Abstract
Whilst ∼93% of domain superfamilies appear to be relatively structurally and functionally conserved based on the available data from the CATH-Gene3D domain classification resource, the remainder are much more diverse. In this review, we consider how domains in some of the most ubiquitous and promiscuous superfamilies have evolved, in particular the plasticity in their functional sites and surfaces which expands the repertoire of molecules they interact with and actions performed on them. To what extent can we identify a core function for these superfamilies which would allow us to develop a ‘domain grammar of function’ whereby a protein's biological role can be proposed from its constituent domains? Clearly the first step is to understand the extent to which these components vary and how changes in their molecular make-up modifies function.
Current Opinion in Genetics & Development 2015, 35:40–49
This review comes from a themed issue on Genomes and evolution
Edited by Antonis Rokas and Pamela S Soltis
For a complete overview see the Issue and the Editorial
Available online 3rd November 2015
http://dx.doi.org/10.1016/j.gde.2015.09.005
0959-437X/© 2015 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
Families of proteins arise through speciation (orthologous relatives) and through duplication of genes during evolution (paralogous relatives) and it is the paralogues that are most likely to diverge, although not necessarily [1]. By classifying families, superfamilies and collating information on their protein structures, sequences and functions, we can explore how relatives diverge and understand the molecular mechanisms underlying any functional changes [2]. Such insights are essential for inheriting properties between relatives to cope with the huge dearth in experimental annotations. For example, an inspection of the experimental annotations in the UniProtKB/Swiss-Prot sequence database (June 2015) reveals that less than 15% of human proteins have detailed functional characterisation and only 4% have known structures. They are also essential for understanding whether genetic variations are likely to be tolerated and affect function.
Many resources now exist for classifying protein families, some of which consider the entire protein (e.g., PANTHER [3], HAMAP [4], TIGRFAMs [5] and SFLD [6]) whilst others classify the domain components (e.g., Pfam [7], SMART [8], PRINTS [9], InterPro [10], CDD [11], CATH [12], SCOP [13] and ECOD [14]) generally considered to be evolutionary independent modules having distinct functional properties [15]. Some resources like PhyloFacts [16] also provide classification of both full-length proteins and domains. At least two thirds of eukaryotic and more than a half of prokaryotic proteins are composed of multiple domains [17] and the most highly populated domain superfamilies are universal to all kingdoms of life or major clades or branches [18]. Therefore, whilst studies have suggested that there may be approximately 100 thousand protein families [16, 19] many proteins can be decomposed into common constituent domains derived from a more limited repertoire of ∼15,000 superfamilies [19]. Within a protein, the different domains tend to have different roles, which when combined make up the general function of that protein. Therefore, by understanding the different functional roles that domains possess we can start to build up a ‘domain grammar of function’ [20]. Interestingly, a few hundred of these domain superfamilies’ dominate nature, accounting for nearly two thirds of all known domains [21]. It is in these superfamilies that we see the most diversity (see Figure 1) and this is largely reflected in their binding properties and/or their ability to metabolise diverse substrates.
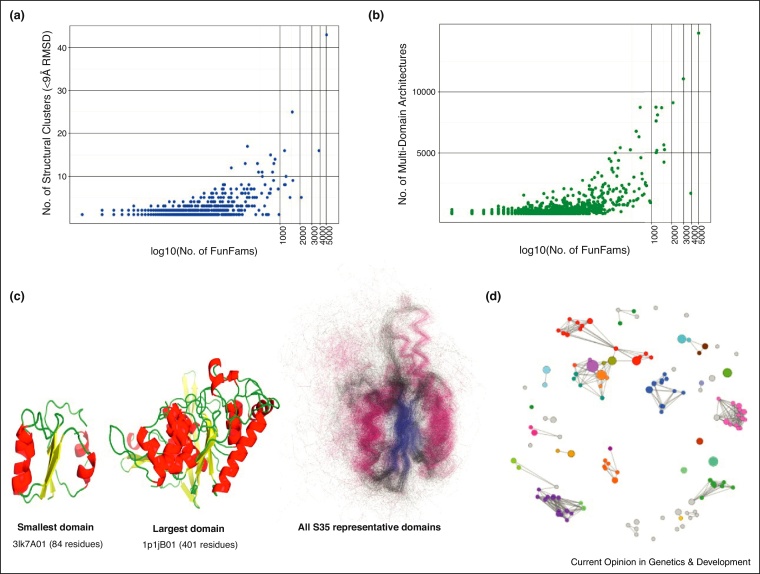
Figure 1.
Diversity in protein domain superfamilies. (a) Correlation of structural diversity with functional diversity in CATH domain superfamilies. Each point represents a CATH superfamily. Structural diversity is given by the number of distinct Structurally Similar Groups (SSGs) in which relatives superpose with <9 Å RMSD. Functional diversity is given by the number of functional families (FunFams), identified using HMM based strategies [22], and is plotted in the logarithmic (log10) scale. (b) Correlation of multi-domain architecture (MDA) diversity with functional diversity in CATH domain superfamilies in the logarithmic (log10) scale. Each point represents a CATH superfamily. MDA diversity is given by the number of different multi-domain architectures containing one or more superfamily domains. (c) Structural diversity in the highly populated ‘NAD(P)-binding Rossmann-like’ superfamily (CATH 3.40.50.720). The figure shows structures of the smallest and largest domain in the superfamily. On the far right is the superposition of all non-redundant superfamily members to highlight the conserved structural core. (d) Visualization of functional diversity in the HUP domain superfamily (CATH 3.40.50.620) using Cytoscape [80] networks. The nodes (represented as circles) represent functional families and the edges represent HMM-based family similarities. Each colour denotes a unique Enzyme Commission (EC) number and grey nodes indicate FunFams without any EC number [22].
In this review we use the CATH-Gene3D domain classification, currently the most comprehensive structure-based superfamily resource, to assess the extent of divergence across protein domain ‘superfamily space’ and review the mechanisms of divergence revealed by detailed studies of specific families undertaken by us and other groups.
Capturing information on structural and functional diversity within superfamilies
Specialised manually curated structure-based classifications like SFLD [6], TEED [23], CYPED [23], LccED [24] and ESTHER [25] provide valuable insights into the diversity of selected enzyme superfamilies and there have been several elegant studies of large, diverse superfamilies in the Structure Function Linking database (SFLD) resource [26, 27•]. However, relatively few superfamilies have been explored in such detail because of the limited experimental data. Since relatives sharing structural and functional properties experience similar constraints on their sequences to preserve these properties, one way to explore diversity across ‘superfamily space’ is to exploit the much more prolific sequence data that is available [22, 23, 28••].
By appropriately clustering relatives with similar sequence properties, several resources [6, 16, 19] classify specific ‘functional families’. Approaches range from pairwise comparisons [6] to more sophisticated profile-based analyses [22] that can also be used to detect key residue sites differing between the functional families. Whilst residues important for folding or stability tend to be conserved across the whole superfamily, positions only conserved in certain functional families (specificity determining positions or SDPs) are often under positive selection and associated with distinct functional properties [29, 30] (see Figure 2(a)). SDPs can be associated with a wide variety of protein sites. For example, in addition to mutations in the ligand binding pocket, diversity in the Metabotropic Glutamate Receptors is conferred by SDPs in allosteric sites, the dimerization interface and the hinge region [31•]. Similarly the functional specificity of signalling proteins like the Ras superfamily involves mutations in the nucleotide-binding pocket and interfaces co-ordinating the communication between the nucleotide and membrane-binding regions [32].
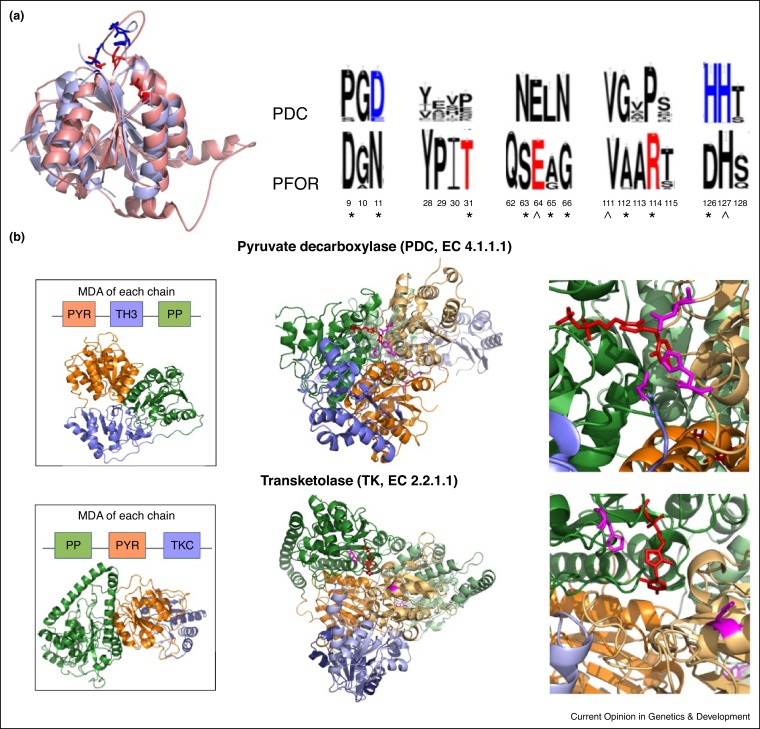
Figure 2.
Functional diversity in the Thiamine pyrophosphate (TPP)-dependant enzyme superfamily (CATH 3.40.50.970) due to: (a) changes in residues. The superposition of the PYR domains of the Pyruvate decarboxylase (PDC, EC 4.1.1.1) (shown in blue) and Pyruvate:ferrodoxin oxidoreductase (PFOR, EC 1.2.7.1) (shown in red) structures highlights the differences in their catalytic residues (shown as sticks). The specificity-determining positions (SDPs, indicated by an asterisk) around the known catalytic residues are displayed in sequence logos corresponding to the PDC and PFOR functional family in CATH-Gene3D. The catalytic residues are shown in blue for PDC and in red for PFOR and the conserved residues are indicated by a caret (^). The positions are numbered according to the corresponding residue in PDB 1PVD. (b) Changes in domain context. Pyruvate decarboxylase (PDC, EC 4.1.1.1) and transketolase (TK, EC 2.2.1.1) in the TPP-dependant superfamily both consist of two chains comprising two TPP domains – PP and PYR (chains are represented by darker and lighter shades of each constituent domain colour). The left hand image shows the difference in multi-domain architectures and 3D arrangements for these two proteins. The middle image shows the different dimeric assemblies that the proteins form. The right image zooms in on the active sites. The TPP molecule is shown in red and the catalytic residues are shown in magenta. Catalytic residues are contributed from the PP domain of one subunit and the PYR of the other subunit. In TK the size of the active site pocket is larger.
For exploring superfamily diversity in the CATH-Gene3D resource, we have used an approach that searches for SDPs to distinguish between different functional clusters [22]. This approach sub-classifies the ∼2700 CATH-Gene3D superfamilies into ∼110,000 functional families by optimal partitioning of hierarchical clustering trees for each superfamily, based on identifying characteristic patterns of differentially conserved positions (SDPs) and conserved positions between different functional groups, all of which have at least one relative with an experimental functional annotation in the Gene Ontology (GO) [33]. Whilst validation suggests that these functional groups are reasonably effective in transferring experimental annotations between relatives, there is still considerable room for improvement, as suggested by the results of a recent international large-scale protein function prediction assessment [34]. However, functional family classification does shed light on superfamily diversity, revealing that for only 7% (∼200) of these superfamilies, sequence change is associated with very significant diversity in structure, function and protein context (see Figure 1) while the remaining ∼93% of the superfamilies appear to have structurally and functionally conserved relatives.
Functional diversity in binding and enzyme superfamilies — ‘molecular tinkering’
Of the 200 most diverse domain superfamilies, each of which have 100 or more functional families and account for ∼50% of all CATH-Gene3D domains, ∼95% of these are superfamilies directly or indirectly associated with enzymatic activity and many of the remainder have relatives with binding activity. Whilst detailed studies of some superfamilies have characterised considerable structural divergence modifying functional site features ([35, 36], see also below), just small changes associated with residue mutations in a binding or active site can alter the shape, physicochemical and electrostatic characteristics significantly, modifying ligand specificities in binding proteins and affecting substrate specificities, chemistries and catalytic efficiencies in enzymes. The Nuclear Receptor superfamily shows amazing diversity in the ligand binding cavity brought about by such mutations, driven by strong divergent selection and adaptive positive selection [37]. Similarly, in the Tubulin superfamily, many of the positively selected sites are found at or adjacent to functionally important sites [38].
In enzymes, considerable sequence divergence can occur in the active sites. In nearly 55% of 101 experimentally well-annotated enzyme superfamilies (accounting for almost 50% of all enzyme sequences in CATH-Gene3D) dramatic changes in catalytic machinery occur [39]. However, in support of previous studies of Babbitt and co-workers [28••] which reported that many relatives in SFLD superfamilies share a common mechanistic step, 40% of these superfamilies have one or two catalytic residues common to all functional families. In some cases catalytic residues with similar physicochemical properties are located at similar 3D locations even though they are in different positions in the sequence (see Figure 2(a)). Thus, frequently some aspect of the chemistry is conserved and analyses based on phylogenetic trees derived from structure-based alignments of CATH-Gene3D superfamilies confirm, on a much larger scale than early studies [2], that most superfamily diversity is associated with changes in substrate specificity [40•], suggesting that it is hard to change the chemistry presumably because of the complex sequence of mutations needed to create a new arrangement of catalytic residues with the correct spatial relationships.
However, dramatic changes in chemistry can occur, such as in the Enolase superfamily [41, 42], Aldolase Class I superfamily [43••] (see Figure 3) and DRE-TIM metallolyase superfamily [44], and sometimes the same catalytic core is used for very different reactions. For example, many diverse enzymes (peptidases, thioesterases, lipases) in the α/β-hydrolase superfamily use the same catalytic triad (Ser-His-Asp) for different types of bond changes [25]. Diversity can also result from loss or changes to metal ions bound by relatives [45•] for example in paraoxonase-1 where an alternative binding mode of the catalytic calcium ion appears to initiate divergence in enzymatic activity [46] and other cases where alterations from the “native” metal of a metalloenzyme have been seen to promote promiscuity [47].
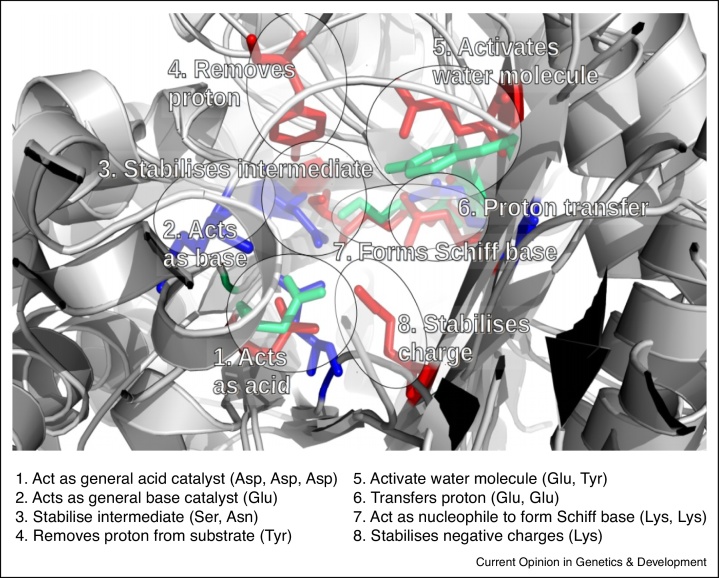
Figure 3.
Different catalytic machinery performing same enzymatic chemistry. The three domains shown in this figure use different catalytic machineries to perform the same enzymatic reaction (EC 4.1.2.13). Each domain belongs to a different functional family in the Aldolase Class I superfamily (CATH 3.20.20.70). On the figure different regions in the active site are assigned to clusters 1–8. The catalytic residues in each cluster are reported to have the same functional properties, summarised on the figure. Each colour represents the catalytic residues of a different domain: red is 1aldA00, blue is 1b57A00, and green is 1ok4A00. The remaining portions of the three domains are coloured grey. The same catalytic residue is used by two or more domains in clusters 1, 6, and 7. Different catalytic residues are used in clusters 3 and 5 but still show enough physicochemical similarity to provide the same functionality. The proteins have different catalytic rates which may reflect their different catalytic machineries [51].
Interestingly, in some enzyme superfamilies, functional families with significantly different catalytic machineries have highly similar functions and substrates, suggesting either convergence within the superfamily or evolutionary drift from a common functional ancestor along different routes, that is, perhaps a trajectory to a less efficient enzyme with subsequent mutations restoring the activity or even resulting in a more efficient form of the enzyme. It is difficult to distinguish these cases without robust phylogenetic analyses. Such studies on Rubisco, an abundant protein important for carbon fixation, show that a more efficient form of Rubisco has emerged by convergent evolution more than 62 times in harsh environments, and structure-based analyses reveal mutations in the active site loop and secondary shell, where they possibly influence rearrangements of the active site; also at interfaces in the oligomer suggesting a role in allostery [48•].
Enzyme superfamilies showing the greatest versatility in CATH-Gene3D, frequently adopt alpha/beta structures, two thirds having TIM or Rossmann folds. As Tawfik and his colleagues have reported in a recent publication, these structures tend to have regular, well-packed structural cores and the catalytic residues mainly locate to loops largely detached from these cores and therefore perhaps better able to tolerate the destabilising effects of mutations [49•, 50••].
Diversity in protein superfamilies can also arise from mutations in protein interfaces. Furthermore, relatives can exploit completely different surfaces in their protein interactions. Large-scale studies comparing CATH-Gene3D functional families showed that in 645 highly versatile superfamilies, cumulative binding sites from diverse relatives covered most of the protein surface and were associated with a wide range of protein partners [52•]. However, sometimes the same interface is exploited but by different partners. In the two Dinucleotide Binding Domains Flavoproteins (tDBDF) superfamily, the diversity of reactions carried out by relatives is achieved by different protein partners acting as electron acceptors and interacting with the same face of the tDBDF domain [53]. Paralogous relatives are more likely to bind different protein partners [54] and this is a significant effect in the beta-propellor superfamilies, whose structures contain repeating WD40 sub-domains, and in which human paralogues have multiple distinct surfaces interacting with a very wide variety of proteins, peptides or nucleic acids [55].
Structural mechanisms of superfamily divergence
Although only 10% of the CATH-Gene3D functional families have structural representatives, this data can help identify superfamilies capable of great structural plasticity where relatives display considerable diversity due to extensive residue insertions and repetitions or inserted structural motifs [56, 57]. For ∼160 CATH superfamilies, accounting for half of all known domains in CATH-Gene3D, at least a two-fold variation in the size is observed between the most diverse domains [58]. However, analyses of selected superfamilies [35, 59] and more recent large-scale studies have shown that the structural core (generally 40–50% of the domain) is highly conserved even for relatives separated by billions of years [57] (see Figure 1). Long residue inserts in diverse relatives generally adopt secondary structure features that form structural decorations to this core and can be associated with modified functions, for example, by altering active site geometry and thereby changing substrate specificity (see Figure 2(a)), or altering surface features and thereby changing protein interaction partners [52•]. In the Thiamine pyrophosphate (TPP)-dependant superfamily, different functional families have varying inserts forming small additional secondary structure features that reshape the active site for different substrates (see Figure 2(b)). In the HUP domain superfamily, also, quite extensive structural embellishments extend the active site [35]. Insertions of motifs or sub-domains can also result from gene fusions, for example, in the Haloalkanoic Acid Dehalogenase (HAD) superfamily where they provide diverse specificity determinants for a broad range of substrates [60••].
Dramatic structural rearrangements can also arise from variations in repeating units. In the Vicinal Oxygen Chelate (VOC) superfamily, members share a common βαβββ subdomain that is organized into different topological (or domain-swapped) combinations in different relatives that maximizes the catalytic versatility of the metal center [61]. These and other structural changes such as circular permutations and rearrangements in β-sheet topologies can sometimes transform the fold [62] as well as modifying the function [50••].
Diversity can also emerge from changes in less structured regions, for example, repeats giving rise to low-complexity regions (LCRs), such as polyalanine or polyglutamine runs. These often evolve rapidly and can have a major influence on the transcriptional activity of the protein [63]. Similarly, variations in (Gly)n -X repeats in glycine rich domains have been observed to alter the expression pattern, modulation and sub-cellular localization of relatives in some plant families [64].
Superfamily diversity arising from different multi-domain contexts
Gene fusions are another evolutionary mechanism conferring diversity as they can significantly alter the context of a domain (i.e., by changing the multi-domain architecture (MDA) of the protein), thereby modifying its molecular function and biological role. Domains have been frequently duplicated and shuffled within genomes, during evolution, with fusions being more frequent and generally occurring at N or C termini [65]. For 92% of the 200 most diverse superfamilies in CATH-Gene3D superfamilies, that is, those having the highest number of functional families, relatives occur in more than 100 different multi-domain contexts [21] (Figure 1(b)). Changes in domain partners may not necessarily alter the function of the domain but change the context in which it operates, for example, locating it in different protein complexes and/or pathways. For example, early studies demonstrated the recruitment of domain relatives to different metabolic pathways for the chemistry they bring [66].
However, changes in domain partners can also alter specificity. For example, in the highly diverse Thiamine pyrophosphate (TPP)-dependant enzyme superfamily changes in domain partnership alter the size and physicochemical properties of the active site pocket (see Figure 2(b)), enabling a huge range of substrates, products and stereo-selectivity [67]. Different oligomerisation states also effectively change the domain context. Again, in the TPP superfamily, various oligomerisation states have evolved in different species. Whilst some may be associated with enhanced stability, others clearly influence active site characteristics by changing the positioning of the domains providing catalytic residues (see Figure 2(b)).
Diversity in superfamilies due to promiscuity
Diversity within a superfamily can also be the result of individual relatives having multiple functions. For example, relatives can have multiple catalytic activities not necessarily of equal efficiency, as in promiscuous enzymes; or moonlighting functions whereby proteins perform completely different functions to their native activity sometimes involving different sites [68, 69]. Promiscuity can be the starting point for the evolution of a new function [49•, 50••]. Under natural selection, promiscuous enzymes can give rise to specialist enzymes by a variety of different mechanisms - protein dynamics (e.g., changes in conformational dynamics have converted a promiscuous generalist beta-lactamase to a penicillin-specific beta-lactamase, without significant changes in the structure of the active site [70]), domain insertions (e.g., HAD superfamily [36, 60••]), rearrangements in the catalytic metal ions [71] and binding of alternative cofactors [72].
An increasing number of proteins are now known to moonlight and these activities can be induced by oligomerisation, cellular localization, differential expression and substrate concentration. For example, Albaflavenone monooxygenase in the Cytochrome P450 superfamily, also functions as a Terpene synthase, an activity not observed in any other superfamily member. The catalytic machineries for the two enzymatic reactions are located in distinct pockets on the domain and the reactions are carried out at different pHs [73].
Conclusions
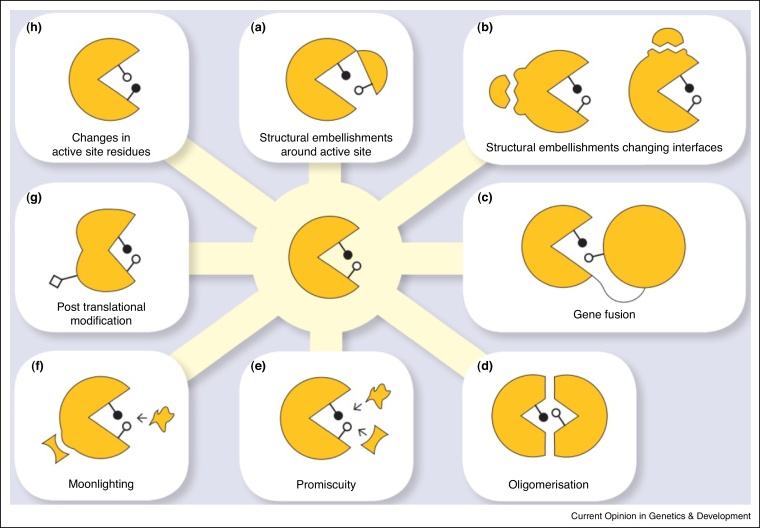
In most large diverse superfamilies, functional diversity results from a combination of different molecular mechanisms (Figure 4). For example, in the PD-(D/E)XK Phosphodiesterase superfamily there are structural embellishments to the core, domain swapping events, active site residue variations and changes in MDA [74]. Similarly, in the Ribonuclease H-like (RNHL) superfamily [75], and many other families discussed above.
Figure 4.
Functional diversity of proteins can arise due to one or more of the following mechanisms: (a) Structural embellishments around active site, (b) Structural embellishments changing interfaces, (c) Gene fusion, (d) Oligomerisation, (e) Promiscuity, (f) Moonlighting, (g) Post-translational modification and (h) Changes in active site residue. Note that for the mechanism panels (a), (c) and (d), one of the enzyme active site residue is contributed by its domain partner.
Experimental data on functional diversity grows slowly as detailed studies are time-consuming and expensive, however, classifying the millions of sequences accumulating in public repositories like UniProt into putative functional families can reveal subtle changes in conservation patterns that suggest shifts in binding specificities or catalytic machineries. These data can guide experiments to focus on unusual relatives and more comprehensively landscape the functional repertoires of the most versatile superfamilies. For example, sequence similarity networks based on protein families can help in providing a comprehensive summary of sequence, structure and function relationships in a functionally diverse superfamily. Recent studies [27•, 60••, 76] of such networks derived from curated family classification for three functionally diverse superfamilies in SFLD have been used to aid in target selection for interesting targets for experimental characterisation. The availability of automated functional classifications of superfamilies will ultimately guide experimental validation using high-throughput approaches and aid in improving the functional annotation of genomes. This will be especially important for large diverse superfamilies.
Only ∼63% of the 25 million domain sequences in CATH-Gene3D can be assigned to an experimentally annotated functional family and less than 10% of these families have a known structure, so there may be much more diversity to discover. Certainly analyses of microbial communities hint at exciting novel chemistries [77, 78]. Although the lack of data hinders our understanding, most studies of enzyme superfamilies, even those that are mechanistically very diverse, suggest that chemistry is usually preserved or there is conservation of a specific partial reaction among all relatives and that it is substrate specificity that is much more likely to change [28••]. Furthermore, the relative success of domain-based strategies for protein function prediction [22, 79] suggests that a general functional role is conserved across most domain superfamilies and that diversity largely results from exploitation of that role on multiple ligands or substrates, and in multiple contexts. In other words, the structural diversity observed in promiscuous superfamilies is more frequently associated with changes that reflect different domain contexts or changes in substrate specificity rather than dramatic changes in the functional role. This suggests that for many domain superfamilies’ a domain grammar of function can be applied.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Altenhoff A.M., Studer R.A., Robinson-Rechavi M., Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd A.E., Orengo C.A., Thornton J.M. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 3.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedruzzi I., Rivoire C., Auchincloss A.H., Coudert E., Keller G., De Castro E., Baratin D., Cuche B.A., Bougueleret L., Poux S. HAMAP in 2013, new developments in the protein family classification and annotation system. Nucleic Acids Res. 2013;41:D584–D589. doi: 10.1093/nar/gks1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akiva E., Brown S., Almonacid D.E., Barber A.E., Custer A.F., Hicks M.A., Huang C.C., Lauck F., Mashiyama S.T., Meng E.C. The structure–function linkage database. Nucleic Acids Res. 2013;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I., Doerks T., Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attwood T.K., Coletta A., Muirhead G., Pavlopoulou A., Philippou P.B., Popov I., Romá-Mateo C., Theodosiou A., Mitchell A.L. The PRINTS database: a fine-grained protein sequence annotation and analysis resource—its status in 2012. Database. 2012;2012:bas019. doi: 10.1093/database/bas019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M.K., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Lanczycki C.J. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreeva A., Howorth D., Chandonia J.-M., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2007;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H., Schaeffer R.D., Liao Y., Kinch L.N., Pei J., Shi S., Kim B.-H., Grishin N.V. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 2014;10:e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponting C.P., Russell R.R. The natural history of protein domains. Annu Rev Biophys Biomol Struct. 2002;31:45–71. doi: 10.1146/annurev.biophys.31.082901.134314. [DOI] [PubMed] [Google Scholar]
- 16.Afrasiabi C., Samad B., Dineen D., Meacham C., Sjölander K. The PhyloFacts FAT-CAT web server: ortholog identification and function prediction using fast approximate tree classification. Nucleic Acids Res. 2013;41:W242–W248. doi: 10.1093/nar/gkt399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J.-H., Batey S., Nickson A.A., Teichmann S.A., Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 18.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 19.Levitt M. Nature of the protein universe. Proc Natl Acad Sci. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashton M., Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Lees J.G., Lee D., Studer R.A., Dawson N.L., Sillitoe I., Das S., Yeats C., Dessailly B.H., Rentzsch R., Orengo C.A. Gene3D: multi-domain annotations for protein sequence and comparative genome analysis. Nucleic Acids Res. 2014;42:D240–D245. doi: 10.1093/nar/gkt1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S., Lee D., Sillitoe I., Dawson N., Lees J., Orengo C. Functional classification of CATH superfamilies: a domain-based approach for protein function annotation. Bioinformatics. 2015:btv398. doi: 10.1093/bioinformatics/btv398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pleiss J. Systematic analysis of large enzyme families: identification of specificity- and selectivity-determining hotspots. ChemCatChem. 2014;6:944–950. [Google Scholar]
- 24.Sirim D., Wagner F., Wang L., Schmid R.D., Pleiss J. The laccase engineering database: a classification and analysis system for laccases and related multicopper oxidases. Database. 2011;2011:bar006. doi: 10.1093/database/bar006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenfant N., Hotelier T., Velluet E., Bourne Y., Marchot P., Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2012;41:423–429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlt J.A., Babbitt P.C., Jacobson M.P., Almo S.C. Divergent evolution in enolase superfamily: strategies for assigning functions. J Biol Chem. 2012;287:29–34. doi: 10.1074/jbc.R111.240945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Mashiyama S.T., Malabanan M.M., Akiva E., Bhosle R., Branch M.C., Hillerich B., Jagessar K., Kim J., Patskovsky Y., Seidel R.D. Large-scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere. PLoS Biol. 2014;12:e1001843. doi: 10.1371/journal.pbio.1001843. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors have used computational approaches to identify new targets in the cytosolic glutathione transferase superfamily which were then characterised experimentally. A network-based, large-scale study of this superfamily using this new and previously reported annotation data, provides valuable insights about how function is modulated by changes in sequence or structure.
- 28••.Brown S.D., Babbitt P.C. New insights about enzyme evolution from large-scale studies of sequence and structure relationships. J Biol Chem. 2014;289:30221–30228. doi: 10.1074/jbc.R114.569350. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review on the functional evolution of enzymes using large scale studies of functionally diverse enzyme superfamilies. The authors report the value of such large scale studies in providing a better understanding of structure–function relationships of enzymes and aiding the functional annotation of proteins.
- 29.Rausell A., Juan D., Pazos F., Valencia A. Protein interactions and ligand binding: from protein subfamilies to functional specificity. Proc Natl Acad Sci U S A. 2010;107:1995–2000. doi: 10.1073/pnas.0908044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty A., Chakrabarti S. A survey on prediction of specificity-determining sites in proteins. Brief Bioinform. 2014;16:71–88. doi: 10.1093/bib/bbt092. [DOI] [PubMed] [Google Scholar]
- 31•.Kang H.J., Wilkins A.D., Lichtarge O., Wensel T.G. Determinants of endogenous ligand specificity divergence among metabotropic glutamate receptors. J Biol Chem. 2015;290:2870–2878. doi: 10.1074/jbc.M114.622233. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors have used a combined approach to identify specificity-determining residues responsible for functional divergence of the metabotropic glutamate receptors involving computational analysis, mutagenesis and biochemical assays. Specificity-determining residues were found to be involved in the ligand-binding pocket, allosteric sites or at the loops near dimerization and interlobe hinge region.
- 32.Rojas A.M., Fuentes G., Rausell A., Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol. 2012;196:189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radivojac P., Clark W.T., Oron T.R., Schnoes A.M., Wittkop T., Sokolov A., Graim K., Funk C., Verspoor K., Ben-Hur A. A large-scale evaluation of computational protein function prediction. Nat Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dessailly B.H., Redfern O.C., Cuff A.L., Orengo C.a. Detailed analysis of function divergence in a large and diverse domain superfamily: toward a refined protocol of function classification. Structure. 2010;18:1522–1535. doi: 10.1016/j.str.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandya C., Farelli J.D., Dunaway-Mariano D., Allen K.N. Enzyme promiscuity: engine of evolutionary innovation. J Biol Chem. 2014;289:30229–30236. doi: 10.1074/jbc.R114.572990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bridgham J.T., Eick G.N., Larroux C., Deshpande K., Harms M.J., Gauthier M.E.A., Ortlund E.A., Degnan B.M., Thornton J.W. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010;8:e1000497. doi: 10.1371/journal.pbio.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z., Liu H., Luo Y., Zhou S., An L., Wang C., Jin Q., Zhou M., Xu J.-R. Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Sci Rep. 2014;4:6746. doi: 10.1038/srep06746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furnham N., Dawson N.L., Rahman S.A., Thornton J.M., Orengo C.A. Large-scale analysis exploring evolution of catalytic machineries and mechanisms in enzyme superfamilies. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.11.010. (in revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Furnham N., Sillitoe I., Holliday G.L., Cuff A.L., Laskowski R.A., Orengo C.A., Thornton J.M. Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLoS Comput Biol. 2012;8:e1002403. doi: 10.1371/journal.pcbi.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]; A large-scale analysis of 276 domain enzyme superfamilies using structural, sequence, phylogeny and biochemical analyses to explore how novel enzyme chemistry can evolve through different evolutionary routes.
- 41.Wichelecki D.J., Balthazor B.M., Chau A.C., Vetting M.W., Fedorov A.A., Fedorov E.V., Lukk T., Patskovsky Y.V., Stead M.B., Hillerich B.S. Discovery of function in the enolase superfamily: d-mannonate and d-gluconate dehydratases in the d-mannonate dehydratase subgroup. Biochemistry. 2014;53:2722–2731. doi: 10.1021/bi500264p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groninger-Poe F.P., Bouvier J.T., Vetting M.W., Kalyanaraman C., Kumar R., Almo S.C., Jacobson M.P., Gerlt J.A. Evolution of enzymatic activities in the enolase superfamily: galactarate dehydratase III from Agrobacterium tumefaciens C58. Biochemistry. 2014;53:4192–4203. doi: 10.1021/bi5005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Martinez Cuesta S., Furnham N., Rahman S.A., Sillitoe I., Thornton J.M. The evolution of enzyme function in the isomerases. Curr Opin Struct Biol. 2014;26:121–130. doi: 10.1016/j.sbi.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; The reports a comprehensive analysis of the isomerase Enzyme Commission (EC) class of enzymes to explore how its members have functionally diverged in their enzyme chemistry throughout evolution. Tools were used to compare enzyme reaction data and evolutionary data, and the authors report that isomerases are more likely to evolve new enzyme chemistry in a different EC class than another type of isomerase enzyme chemistry.
- 44.Casey A.K., Hicks M.A., Johnson J.L., Babbitt P.C., Frantom P.A. Mechanistic and bioinformatic investigation of a conserved active site helix in α-isopropylmalate synthase from Mycobacterium tuberculosis, a member of the DRE-TIM metallolyase superfamily. Biochemistry. 2014;53:2915–2925. doi: 10.1021/bi500246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Sugrue E., Fraser N.J., Hopkins D.H., Carr P.D., Khurana J.L., Oakeshott J.G., Scott C., Jackson C.J. Evolutionary expansion of the amidohydrolase superfamily in bacteria in response to the synthetic compounds molinate and diuron. Appl Environ Microbiol. 2015;81:2612–2624. doi: 10.1128/AEM.04016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that new functions evolve in the functionally diverse amidohydrolase superfamily in response to changes in environmental conditions due to a transition of binuclear to mononuclear metal ion coordination at the active site.
- 46.Ben-David M., Wieczorek G., Elias M., Silman I., Sussman J.L., Tawfik D.S. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J Mol Biol. 2013;425:1028–1038. doi: 10.1016/j.jmb.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Daumann L.J., McCarthy B.Y., Hadler K.S., Murray T.P., Gahan L.R., Larrabee J.A., Ollis D.L., Schenk G. Promiscuity comes at a price: catalytic versatility vs efficiency in different metal ion derivatives of the potential bioremediator GpdQ. Biochim Biophys Acta. 2013;1834:425–432. doi: 10.1016/j.bbapap.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 48•.Studer R.A., Christin P.A., Williams M.A., Orengo C.A. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci U S A. 2014;111:2223–2228. doi: 10.1073/pnas.1310811111. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that the evolution of RubisCO, the enzyme responsible for fixation of CO2 during photosynthesis, occur by ancestral destabilizing mutations near the active site or inter-subunit interfaces which are involved in enhanced activity of the enzyme.
- 49•.Dellus-Gur E., Toth-Petroczy A., Elias M., Tawfik D.S. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]; The authors hypothesise that the key to enzymes evolving new functions is the position of the active site residues in relation to the structural scaffold. Folds that support numerous functions, such as the TIM barrel and Rossmann folds, are reported to have active site residues in loop regions that are separated from a well-packed, highly stable scaffold. Folds that support only a single function, such as dihyrofolate reductase, are shown to have the majority of their active site residues within the scaffold.
- 50••.Tóth-Petróczy Á., Tawfik D.S. The robustness and innovability of protein folds. Curr Opin Struct Biol. 2014;26:131–138. doi: 10.1016/j.sbi.2014.06.007. [DOI] [PubMed] [Google Scholar]; A review on the relationship between functional diversity and protein structure architecture. It explores why relatively few folds have the ability to support numerous functions. A protein's polarity — a term used to capture the low connectivity between a protein's scaffold and its active site — is reported to be a key feature in functional divergence.
- 51.Wittig U., Kania R., Golebiewski M., Rey M., Shi L., Jong L., Algaa E., Weidemann A., Sauer-Danzwith H., Mir S. SABIO-RK—database for biochemical reaction kinetics. Nucleic Acids Res. 2012;40:D790–D796. doi: 10.1093/nar/gkr1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Dessailly B.H., Dawson N.L., Mizuguchi K., Orengo C.A. Functional site plasticity in domain superfamilies. Biochim Biophys Acta – Proteins Proteomics. 2012;1834:874–889. doi: 10.1016/j.bbapap.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; A quantitative analysis of functional site diversity in CATH domain superfamilies. The authors report that for functionally diverse superfamilies, protein-protein interfaces are typically very versatile in their 3D surface location although there does seem to be a preferential site common to many relatives. Catalytic sites on the other hand are found to be the most stringent in their location.
- 53.Ojha S., Meng E.C., Babbitt P.C. Evolution of function in the ‘two dinucleotide binding domains’ flavoproteins. PLoS Comput Biol. 2007;3:e121. doi: 10.1371/journal.pcbi.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid A.J., Ranea J.A., Orengo C.A. Comparative evolutionary analysis of protein complexes in E. coli and yeast. BMC Genomics. 2010;11:79. doi: 10.1186/1471-2164-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu C., Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandhya S., Rani S.S., Pankaj B., Govind M.K., Offmann B., Srinivasan N., Sowdhamini R. Length variations amongst protein domain superfamilies and consequences on structure and function. PLoS ONE. 2009;4:e4981. doi: 10.1371/journal.pone.0004981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuff A., Redfern O.C., Greene L., Sillitoe I., Lewis T., Dibley M., Reid A., Pearl F., Dallman T., Todd A. The CATH hierarchy revisited — structural divergence in domain superfamilies and the continuity of fold space. Struct Des. 2009;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reeves G.A., Dallman T.J., Redfern O.C., Akpor A., Orengo C.A. Structural diversity of domain superfamilies in the CATH database. J Mol Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 59.Galperin M.Y., Koonin E. V: Divergence and convergence in enzyme evolution. J Biol Chem. 2012;287:21–28. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Huang H., Pandya C., Liu C., Al-Obaidi N.F., Wang M., Zheng L., Keating S.T., Aono M., Love J.D., Evans B. Panoramic view of a superfamily of phosphatases through substrate profiling. Proc Natl Acad Sci. 2015;112:E1974–E1983. doi: 10.1073/pnas.1423570112. [DOI] [PMC free article] [PubMed] [Google Scholar]; A large-scale activity profiling of the haloalkanoic acid dehalogenase (HAD) superfamily revealed a high degree of substrate ambiguity among the superfamily members and enabled inferred functional annotation for ∼35% of the superfamily. The study also suggested that domain insertions to the core catalytic Rossmann fold may drive the evolution of new functions, that is, increased substrate range in this superfamily.
- 61.He P., Moran G.R. Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J Inorg Biochem. 2011;105:1259–1272. doi: 10.1016/j.jinorgbio.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Sadreyev R.I., Kim B.-H., Grishin N.V. Discrete–continuous duality of protein structure space. Curr Opin Struct Biol. 2009;19:321–328. doi: 10.1016/j.sbi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radó-Trilla N., Arató K., Pegueroles C., Raya A., de la Luna S., Albà M.M. Key role of amino acid repeat expansions in the functional diversification of duplicated transcription factors. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mangeon A., Junqueira R.M., Sachetto-Martins G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav. 2010;5:99–104. doi: 10.4161/psb.5.2.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buljan M., Bateman A. The evolution of protein domain families. Biochem Soc Trans. 2009;37:751–755. doi: 10.1042/BST0370751. [DOI] [PubMed] [Google Scholar]
- 66.Teichmann S.A., Rison S.C.G., Thornton J.M., Riley M., Gough J., Chothia C. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J Mol Biol. 2001;311:693–708. doi: 10.1006/jmbi.2001.4912. [DOI] [PubMed] [Google Scholar]
- 67.Vogel C., Pleiss J. The modular structure of ThDP-dependent enzymes. Proteins. 2014;82:2523–2537. doi: 10.1002/prot.24615. [DOI] [PubMed] [Google Scholar]
- 68.Copley S.D. An evolutionary biochemist's perspective on promiscuity. Trends Biochem Sci. 2015;40:72–78. doi: 10.1016/j.tibs.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kainulainen V., Korhonen T. Dancing to another tune—adhesive moonlighting proteins in bacteria. Biology (Basel) 2014;3:178–204. doi: 10.3390/biology3010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou T., Risso V.A., Gavira J.A., Sanchez-Ruiz J.M., Ozkan S.B. Evolution of conformational dynamics determines the conversion of a promiscuous generalist into a specialist enzyme. Mol Biol Evol. 2015;32:132–143. doi: 10.1093/molbev/msu281. [DOI] [PubMed] [Google Scholar]
- 71.Baier F., Tokuriki N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J Mol Biol. 2014;426:2442–2456. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 72.Baier F., Chen J., Solomonson M., Strynadka N.C.J., Tokuriki N. Distinct metal isoforms underlie promiscuous activity profiles of metalloenzymes. ACS Chem Biol. 2015;10:1684–1693. doi: 10.1021/acschembio.5b00068. [DOI] [PubMed] [Google Scholar]
- 73.Lamb D.C., Waterman M.R. Fifty Years of Cytochrome P450 Research. Springer; 2014. Fifty years of cytochrome P450 research: examples of what we know and do not know; pp. 43–71. [Google Scholar]
- 74.Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majorek K.A., Dunin-Horkawicz S., Steczkiewicz K., Muszewska A., Nowotny M., Ginalski K., Bujnicki J.M. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014;42:4160–4179. doi: 10.1093/nar/gkt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao S., Sakai A., Zhang X., Vetting M.W., Kumar R., Hillerich B., San Francisco B., Solbiati J., Steves A., Brown S. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. Elife. 2014;3:e03275. doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uchiyama T., Miyazaki K., Yaoi K. Characterization of a novel β-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J Biol Chem. 2013;288:18325–18334. doi: 10.1074/jbc.M113.471342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milshteyn A., Schneider J.S., Brady S.F. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol. 2014;21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang H., Gough J. A domain-centric solution to functional genomics via dcGO Predictor. BMC Bioinformatics. 2013;14:S9. doi: 10.1186/1471-2105-14-S3-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shannon P., Markiel A., Ozier O., Baliga N.S., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]