–8 μm), isomorphic/isotropic. Anterior part of the stoma (Cheilostom) cuticularized; gymnostom together with prostegostom and mesostegostom forming main part of the stoma, cylinder-like and slightly funnel-shaped; metastegostom ridges present, but lacking teeth or denticles; telostegostom short (Fig 2D and 2E). Anterior part of pharynx (= pro- + meta-corpus) slightly longer than posterior (= isthmus + basal bulb). Metacorpus muscular, forming swollen median bulb (25–52 μm in diam.). Isthmus distinct. Nerve-ring encircling posterior region of isthmus. Basal bulb pyriform (39–71 μm in diam.) with valvular apparatus, but posterior structure of haustrulum obscure in many specimens. Pharyngo-intestinal valves (cardia) present. Excretory pore observed behind the base of basal bulb by light microscopy in a few females, but only ca 106 μm from anterior end revealed by SEM examination for one male (Fig 3D). Measurements are listed in Table 1.
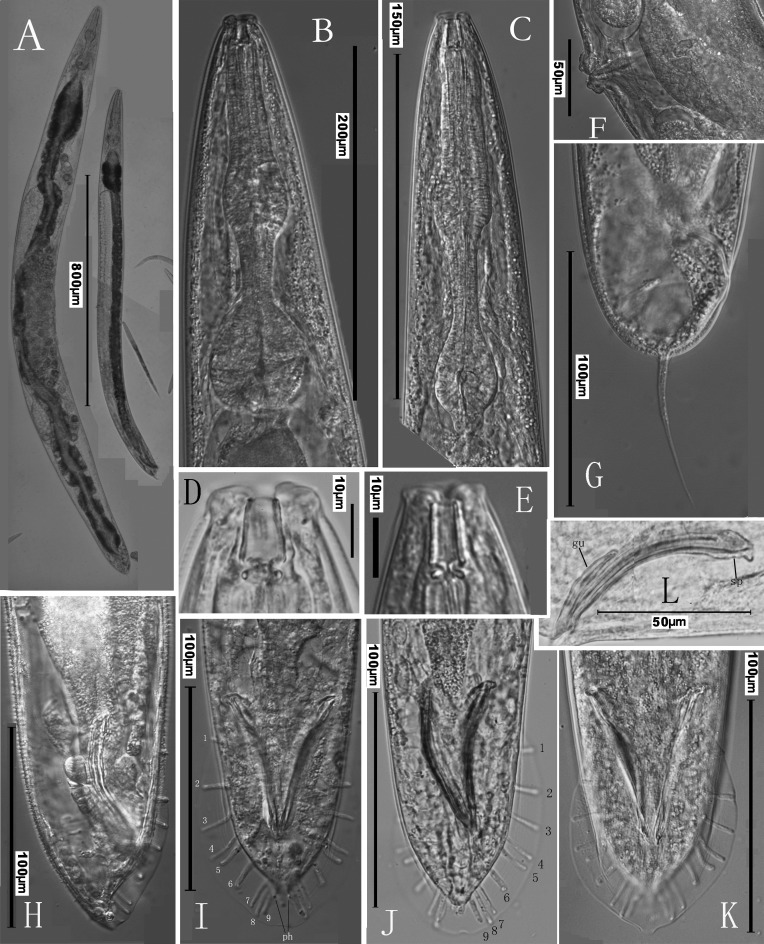
Fig 2. Micrographs of Phasmarhabditis huizhouensis sp. nov.
(A) Overview of female (left) and male (right) adults. (B) Pharyngeal region of an adult female. (C) Pharyngeal region of an adult male. (D) Anterior end (stoma) of a female. (E) Anterior end (stoma) of a male. (F) Vulva. (G) Female tail. (H) Lateral view of male tail. (I) and (J) Ventral view of male tails showing ten pairs of bursal papillae [nine pairs of genital papillae (1–9) and one terminal pair of phasmids (ph) (I)]. (K) Ventral view of the male caudal bursa. (L) Spicules (sp) and gubernaculum (gu).
Table 1. Morphometrics of Phasmarhabditis huizhouensis sp. nov. [20].
All measurements are in μm and shown in the form: mean ± s.d. (range).
| Character | Females | Males |
|---|---|---|
| (n = 16) | (n = 14) | |
| L | 1875.19 ± 340.84 (1333.07–2341.16) | 1282.11±247.97 (907.89–1669.21) |
| a | 13.97 ± 0.94 (12.61–15.97) | 14.69±1.44 (12.11–17.3) |
| b | 8.33±1.05 (6.39–10.47) | 6.6±0.79 (5.3–7.76) |
| c | 20.55±3.3 (15.5–26.86) | 26.83±3.54 (20.85–34.27) |
| c’ | 1.61±0.17 (1.28–2.02) | 1.14±0.18 (0.90–1.47) |
| V | 52.41±0.85 (50.78–54.01) | – |
| Stoma length | 19.68±1.98 (16.84–23.49) | 17.8±1.78 (15.1–20.47) |
| Stoma diam. 1 | 6.53±0.47 (5.91–7.89) | 5.66±0.48 (5.03–6.49) |
| Pharynx length | 214.59±19.62 (183.93–251.02) | 189.04±12.01 (168.84–211.45) |
| Anterior part of pharynx length | 117.17±10.34 (101.03–138.93) | 104.54±5.91 (91.5–112.1) |
| Posterior part of pharynx length | 99.07±10.2 (84.72–117.17) | 85.73±7.3 (78.39–103.03) |
| Distance from anterior end to | 220.2±19.98 (188.55–255.89) | 195.43±12.71 (176.88–222.22) |
| the base of pharynx | ||
| Diam. of median bulb | 37.85±7.11 (24.16–51.68) | 28.94±2.98 (25.17–34.23) |
| Diam. of terminal bulb | 53.24±9.64 (39.06–71.14) | 40.91±6.41 (27.85–52.68) |
| Body diam. | 134.38 ± 23.95 (85.5–171) | 88.09±15.45 (65.14–113.99 |
| Vulva body diam. | 133.45 ± 20.98 (85.5–162.85) | – |
| Vulva to anus distance | 800.22±169.49 (518.22–1053.37) | – |
| Anal body diam. | 57.22 ± 5.89 (42.62–67.11) | 41.02±6.99 (30.87–55.17) |
| Tail length | 91.14±6.46 (80.81–105.58) | 47.28±8.06 (35.54–61.41) |
| Length of female pointed tip | 56.03±4.99 (48.32–63.97) | – |
| Spicule length | – | 69.96±5.62 (61.07–81.88) |
| Gubernaculum length | – | 35.44±3.07 (29.9–41) |
| Gubernaculum length as % spicule length | – | 50.4±2.03 (48–55) |
1 Measured in the middle of the stoma; diam. denotes diameter.
Female
Body robust, tapering gradually to a blunt anterior end. Gonad didelphic, amphidelphic; anterior branch on the right of intestine, but posterior arm to the left. Vulva located near the middle of entire body (V = 51–54) with lips protruding (Fig 2F). Paired ovaries reflexed opposite to the vulva. Germ cells in both distal parts of ovaries arranged in a central rachis with proximal cells in elongation. Maturing oocytes proximally located in a single row at reflexed regions of both ovaries. Oviduct relatively long (127–221 μm). One or more aggregations of sperm cells visible occasionally but small (24–53 μm × 20–43 μm), each containing two or more sperm cells. Uteri often with numerous embryos irregularly arranged. Many embryos usually carrying unhatched larvae. Both distal parts of uteri (20–69 μm × 36–73 μm) filled with sperm cells, sometimes visible. Tail short and cupola-shaped with a slender pointed tip in the middle (Fig 2G). The pointed tip varying in length from 48 to 64 μm. Phasmids ‘rod-like’ papilliform in SEM (1.6–2.2 μm in diam.), flanking the junction between the cupola-shaped tail end and its slender tip (Fig 3I–3K).
Male
Males of Phasmarhabditis huizhouensis sp. nov. are common in culture. Testis single-armed, located on right of intestine with anterior end reflexed ventrally. The reflexed part approximately extending for 300–500 μm. Spermatogonia arranged in two to four rows; maturing spermatocytes situated proximally in two to three rows; then spermatids in multiple rows present in remainder of testis. Caudal region enveloped by an open medium-developed peloderan (tail-encompassing) bursa (88–95 μm × 69–78 μm) (Fig 3L and 3M). 10 pairs of bursal papillae present, comprising 9 pairs of genital papillae (GP) or rays (1+1+1+2+1+3) and 1 terminal pair of papilliform phasmids (ph) (Fig 2I). GP1, GP2, and GP3 precloacally standing; GP4 and GP5 cloacally situated; the rest post-clocacally located. GP1, GP2, GP3, GP4, and GP8 extending to the velum edge or near it. The sensory tip of GP5 lying to the dorsal surface; GP9 also appears dorsal (Fig 3L). An additional pair of small cone-shaped papillae flanking the cloaca (Fig 3L). Spicules 61–82 μm long, paired, separated, but in “Y” shape from ventral view when jointed by gubernaculum. Each spicule shaft slightly expanded in anterior end then gradually tapered to a distal small forked terminus (an obtuse tip divided by shallow indentation) (Fig 2L). Gubernaculum short, only half spicule length. Phasmids in fine cone shape, located posterior to GP9 where flanking the tail terminal tip (Fig 2I). Tail short conoid, tapered to a pointed terminus.
Type locality
The type specimens of Phasmarhabditis huizhouensis sp. nov. were derived from Strain ZZY0412, which was established from individuals isolated from rotting leaves in Kowloon Peak of Huidong County, Huizhou City, Guangdong Province, P.R. China (22°59′N, 114°43′E) by rearing on NGM agar plate seeded with colibacillus Escherichia coli strain OP50.
Type materials
Fifteen permanent slides containing paratype males and females of Phasmarhabditis huizhouensis sp. nov. strain ZZY0412 were deposited in the United States Department of Agriculture Nematode Collection (USDANC), Beltsville, MD, USA (T-6557p to T-6571p). Additional specimens fixed in 3% formalin were deposited in the laboratory of the Department of Biology, Hong Kong Baptist University, Hong Kong, China.
Living worms of the type isolate ZZY0412 were cryogenically preserved and deposited in the Department of Biology, Hong Kong Baptist University, Hong Kong, China.
Diagnosis and relationships
Phasmarhabditis huizhouensis sp. nov. is characterized by the following features, including gonochoristic reproductive mode, large female body size (L = 1.3–2.3 mm; ‘a’ value = 12.6–16), longitudinally striated lateral field with 3 distinct ridges/bands, relatively short and wide buccal capsule with simple metastegostom ridges, and a rhabditiform pharynx. Female didelphic-amphidelphic reproductive system carrying numerous embryos, vulva mid-body located (V = 51–54); a short cupola-shaped tail end conjoining a slender pointed tip which is flanked by a pair of ‘rod-like’ phasmids. Male open peloderan bursa medium-developed that is supported by 9 pairs of genital papillae and 1 terminal pair of phasmids; spicules paired, with small forklike distal termini (obtuse tips divided by shallow indentation) and a short gubernaculum.
Phasmarhabditis huizhouensis sp. nov. morphologically belongs to the genus because of its large female body size, relatively short and wide buccal capsule, longitudinally striated lateral field, female ‘rod-like’ phasmids, and male open peloderan bursa as well as its 10 pairs of bursal papillae. Within the genus Phasmarhabditis, P. huizhouensis sp. nov. is clearly distinguished from P. hermaphrodita, P. neopapillosa and P. tawfiki by the female/hermaphroditic tail shape (a short cupola joining a slender pointed tip vs. a relatively short conoid gradually tapered to a pointed terminus) [14, 17]. P. huizhouensis sp. nov. shares natural habitats (e.g. isolated from decaying organic substances) and the female tail shape (cupola-shaped with a slender tip) with P. papillosa, but the new species can be differentiated from P. papillosa by a significantly longer spicule length (61–82 μm vs. ca 30 μm for P. papillosa, as measured from Fig 1B [15]) and a less-developed peloderan bursa (88–95 μm × 69–78 μm vs. 141 μm × 107 μm—the size for P. papillosa in Fig 1B [15]). In addition to these Phasmarhabditis species, there are remaining taxa of P. incilaria, P. mairei and P. pellio in the genus Pellioditis sensu Sudhaus [3]. P. huizhouensis sp. nov. is different from P. pellio in female body size: L = 1.3–2.3 mm vs. 0.79–0.90 mm, and male spicule length: 61–82 μm vs. only 21–26 μm for P. pellio [34]. The new species differs from P. incilaria in the stoma morphology (without ornamentation on metastegostom ridges vs. bearing bristle-like denticles) [5]. The description of P. mairei is unavailable and this species was considered to be ‘species inquirenda’ by Andrássy [5]. Within the genus Pellioditis sensu Andrássy [5], P. huizhouensis sp. nov. is distinguished from all species in the genus by the typology of stoma (relatively short and wide without ornamentation on glottoid apparatus vs. at least four times as long as its width with warts on metastegostom ridges).
Molecular analysis
The best scores from the query of 1633-bp SSU gene fragment of P. huizhouensis sp. nov. against the GenBank database are the sequences from 3 isolates of unidentified Phasmarhabditis species, 6 isolates of P. hermaphrodita and 1 isolate of P. neopapillosa. SSU sequence alignments between P. huizhouensis sp. nov. and the most similar isolates revealed that the minimal sequence divergence is from an unidentified species ITD046 from North America (KM510210) [8], with 2.88% divergence over complete coverage. The four US P. hermaphrodita isolates of ITD290 (KM510209), ITD272 (KM510208), ITD207 (KM510207), and ITD056 (KM510206) [8] have a uniform divergence of 3.55% over complete coverage. The sequence of P. neopapillosa (FJ516754) gives 95% coverage yielding 2.94% divergence. The best match to the 902-bp D2-D3 sequence of P. huizhouensis sp. nov. from GenBank is another unidentified Phasmarhabditis species EM434 (EU195967), with 5.78% divergence over 95% coverage. D2-D3 sequence alignments between P. huizhouensis sp. nov. and the four US P. hermaphrodita isolates of ITD290 (KM510296), ITD272 (KM510195), ITD207 (KM510194), and ITD056 (KM510193) [8], yielded only 59% coverage with a uniform divergence of 7.84%. Comparisons of the sequences of the SSU and D2-D3 domain of the LSU rDNA genes between P. huizhouensis sp. nov. and its relatives further support that our rhabditid nematode is a novel species.
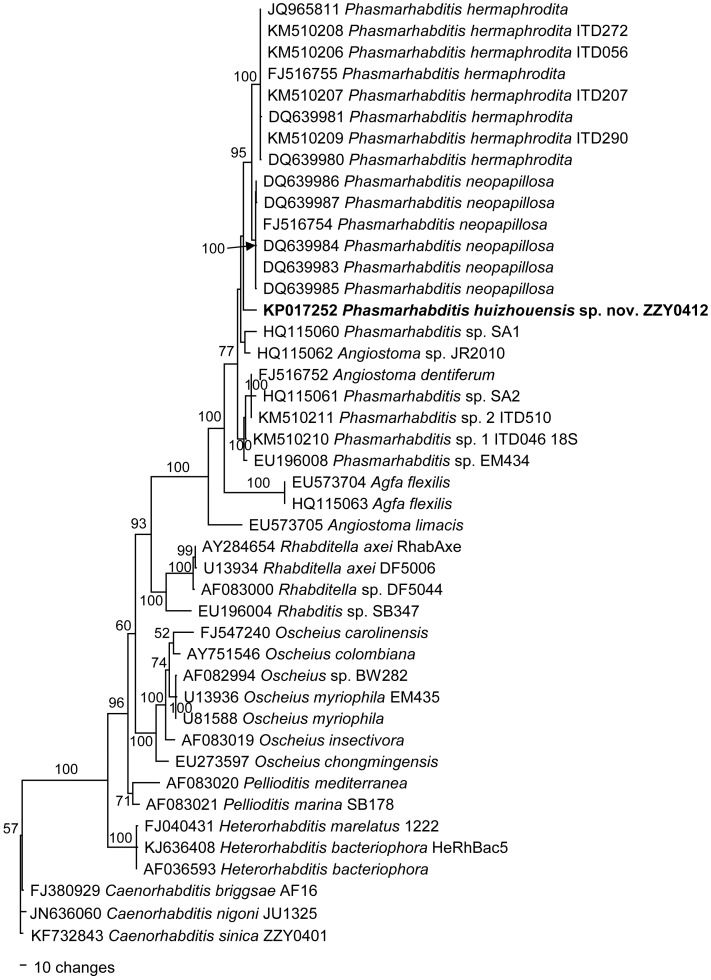
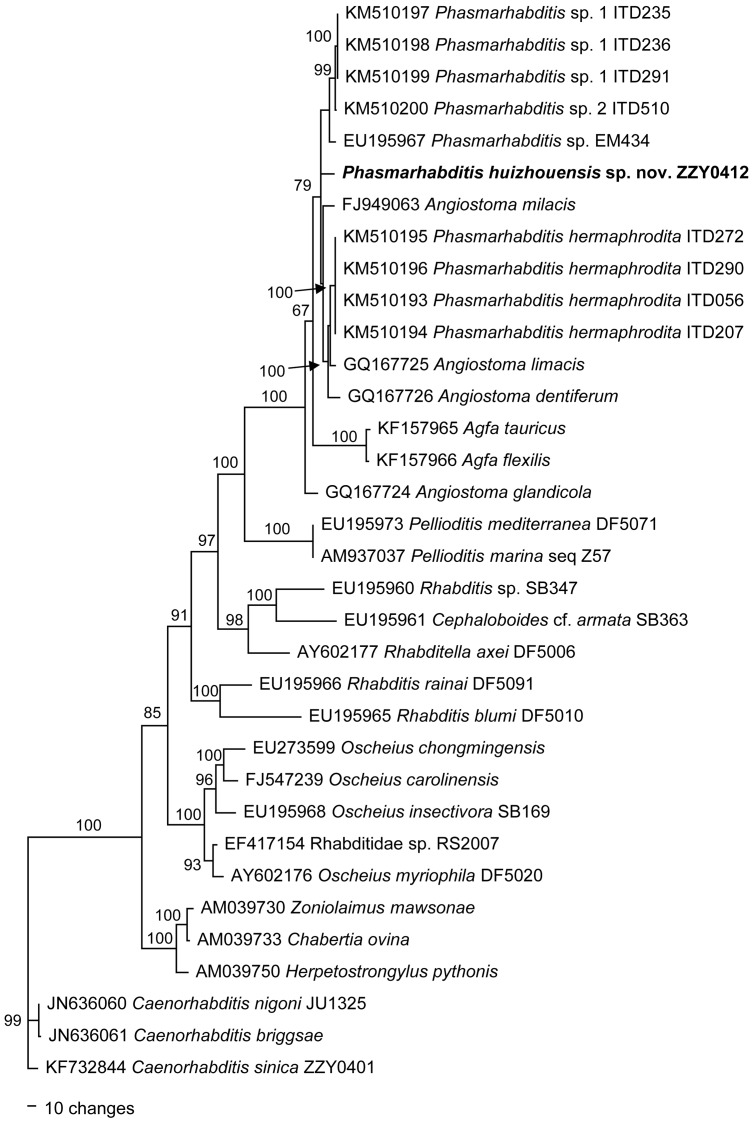
Fig 4 presents a phylogenetic tree based on the near-full-length SSU rDNA sequences from a multiple alignment of 1636 total characters under the GTR+I+G model [-inL = 8287.9473; AIC = 16595.8945; freq A = 0.2517; freq C = 0.2001; freq G = 0.2626; freq T = 0.2855; R(A-C) = 0.898; R(A-G) = 3.775; R(A-T) = 2.795; R(C-G) = 0.6581; R(C-T) = 6.1285; R(G-T) = 1; Pinvar = 0.33; Shape = 0.6046]. Using Caenorhabditis sinica as an outgroup taxon, this tree inferred two monophyletic clades. One clade includes Heterorhabditis species and the other encompasses all remaining taxa. P. huizhouensis sp. nov. is in a highly supported monophyletic clade with other species of Phasmarhabditis, 2 isolates of Agfa flexilis and 3 isolates of Angiostoma spp. This clade is sister to Rhabditella. P. huizhouensis sp. nov. is different from P. hermaphrodita, P. neopapillosa and other unidentified Phasmarhabditis species. Fig 5 presents a phylogenetic tree based on the D2-D3 LSU rDNA sequences from a multiple alignment of 940 total characters under the GTR+I+G model [-inL = 6848.9263; AIC = 13717.8525; freqA = 0.237; freqC = 0.1867; freqG = 0.3007; freqT = 0.2757; R(A-C) = 0.463; R(A-G) = 2.4133; R(A-T) = 1.6894; R(C-G) = 0.5068; R(C-T) = 4.3753; R(G-T) = 1; Pinvar = 0.3433; Shape = 1.0758]. Using C. sinica as an outgroup taxon, P. huizhouensis sp. nov. is in a highly supported monophyletic clade with many isolates of Phasmarhabditis, 2 isolates of Agfa and 4 isolates of Angiostoma. This clade is sister to Pellioditis. The phylogenetic inferences based on two rDNA genes are largely consistent with each other. Both phylogenies reveal that P. huizhouensis sp. nov. is grouped with other nominal Phasmarhabditis species but in a unique branch.
Fig 4. Bayesian tree inferred from SSU rDNA sequences.
Posterior probability values exceeding 50% are given on appropriate clades. GenBank and strain ID associated with DNA sequences are shown on the left and right of the species name respectively.
Fig 5. Bayesian tree inferred from LSU D2-D3 rDNA sequences.
Posterior probability values exceeding 50% are given on appropriate clades. GenBank and strain ID associated with DNA sequences are shown on the left and right of the species name respectively.
Discussion
The interrelationship between the genera Phasmarhabditis and Pellioditis remains unresolved, but in accord with both revisions by Andrássy (1983) and Sudhaus (2011) our study supported that the ‘Papillosa’ group of four known species comprising P. papillosa, P. hermaphrodita, P. neopapillosa and P. tawfiki, together with the new member of P. huizhouensis sp. nov. constitute a sound taxonomic group, based on an amalgamation of morphological and molecular data. The group is characterized by relatively short and wide buccal capsule, longitudinally striated lateral field with 3 or more ridges, relatively short female/hermaphroditic tail with prominent phasmids projecting from body contour, and male open peloderan bursa with 10 pairs of bursal papillae. However, the systematic relationships between these Phasmarhabditis taxa and their relatives in the subfamily Peloderinae are poorly understood, for example, two species of this subfamily Pellioditis marina and P. mediterranea sensu Andrássy [5] included by both phylogenetic trees (Figs 4 and 5) have been recently proposed fortransfer into another genus (Litoditis Sudhaus, 2011) [3]. This volatility of nematode systematics is a result of the scarcity of informative morphological/anatomical characters and a lack of DNA barcode sequences. The taxonomy of the whole Peloderinae group needs more work. Within the genus Phasmarhabditis, P. huizhouensis sp. nov. can be easily distinguished from other species except for P. papillosa by the female/hermaphroditic tail shape. P. huizhouensis sp. nov. is a morphologically cryptic species of P. papillosa, lacking typical autapomorphies except in males. The basis on which the former can be convincingly differentiated from P. papillosa are solely male caudal characters derived from available drawings for P. papillosa. Because of the previous inadequate morphological study and absence of DNA sequences for P. papillosa, further comparison at morphological as well as molecular levels between P. huizhouensis sp. nov. and P. papillosa is necessary when P. papillosa is recollected and sequenced. The excretory pore position of P. huizhouensis sp. nov. is significantly variable between females and a male observed in this study, which suggests the variability of excretory pore position for this species. However, a possibility of misidentification is unavoidable given the microscopic observation on the robust female adults in low magnifications. Thus, the excretory pore position of this species needs re-examination in the future.
Scanning electron micrographs of Phasmarhabditis hermaphrodita isolate ITD272 [8] and P. huizhouensis sp. nov. (Fig 3) reflect a number of apomorphic features shared by both species, e.g. the lip region characters (a triangular mouth opening formed by three pairs of lips with 6 labial and 4 cephalic sensilla, as well as small amphids) and lateral field morphology (longitudinally striated with 3 obvious middle-ridges). Female/hermaphroditic tail shapes are clearly distinct in the species but exhibit some degree of homology, i.e., a short conoid gradually tapered to a pointed terminus with prominent phasmids (see Fig 1F [8]) vs. a short cupola sharply to a pointed tip with ‘rod-like’ phasmids for P. huizhouensis sp. nov. (Fig 3I). Morphological features combined with molecular evidence are sufficiently for supposing a close systematic affinity in generic level between P. huizhouensis sp. nov. and P. hermaphrodita. However, other relatives of P. huizhouensis sp. nov. inferred from phylogenetic trees, such as Angiostoma and Agfa nematodes, are distantly related to the new species taxonomically. The genera belong to the distinct families Angiostomatidae and Agfidae respectively [35], and are clearly different from P. huizhouensis sp. nov. morphologically. P. huizhouensis sp. nov. differs from Angiostoma species in the type of stoma and esophagus (the former has a typically rhabditiform pharynx and a wide buccal capsule; the buccal capsule of Angiostoma spp. is wide or narrow which is conjoined by a pharynx with indistinct isthmus and a small-valvate or valveless basal bulb), cephalic sensory traits (6 labial as well as 4 cephalic sensory papillae vs. 6+6 papillae at the cephalic end), lateral field morphology (marked with longitudinal ridges vs. presence of lateral alae or lacking), and male caudal configuration (enveloped by an open peloderan bursa vs. an open leptoderan bursa or differing in the patterning of bursal papillae) [36–38]. P. huizhouensis sp. nov. is easily differentiated from Agfa flexilis by the morphology of the pharynx (a typically rhabditiform that is valvulated in the basal bulb for P. huizhouensis sp. nov. vs. a much longer one without valvular apparatus for A. flexilis) [39]. Nevertheless, molecular phylogenetic inference based on SSU rDNA sequences reveals that Phasmarhabditis isolates, Angiostoma species. and A. flexilis constitute a well-supported monophyletic clade (Fig 4), which conforms to a previous phylogenetic study among these genera [40]. Phylogenetic analysis based on the D2-D3 domain of LSU rDNA sequences in this study confirmed the close molecular relationship between these taxa (Fig 5). The reason is partly owing to the number of MOTUs (Molecular Operational Taxonomic Unit) which is insufficient to infer phylogenetic relationship between these genera, but the results reflect the inconsistencies between molecular phylogeny and traditional morphological classification. Revision of the systematics of the order Rhabditida based on previous morphological studies is needed.
Phasmarhabditis huizhouensis sp. nov. was isolated from rotting plant tissues, but is phylogenetically related to the slug-parasite P. hermaphrodita and other parasitic nematodes of Angiostoma spp. and Agfa species (Figs 4 and 5), suggesting that P. huizhouensis sp. nov. may be a facultative parasite. The ecology and host associations of P. huizhouensis sp. nov. remain unknown. It would be worthwhile to explore the potential of this Phasmarhabditis nematode to parasitize slugs as a local bio-control agent. Mass production of its dauer larvae would be essential if this species is proved to be capable of controlling slugs. In this study, the C. elegans bacterial food of E. coli OP50 was successfully used for culture of P. huizhouensis sp. nov., but no dauer stage was observed, suggesting that this bacterium may not be the best kind of food for this phasmarhabditid nematode, given that the growth rate of Phasmarhabditis species and its reproductive capacity for dauer larvae are strongly influenced by bacterial species [41]. The most suitable type of bacterial food for dauer larval formation and for mass production of P. huizhouensis sp. nov. require further study.
Acknowledgments
The first author is very grateful to Dr. Junbiao Dai (School of Life Sciences, Tsinghua University) for hosting Ren-E Huang as a postdoctoral fellow in his lab with generous support. We thank Dr. Wai Shing Chung and Yifei Qiu (both from Hong Kong Baptist University) for their kind help with sample preparations. We also thank for the constructive comments from the reviewers and a critically review by the editor that significantly helped improve this manuscript.
Data Availability
The new DNA sequences are available from the GenBank database (accession numbers are KP017252 and KP017253 respectively).
Funding Statement
This study was funded by the National Natural Science Foundation of China (http://www.nsfc.gov.cn/) (Grant No. 31201700) to REH, and by the Early Career Scheme (ECS) and General Research Fund (GRF) of the Research Grants Council (RGC) of Hong Kong (http://www.ugc.edu.hk/eng/rgc/) (Grant No. HKBU263512 and HKBU12103314 respectively) to ZZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sudhaus W. Vergleichende untersuchungen zur phylogenie, systematik, Ökologie, biologie und ethologie der Rhabditidae (Nematoda). Zoologica. 1976; 43: 1–229. [Google Scholar]
- 2. Sudhaus W, Fitch D. Comparative studies on the phylogeny and systematics of the Rhabditidae (Nematoda). J Nematol. 2001; 33: 1–70. [PMC free article] [PubMed] [Google Scholar]
- 3. Sudhaus W. Phylogenetic systematisation and catalogue of paraphyletic “Rhabditidae” (Secernentea, Nematoda). J Nematode Morphol Syst. 2011; 14: 113–178. [Google Scholar]
- 4. Andrássy I. Evolution as a basis for the systematization of nematodes Budapest, London, San Francisco, Melbourne, Pitman; 1976. [Google Scholar]
- 5. Andrássy I. A taxonomic review of the suborder Rhabditina (Nematoda: Secernentia) Paris, France, ORSTOM; 1983. [Google Scholar]
- 6. Bonaparte CLJL . Iconographia della fauna italica per le quattro classi degli animali vertebrati Tomo II. Amphibi Fascicolo: Rome, Italy, Salviucci; 1838. [Google Scholar]
- 7. Sánchez-Herráiz MJ, Barbadillo LJ, Machordom A, Sánchez B. A new species of pelodytid frog from the Iberian Peninsula. Herpetologica. 2000; 56: 105–118. [Google Scholar]
- 8. Tandingan De Ley I, McDonnell RD, Lopez S, Paine TD, De Ley P. Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae), a potential biocontrol agent isolated for the first time from invasive slugs in North America. Nematology. 2014; 16: 1129–1138. [Google Scholar]
- 9. Wilson MJ, Glen DM, George SK. The rhabditid nematode Phasmarhabditis hermaphrodita as a potential biological control agent for slugs. Biocontrol Sci Techn. 1993; 3: 503–511. [Google Scholar]
- 10. Wilson MJ, Glen DM, Hamacher GM, Smith JU. A model to optimise biological control of slugs using nematode parasites. Appl Soil Ecol. 2004; 26: 179–191. [Google Scholar]
- 11. Rae RG, Robertson JF, Wilson MJ. Organic slug control using Phasmarhabditis hermaphrodita . Aspects of Applied Biology. 2006; 79: 211–214. [Google Scholar]
- 12. Rae RG, Verdun C, Grewal PS, Robertson JF, Wilson MJ. Biological control of terrestrial molluscs using Phasmarhabditis hermaphrodita—progress and prospects. Pest Manag Sci. 2007; 63:1153–1164. 10.1002/ps.1424 [DOI] [PubMed] [Google Scholar]
- 13. Ross JL, Ivanova ES, Severns PM, Wilson MJ. The role of parasite release in invasion of the USA by European slugs. Biol Invasions. 2010; 12: 603–610. [Google Scholar]
- 14. Hooper DJ, Wilson MJ, Rowe JA, Glen DM. Some observations on the morphology and protein profiles of the slug-parasitic nematodes Phasmarhabditis hermaphrodita and P. neopapillosa (Nematoda: Rhabditidae). Nematology. 1999; 1: 173–182. [Google Scholar]
- 15. Mengert H. Nematoden und Schnecken. Zeitschrift für Morphologie und Ökologie der Tiere. 1953; 4: 311–349. [Google Scholar]
- 16. Poinar GO, Jr. Origins and phylogenetic relationships of the entomophilic rhabditids, Heterorhabditis and Steinernema . Fundam appl Nematol. 1993; 16: 333–338. [Google Scholar]
- 17. Azzam KM. Description of the nematode Phasmarhabditis tawfiki n. sp. isolated from Egyptian terrestrial snails and slugs. J Egypt Ger Soc Zool. 2003; 42: 79–87. [Google Scholar]
- 18. Ross JL, Ivanova ES, Sirgel WF, Malan AP, Wilson MJ. Diversity and distribution of nematodes associated with terrestrial slugs in the Western Cape Province of South Africa. J Helminthol. 2012; 86: 215–221. 10.1017/S0022149X11000277 [DOI] [PubMed] [Google Scholar]
- 19. Zaborski ER, Gittenger LAS, Roberts SJ. A possible Phasmarhabditis sp. (Nematoda: Rhabditidae) isolated from Lumbricus terrestris (Oligochaeta: Lumbricidae). J Invertebr Pathol. 2001; 77: 284–287. [DOI] [PubMed] [Google Scholar]
- 20. de Man JG. Die Einheimischen, frei in der reinen Erde und im süssen Wasser lebende Nematoden. Vorläufiger Bericht und descriptiv-systematischer Theil. Tijdschrift der Nederlandsche dierkundige Vereeniging. 1880; 5: 1–104. [Google Scholar]
- 21. Huang R-E, Ren X, Qiu Y, Zhao Z. Description of Caenorhabditis sinica sp. n. (Nematoda: Rhabditidae), a nematode species used in comparative biology for C. elegans . PLoS ONE. 2014. November 6; 9(11): e110957 10.1371/journal.pone.0110957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryss AY. Express technique to prepare permanent collection slides of nematodes. Zoosystematica Rossica. 2003; 11: 257–260. [Google Scholar]
- 23. Barrière A. and Félix M-A. Isolation of C. elegans and related nematodes WormBook, ed. The C. elegans Research Community, WormBook; 2006. July 17 10.1895/wormbook.1.115.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Floyd R, Abebe E, Papert A, Blaxter M. Molecular barcodes for soil nematode identification. Mol Ecol. 2002; 11: 839–850. [DOI] [PubMed] [Google Scholar]
- 25. Haber M, Schungel M, Putz A, Muller S, Hasert B, Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol Biol Evol. 2005; 22: 160–173. [DOI] [PubMed] [Google Scholar]
- 26. da Silva NRR, da Silva MC, Genevois VF, Esteves AM, De Ley P, Decraemer W, et al. Marine nematode taxonomy in the age of DNA: the present and future of molecular tools to assess their biodiversity. Nematology. 2010; 12: 661–672. [Google Scholar]
- 27. Blaxter ML, Deley P, Garey JR, Liu LX, Scheldemann P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998; 392: 71–75. [DOI] [PubMed] [Google Scholar]
- 28. Nadler SA, Carreno RA, Adams BJ, Kinde H, Baldwin JG, Mundo-Ocampo M. Molecular phylogenetics and diagnosis of soil and clinical isolates of Halicephalobus gingivalis (Nematoda: Cephalobina: Panagrolaimoidea) an opportunistic pathogen of horses. Int J Parasitol. 2003; 33: 1115–1125. [DOI] [PubMed] [Google Scholar]
- 29. Nadler SA, Hudspeth DSS. Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): Implications for morphological evolution and classification. Mol Phylogenet Evol. 1998; 10: 221–236. [DOI] [PubMed] [Google Scholar]
- 30. Thomas WK, Vida JT, Frisse LM, Mundo Ocampo M, Baldwin JG. DNA sequences from formalin-fixed nematodes: integrating molecular and morphological approaches to taxonomy. J Nematol. 1997; 29: 250–254. [PMC free article] [PubMed] [Google Scholar]
- 31. Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998; 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 32. Huelsenbeck JP, Ronquist F. MR BAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17: 1754–1755. [DOI] [PubMed] [Google Scholar]
- 33. Larget B, Simon DL. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol. 1999; 16: 750–759. [Google Scholar]
- 34. Stock SP, Camino NB. Pellioditis pellio (Schneider) (Nematoda: Rhabditidae) parasitizing Scaptia (Scaptia) lata (Guerin-meneville) (Diptera: Tabanidae). Mem Inst Oswaldo Cruz Rio de Janeiro. 1991; 86: 219–222. [Google Scholar]
- 35. Ivanova ES, Panayotova-Pencheva MS, Spiridonov SE. Observations on the nemaotode fauna of terrestrial molluscs of the Sofia area (Bulģaria) and the Crimea peninsula (Ukraine). Russ J Nematol. 2013; 21: 41–49. [Google Scholar]
- 36. Ivanova ES, Wilson MJ. Two new species of Angiostoma Dujardin, 1845 (Nematoda: Angiostomatidae) from British terrestrial molluscs. Syst Parasitol. 2009; 74: 113–124. 10.1007/s11230-009-9200-z [DOI] [PubMed] [Google Scholar]
- 37. Ivanova ES, Spiridonov SE. Angiostoma glandicola sp. n. (Nematoda: Angiostomatidae): a parasite in the land snail Megaustenia sp. from the Cat Tien Forest, Vietnam. J Helminthol. 2010; 84: 297–304. 10.1017/S0022149X09990678 [DOI] [PubMed] [Google Scholar]
- 38. Ross JL, Malan AP, Ivanova ES. Angiostoma margaretae n. sp. (Nematoda: Angiostomatidae), a parasite of the milacid slug Milax gagates Draparnaud collected near Caledon, South Africa. Syst Parasitol. 2011; 79: 71–76. 10.1007/s11230-011-9294-y [DOI] [PubMed] [Google Scholar]
- 39. Morand S, Hommay G. Redescription de Agfa flexilis (Nematoda: Agfidae) parasite de I’appareil genital de Limax cinereoniger (Gastropoda: Limacidae). Syst Parasitol. 1990; 15: 127–132. [Google Scholar]
- 40. Ross JL, Ivanova ES, Spiridonov SE, Waeyenberge L, Moens M, Nicol GW, et al. Molecular phylogeny of slug-parasitic nematodes inferred from 18S rRNA gene sequences. Mol Phylogenet Evol. 2010; 55: 738–743. 10.1016/j.ympev.2010.01.026 [DOI] [PubMed] [Google Scholar]
- 41. Wilson MJ, Glen DM, Pearce JD, Rodgers PB. Monoxenic culture of the slug parasite Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) with different bacteria in áliquid and solid phase. Fundam appl Nematol. 1995; 18: 159–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The new DNA sequences are available from the GenBank database (accession numbers are KP017252 and KP017253 respectively).