Abstract
Irradiation commonly causes pneumocyte senescence, which may lead to severe fatal lung injury characterized by pulmonary dysfunction and respiratory failure. However, the molecular mechanism underlying the induction of pneumocyte senescence by irradiation remains to be elucidated. In the present study, weighted gene co-expression network analysis (WGCNA) was used to screen for differentially expressed genes, and to identify the hub genes and gene modules, which may be critical for senescence. A total of 2,916 differentially expressed genes were identified between the senescence and non-senescence groups following thoracic irradiation. In total, 10 gene modules associated with cell senescence were detected, and six hub genes were identified, including B-cell scaffold protein with ankyrin repeats 1, translocase of outer mitochondrial membrane 70 homolog A, actin filament-associated protein 1, Cd84, Nuf2 and nuclear factor erythroid 2. These genes were markedly associated with cell proliferation, cell division and cell cycle arrest. The results of the present study demonstrated that WGCNA of microarray data may provide further insight into the molecular mechanism underlying pneumocyte senescence.
Keywords: irradiation, pneumocyte senescence, weighted gene co-expression network analysis, gene modules, hub genes
Introduction
Increasing numbers of radioactive sources have been widely used as nuclear-generated energy for engineering and in nuclear weapons (1). As a medical diagnosis and treatment method, radiotherapy is ubiquitously applied in cancer treatment, owing to its significant elevation in patient survival rates (2–4). However, radiotherapy presents latent hazards, which lead to severe toxic effects on the healthy tissues of patients (5). Due to its harmful side-effects, it is necessary to investigate the cellular reaction caused by irradiation at the genetic level.
Cell senescence is one of the common side-effects of lung cancer treatment by radiotherapy. Cell senescence may lead to irreversible cell cycle arrest, which maintains cell viability and metabolically activity, but resistance to apoptosis and proliferation occurs (6). Cell senescence may also result in various pathological changes in different types of cell (7–11). A previous study demonstrated that pneumocyte senescence induced by irradiation may promote severe radiation-induced lung injury (RILI), which is a progressive, life-threatening complication characterized by interstitial infiltrates, dyspnoea and pulmonary dysfunction that can result in respiratory failure (12). At present, there are few potentially effective therapeutics for the treatment of RILI (13). Therefore, elucidating the genetic pathophysiology of pneumocyte senescence may be a useful strategy to prevent patients from developing severe RILI during lung cancer treatment.
Gene expression microarrays allow the measurement of alterations in genetic expression patterns, and facilitate the identification of genes, which are crucial to diseases induced by irradiation (14). Xie et al (15) used a cDNA microarray to analyze miRNA and mRNA expression levels in rat lung tissue samples 3, 12 and 26 weeks following exposure to 24-Gy X-ray irradiation. The results confirmed that the miRNA expression levels were negatively correlated with the mRNA expression levels (16). They also demonstrated that RILI did not develop in a single linear process (16). Chauhan et al (16) identified 67 upregulated and 141 downregulated genes in human lung fibroblast cells 24 h following 0–1.5-Gy X-ray irradiation, compared with the expression profile of untreated lung fibroblasts cells. These genes were involved in cell cycle control/mitosis, chromosome instability and cell differentiation (16). Gene Ontology and pathway enrichment analyses of genes enable the molecular pathogenesis of irradiation to be elucidated.
It is necessary to extract available information by discarding redundant or 'noisy' information from high-throughput data sets. With a systemic biological view, WGCNA is a novel approach, which quantitatively measures the interconnectivity of genes, and reveals the importance of genes within networks. WGCNA is a useful tool for detecting gene modules that maintain genes with similar expression patterns, as well as for identifying disease biomarkers and the functions of genes (17). Furthermore, due to the fact that less false positive correlations are found between genes using WGCNA, it is widely utilized to investigate complex diseases, including endometrial cancer (18), schizophrenia (19) and breast cancer (20).
In the present study, based on the microarray data of pneumocyte senescence induced by irradiation, WGCNA was used to construct a scale-free weighted genetic interaction network comprising specific gene modules that maintain common biological roles in the process of pneumocyte senescence. Moreover, in a given gene module, the present study attempted to identify hub genes as candidate biomarkers and as therapeutic targets for pneumocyte senescence.
Materials and methods
Microarray data, processing and differentially expressed gene filtering
The high-throughput data was deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), which is the predominant public repository for microarray data (21). The transcription profiles of GSE41789 (Affymetrix mouse 430_2 GeneChips; Affymetrix, Inc., Santa Clara, CA, USA), submitted by Citrin et al and updated in 2014 (2), were downloaded from the GEO database. A total of 30 mouse lung tissue samples from the dataset were selected and divided into non-senescence (n=15) and senescence (n=15) subgroups. The non-senescence group (n=15) pneumocytes exhibited no signature features of senescence (senescence-associated β-galactose) following thorax X-ray irradiation (Precision X-Ray, North Branford, CT, USA) at a dose of 0 Gy, whereas the senescence group (n=15) exhibited the signature of pneumocyte senescence following thorax X-ray irradiation at doses of 5 or 17.5 Gy (2).
The multi-microarray raw data of the CEL files were then corrected, quantile normalized, and log2 transformed with the rma function using the Affy package in R 3.0.3 software in Bioconductor (http://www.bioconductor.org/) (22,23). Only the perfectly matched probes were maintained for further analysis, and mismatched probes were discarded. The collapseRows and intersect functions of the WGCNA package were used to combine multiple probes by the highest intensity values. The differentially expressed genes between the non-senescence and senescence groups were identified using Student's t-test with R software, and only genes with P≤0.05 (n=2,916 genes) were considered for subsequent network analysis (24). In addition, the annotation information of the GeneChip was obtained from the GPL1216 microarray platform (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL1261).
Pneumocyte-weighted gene co-expression network construction
The differentially expressed genes were used for weighted gene co-expression network construction in the WGCNA package (25). In the WGCNA package, the discrete adjacency matrix is replaced with the weighted adjacency matrix considering a continuous connection strength ([0,1]) with respect to the β parameter. In the present study, β=9 was selected, according to the scale-free topology criterion proposed by Zhang and Horvath (17). Following the definition of the weighted adjacency matrix for each group (senescence and non-senescence), the co-expression matrix and the topological overlap matrix (TOM) were established. The TOM reflects the relative interconnectivity between two genes according to their degree of shared neighbors across whole network (17). The gene co-expression networks were constructed using the blockwiseModules function in the WGCNA package of the R software.
Pneumocyte module analysis
Module detection is the primary strategy for reducing high-dimensional microarray data (26). In the present study, the differentially expressed genes were considered in performing module detection. Modules were defined as groups of genes with high topological overlap (TO). Using the average linkage hierarchical clustering coupled with the TO, the present study detected the modules of the senescence and non-senescence groups, respectively. The intramodular connectivity, known as the intramodule degree, of the genes was also evaluated (27). The module eigengene (ME), which is defined as the first principal component of a given module, was then calculated. The ME can also be considered as a representative of the gene expression profiles in a module (28). The intramodular Connectivity and signed KME functions in the WGCNA package were used to compute intramodular connectivity and the ME.
Module preservation evaluation
Module preservation statistics supply information about whether the properties of a module in a network are altered under different conditions. For the correlation network, the following composite module preservation statistic: Zsummary score [Zsummary = (Zdensity + Zconnectivity)/2)] was recommended by Langfelder et al (29), and has been used extensively in previous studies (30,31). The Zdensity emphasizes whether the genes in each of the defined modules in the reference network remain highly connected in the experimental network, whereas Zconnectivity identifies whether the connectivity patterns between the genes in the experimental network remain similar, compared with the reference network. Simulations or permutation tests are used to determine the thresholds for Zsummary. A Zsummary value <2 suggests that there is no evidence for module preservation, Zsummary values >2 and <10 indicate weak to moderate evidence of module preservation, and Zsummary values >10 indicate substantial evidence for module preservation. Notably, the Zsummary score is size-dependent, and tends to increase with module size. Another, less size-dependent, preservation statistic is the medianRank, as follows: (medianRankdensity + median-Rankconnectivity)/2, which is recommended to assess module preservation (29). Generally, a module with a lower median rank tends to exhibit more marked preservation than a module with a high median rank. Combining the Zsummary with preservation medianRank was considered in the assessment of module preservation in the present study (29,32). Module preservation was evaluated using the modulePreservation function of the WGCNA package.
Measurement of gene significance (GS) and module membership (MM)
The GS of a gene is defined as the differential expression between non-senescence and senescence groups, determined using a t-test, and is evaluated using the P-value obtained from the t-test, as GSi = -log(pi) (33). The GS for each gene in a module was calculated to quantitatively assess how connected the gene was with senescence. Using the ME, the MM was measured, and was determined as the correlation between the gene expression profile and the ME. The MM is characterized by correlating the expression profile of the i-th gene with the ME of a given module, as follows: MMi = |cor (x(i), ME)|. The i-th gene is not part of the module if MMi is close to 0. In addition, the i-th gene is deemed to have a high level of connectivity to a given module if the MMi is close to 1. The MM is highly correlated with intramodular connectivity, and the highly connected intramodular genes tend to have high MM values correlated with their respective module (17,33).
Hub gene analyses
Certain terminology may be considered prior to identifying the hub genes. Generally, intramodular hub genes exhibit higher biologically significance, compared with global network hub genes. Intramodular connectivity assesses the connection strength among the genes, and intramodular connectivity is considered more reliable than differential expression assessed using Student's t-test (17). The GS incorporates external information into the co-expression network; higher absolute values of GSi indicate higher biological significance of the i-th gene. The MM assesses the correlation between a gene and the ME in a given module. Therefore, an ideal hub gene possesses the highest intra-modular connectivity, highest MM and highest GS within a given module (19,30).
Results
Weighted co-expression network construction with respect to the senescence and non-senescence groups
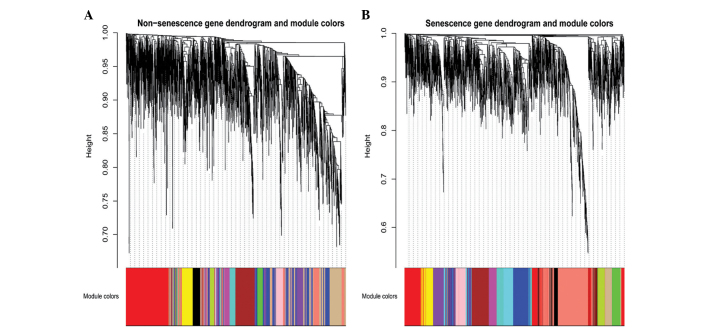
Network construction was restricted to 2,916 differentially expressed genes between the non-senescence and senescence groups, which were determined using t-tests (P≤0.05). The weighted gene co-expression networks were constructed with respect to each group using the WGCNA package of the R software. Based on the TOM, a hierarchical average linkage clustering method was used to cluster the genes into modules. The number of modules was detected using Dynamic Tree Cut (deep split = 2, cut height = 0.99; http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/BranchCut), which is a novel cluster detection technique which uses an iteration of an adaptive process of cluster decomposition and combination until the number of clusters becomes stable. In the senescence group, 13 modules were detected, and in the non-senescence group, 12 modules were detected (Fig. 1). Genes that did not cluster into any of the modules were retained in the grey module in the WGCNA package.
Figure 1.
Gene co-expression modules in the non-senescence and senescence groups. A total of (A) 12 and (B) 13 modules were identified in the non-senescence and senescence groups, respectively. Colors in the horizontal bar represent the different modules.
Screening of specific modules associated with pneumocyte senescence
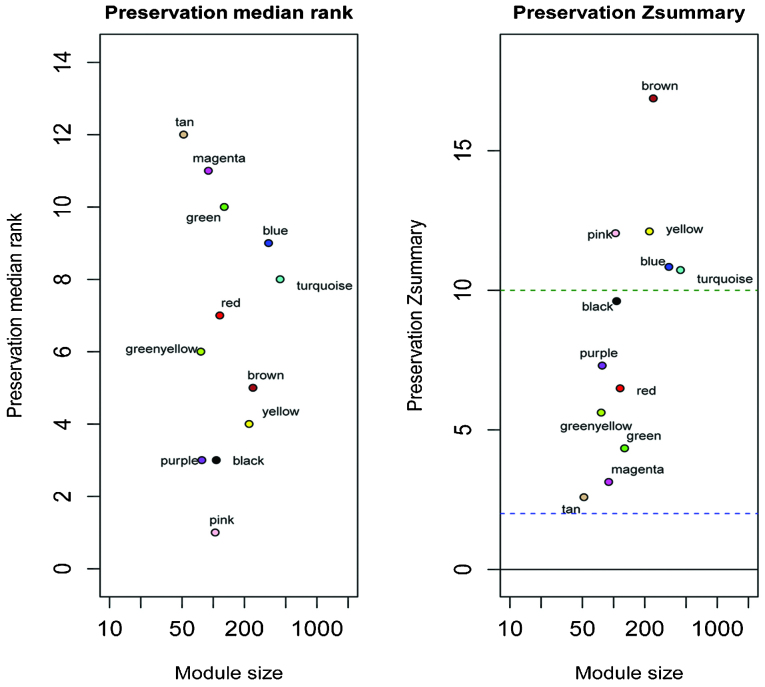
The strategy for screening the gene modules of interest associated with pneumocyte senescence depended on the preservation statistics. Module preservation statistics are based on a permutation test implemented in the modulePreser-vation function of the WGCNA package. The non-senescence network was referred to as the reference network in the preservation statistics. In total, nine gene modules were identified with small Zsummary values and large preservation median rank values, including the blue, brown, green, green/yellow, magenta, red, tan, turquoise and yellow modules. These nine modules exhibited significantly altered intramodular connectivity in the senescence group, compared with the non-senescence group following irradiation exposure (29) (Fig. 2). The salmon-colored module was detected only in the senescence group, and this module was specific to the senescence group exposed to irradiation. Therefore, a total of 10 modules were selected as modules that may be important in cellular senescence. The remaining black, pink and purple gene modules were well-preserved in the two groups, and were excluded from the following analysis.
Figure 2.
Composite preservation statistics of non-senescence in the senescence group. Each point represents a module, labeled with a color. In the preservation Zsummary graph on the right, the blue and green horizontal lines show the thresholds of Zsummary (y-axis) =2 and Zsummary (y-axis) =10, respectively. Zsummary >10 is indicative of strong preservation of the modules. In the preservation median rank graph on the left, the median rank of the modules close to zero indicated a high degree of module preservation. The comprehensive analysis is based on the median rank and Zsummary. The purple, black and pink modules were ultimately considered to be well preserved.
Candidate genes associated with senescence
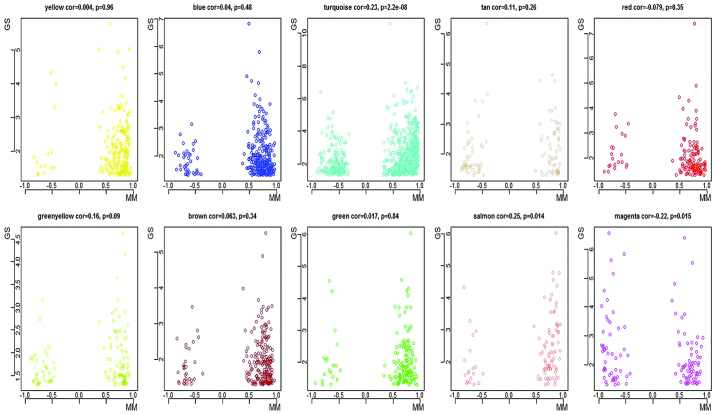
Another aim of the weighted network analysis was to identify the hub genes associated with irradiation-induced senescence. It is well-established that the MM measures the importance of a node (gene) within a network, and that the GS indicates the differential degree of the node under different conditions. A node with maximum connectivity strength is centrally located in the network (17). A marked positive correlation was observed between MM and intramodular connectivity in the modules of interest. However, no correlation was observed between GS and intramodular connectivity, or between GS and MM in the modules of interest (Fig. 3). Therefore, the hub genes were identified in each module of interest, predominantly by relying on high MM values, owing to the weak correlation between the GS and MM. Accordingly, 10 hub genes were identified with respect to the modules of interest, including translocase of outer mitochondrial membrane 70 homolog A (Tomm70a; MM.blue = 0.96; P=0.02), actin filament-associated protein 1 (Afap1; MM.brown = 0.97; P=0.04), Zfp518b (MM.green = 0.94; P=0.03), Tbc1d9b (MM. greenyellow = 0.93; P=0.03), Cd84 (MM.magenta = -0.95; P=0.002), Nuf2 (MM.red = 0.98; P=0.03), B-cell scaffold protein with ankyrin repeats 1 (Bank1; MM.salmon = 0.94; P=0.001), Gm6377 (MM.tan = 0.94; P=0.0009), nuclear factor erythroid 2 (NFE2; MM.turquoise = 0.98; P=0.001), and Slc25a15 (MM.yellow = 0.95; P=0.04).
Figure 3.
Correlation of the MM (x-axis) and the GS (y-axis). The color indicates the module, and the dot indicates the gene within the module. The identification of hub genes was dependent on the presence of high MM values. MM, module membership; GS, gene significance.
Annotation and functional enrichment analysis of co-expressed modules
Gene ontology (GO) functional annotation and enrichment analyses were used to identify the significantly enriched biological terms for genes in the modules of interest (26). GO enrichment analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) software (http://david.abcc.ncifcrf.gov/) by uploading a probe set to DAVID and initiating the Annotation Tool. Functional annotations enriched in the modules of interest are shown in Table I. A total of 10 biological processes were involved in the modules of interest: RNA processing (blue), mesoderm development (brown), apoptotic mitochondrial changes (green), regulation of the hippo signaling cascade (green/yellow), regulation of cell development (magenta), cell division (red), the immune system process (salmon), signal transduction (tan), the immune system process (turquoise) and the phenol-containing compound metabolic process (yellow).
Table I.
Top functional annotations enriched in the cellular senescence-specific modules for term ontology 'Biological process'.
| Module | Term name | P-value |
|---|---|---|
| Blue | RNA processing | 4.42×10−9 |
| Blue | mRNA metabolic process | 1.19×10−6 |
| Brown | Mesoderm development | 6.72×10−5 |
| Brown | Protein localization | 2.18×10−3 |
| Green | Apoptotic mitochondrial changes | 3.20×10−5 |
| Green | Regulation of mitochondrion organization | 5.09×10−5 |
| Green/yellow | Hippo signaling cascade | 6.27×10−5 |
| Green/yellow | Regulation of hippo signaling cascade | 1.59×10−3 |
| Magenta | Regulation of cell development | 2.90×10−4 |
| Magenta | Positive regulation of catalytic activity | 3.64×10−4 |
| Red | Cell division | 5.48×10−11 |
| Red | Cell cycle | 4.78×10−9 |
| Salmon | Immune system process | 5.34×10−7 |
| Salmon | Cell surface receptor signaling pathway | 6.48×10−7 |
| Tan | Signal transduction | 1.48×10−7 |
| Tan | Immune response | 5.23×10−7 |
| Turquoise | Immune system process | 1.56×10−22 |
| Turquoise | Response to wounding | 9.28×10−20 |
| Yellow | Phenol-containing compound metabolic process | 3.20×10−3 |
| Yellow | Neurotransmitter catabolic process | 5.06×10−3 |
Pathway enrichment analysis of co-expression modules
The Kyoto Encyclopaedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) database was used to enrich the biological signaling pathways for the 10 identified modules. The pathway enrichment analysis was implemented using DAVID software by assigning a probe set to KEGG metabolic processes and testing the statistical enrichment of the target gene in KEGG pathways. The most highly enriched pathways in each module were as follows: Cell cycle (P=1.60×10−6) in the blue module; cell cycle (P=1.67×10−3) in the brown module; glycolysis/gluconeogenesis (P=1.50×10−2) in the green module; spliceosome (P=2.22×10−2) in the green/yellow module; steroid biosynthesis (P=6.38×10−3) in the magenta module; proteasome (P=9.40×10−3) in the red module; melanoma (P=9.63×10−3) in the salmon module; renal cell carcinoma (P=1.43×10−2) in the tan module; protein processing in endoplasmic reticulum (ER; P=3.79×10−4) in the turquoise module and linoleic acid metabolism (P=2.92×10−3) in the yellow module (Table II).
Table II.
Most enriched pathways in the cellular senescence-specific modules.
| Module | Pathway name | Number of genes | P-value |
|---|---|---|---|
| Blue | Cell cycle | 12 | 1.60×10−6 |
| Blue | DNA replication | 5 | 3.02×10−4 |
| Brown | Cell cycle | 7 | 1.67×10−3 |
| Brown | mTOR signaling pathway | 4 | 5.73×10−3 |
| Green | Glycolysis/Gluconeogenesis | 3 | 1.50×10−2 |
| Green | Oxidative phosphorylation | 4 | 3.45×10−2 |
| Green/yellow | Spliceosome | 4 | 2.22×10−2 |
| Green/yellow | Pentose and glucuronate interconversions | 2 | 2.41×10−2 |
| Magenta | Steroid biosynthesis | 2 | 6.38×10−3 |
| Magenta | Alzheimer's disease | 4 | 3.73×10−2 |
| Red | Proteasome | 3 | 9.40×10−3 |
| Red | Oxidative phosphorylation | 5 | 1.40×10−2 |
| Salmon | Melanoma | 3 | 9.63×10−3 |
| Salmon | Prostate cancer | 3 | 1.71×10−2 |
| Tan | Renal cell carcinoma | 3 | 1.43×10−2 |
| Tan | Rheumatoid arthritis | 3 | 2.02×10−2 |
| Turquoise | Protein processing in endoplasmic reticulum | 16 | 3.79×10−4 |
| Turquoise | PPAR signaling pathway | 10 | 5.66×10−4 |
| Yellow | Linoleic acid metabolism | 4 | 2.92×10−3 |
| Yellow | Cell cycle | 6 | 6.12×10−3 |
mTOR, mammalian target of rapamycin; PPAR, peroxisome proliferator-activated receptor.
Discussion
In the present study, 10 specific modules and 10 hub genes associated with pneumocyte senescence were identified, of which the salmon module was only observed in the senescence group. The GO enrichment of the salmon module suggested that the immune process may be involved in senescence. Accordingly, Bank1 was identified as a hub gene of this module, and was significantly upregulated (P=0.0007) in the senescence group. Bank1 encodes a B cell-specific scaffold protein, which functions in the B cell receptor (35). Bank1 has been associated with systemic lupus erythematosus and diffuse systemic sclerosis (36,37). Using Bank1 deficient-mice, it has been previously demonstrated that BANK1 acts as a negative regulator of CD40-mediated protein kinase B activation to prevent hyperactive responses. The absence of BANK1 reduced the secretion of interleukin-6 via the p38/mitogen-activated protein kinase 1/2 signaling pathway and decreased the expression levels of translation initiation factor eIF4e in B cells. Notably, high expression levels of eIF4E promotes cell proliferation in carcinoma (38). Whether BANK1 is associated with cellular senescence remains to be fully elucidated, however, these results provide a novel research direction for further investigations to clarify the importance of Bank1 during the cellular senescence process.
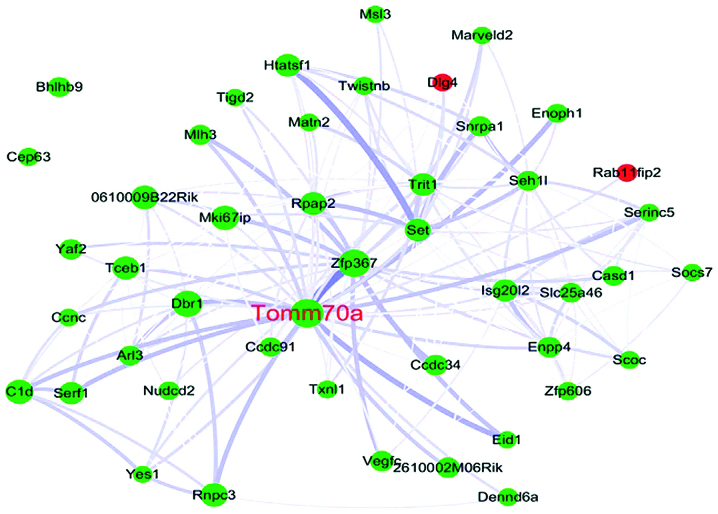
In the present study, the blue module was determined as the RNA-modifying module, based on the GO analysis. Tomm70a, which encodes the TOM70 protein in humans, was selected as the hub gene in the blue module and was downregulated during senescence (P=0.019). TOM70 is a subunit of the outer mitochondrial membrane translocase, which acts as a receptor for hydrophobic pre-proteins targeted to the mitochondria and is involved in importing the majority of mitochondrial proteins from the cytosol into the mitochondria (39). For example, ribosomal protein S3 (rpS3) may be effectively transported into the mitochondria via the interaction between TOM70, heat shock protein (HSP)90 and HSP70. When rpS3 accumulates in the mitochondria, it repairs damaged mitochondrial DNA and decreases the levels of reactive oxygen species (ROS) (40). rpS3 not only mediates cellular anti-apoptosis by binding the p65 protein (41), it also induces interferon (IFN)-β production (42,43). It is well-established that IFN-β inhibits cell proliferation and arrests the cell cycle by targeting the p53 signaling pathway (44). In addition, TOM70 is a member of the TOM machinery, a molecular switch of the phosphatase and tensin homolog-induced putative kinase 1-PARKIN signaling pathway, which clears cell remnants in an autophagy-dependent manner in mitochondria (45). Once this signaling pathway is interrupted, cellular senescence is induced via ROS and the p53 signaling pathway (46). Based on these results, Tomm70a may be a novel hub gene associated with pneumocyte senescence induced by thoracic irradiation (Fig. 4).
Figure 4.
Interaction of gene co-expression patterns in the blue module. For clarity, only certain nodes are presented. The network was visualized using Cytoscape 3.0 software. The node size is proportional to the connectivity. The edge width is proportional to the connection strength. Downregulated genes and upregulated genes are colored green and red, respectively.
In the present study, the brown module was predominantly enriched in the protein localization process by GO analysis. Afap1 was selected as the hub gene of the brown module and was significantly downregulated (P=0.04) in the senescence group. AFAP1 regulates actin filament integrity, podosome formation, focal contacts and cell migration (47). AFAP1 is an essential adaptor protein, which activates c-Src tyrosine kinase by binding and interacting with SH3 (48). c-Src is a proto-oncogene with a wide range of substrates and various functions in processes, including cell proliferation, differentiation, cell cycle, adhesion, invasion and motility (49). Zhang et al (50) demonstrated that the loss of AFAP1 in PC3 prostate cancer cells reduces cell proliferation in vitro. Using an AFAPΔABD expression vector, a previous study reported that AFAP1 activates c-Src and subsequently elevates the expression levels of transcriptional factor activator protein 1 (AP-1) (51). AP-1 modulates a wide range of cellular processes, including proliferation, apoptosis, differentiation and survival (52). Therefore, the present study hypothesized that AFAP1 regulates cell senescence through c-Src or AP-1, however, further investigations are required in order to confirm this hypothesis.
The magenta module was predominantly enriched in the cell development and protein transport GO terms. Cd84 encodes the type I transmembrane glycoprotein CD84, and was significantly upregulated (P=0.002) in the senescence group. CD84 is a member of the signaling lymphocyte activation molecule family, which is expressed in the majority of immune cell populations (53,54). A previous study has indicated that CD84 promotes early-stage chronic lymphocytic leukemia (CLL) survival, and that cell death is induced in CLL following the inhibition of CD84 in vitro and in vivo (55). By measuring the incorporation of 3H thymidine during the final 8 h of a 72-h culture period, the investigators demonstrated that CD84 regulated the proliferation of anti-CD3 monoclonal antibody-stimulated human T cells (56). These studies suggested that CD84 may be associated with the cell cycle and survival of lymphocytes.
Based on the GO analysis in the present study, the red module was denoted the cell cycle module. The evolutionarily conserved gene Nuf2 was significantly downregulated in the senescence group (P=0.03). Nuf2, also known as cell division cycle associated 1 (CDCA1) is a kinetochore protein, which forms a subcomplex with Hec1 and belongs to the larger Ndc80 complex (57). The Nuf2-Hec1 complex connects the plus ends of spindle microtubules to centromeres, and is essential for correct chromosome segregation and genomic stability during mitosis (57,58). In HeLa cells, it has been confirmed that prometaphase is prolonged during the mitosis, if Nuf2 or Hec1 was silenced by RNA interference (59). The interaction of Nuf2 and PTPIP51 recruits microtubules to the kinetochore and ensures proper cellular proliferation, differentiation and apop-tosis (60). In addition, the interaction of CDCA1 and KNTC2 has been reported to induce cell cycle arrest in non-small cell lung carcinoma, as well as in colorectal and gastric cancer (61,62). Therefore, Nuf2 may be a hub gene, as well as a therapeutic target of irradiation-induced tumor and pneumocyte senescence.
In the present study, the turquoise module was associated with immune system and wound healing in the GO analysis. NFE2 was considered the hub gene and was significantly upreg-ulated in the senescence group (P=0.001). NFE2 is a member of the basic-leucine zipper family of heterodimeric transcriptional activators, and comprises p45 and Maf subunits (63). Studies have confirmed that NFE2 is an important transcriptional regulator of hemoglobin biosynthesis, normal platelet function and erythroid differentiation (64,65). In addition, NFE2 increases the expression levels of IL-8 to stimulate CD34+ proliferation and survival in bone marrow stromal cells, and ensure megakaryocyte proliferation and differentiation in primary myelofibrosis (66). In red blood cells (RBCs), p45NFE2 decreases ROS levels in order to protect RBCs from oxidative stress damage (67). Therefore, NFE2 may be involved in inflammatory cell proliferation or senescence during irradiation damage.
The four hub genes identified in the remaining modules included Zfp518b (green), Tbc1d9b (greenyellow), Gm6377 (tan) and Slc25a15 (yellow). To date, no reports regarding the roles of these four hub genes in the cell cycle, proliferation or senescence are available.
The results of the present stead revealed a total of 70 KEGG signaling pathways in the senescence group, which were enriched in the 10 modules of interest. Cell cycle, DNA replication and lysine biosynthesis were most highly enriched in the blue module, whereas the cell cycle and mammalian target of rapamycin (mTOR) signaling pathways were the most highly enriched in the brown module. mTOR is highly conserved across species. Rapamycin, a specific inhibitor of mTOR, mitigates senescence progression in HT-p21 cells (68), normal human fibroblast WI-38 cells (69) and ARPE-19 cells (70). Furthermore, the oncogenic proteins RAF and RAS, which are known to activate the mTOR signaling pathway, have been reported to mediate cellular senescence (71,72). The glycolysis/gluconeogenesis and oxidative phosphorylation signaling pathways were enriched in the green module. Dichloroacetate mitigated senescence by suppressing glycol-ysis, and a study by Liao et al (73) demonstrated the activation of glyceraldehyde-3-phosphate dehydrogenase and the upregu-lation of glycolysis in radiation-induced human breast cancer cell senescence. The spliceosome biological pathway was the most highly enriched in the green-yellow module. In WI-38, WiDr, HeLa and HEK239 cells, the cell cycle is blocked in the G1 and G2/M phases upon the deletion or inhibition of splicing factors by small interfering RNA or inhibitors (74-76). A previous investigation also reported the impairment of spliceosome assembly-promoted cell cycle arrest in the S and G2/M phases (76). The proteasome and oxidative phos-phorylation signaling pathways were the most highly enriched pathways in the red module. The ubiquitination/proteasome signaling pathway is crucial for determining protein fate post-translation (77). p53 is a substrate of Mdm2, which is an E3 ubiquitin ligase of the proteasome signaling pathway, and Mdm2 regulates the stability and activity of p53, ultimately contributing to the process of cellular senescence (76). The mitogen-activated protein kinase (MAPK) signaling pathway was significantly enriched in the salmon module. The MAPK signaling pathway has long been associated with cell senescence. For example, ROS or oncogene-induced senescence enables activation of the p38 MAPK signaling pathway through the activation of McKusick-Kaufman syndrome 3/6, which subsequently promotes the expression of p16 and p53-p21, and increases the DNA damage response (78). This response suppresses cyclin-dependent kinase and ultimately generates cell senescence (79). In the tan module, the ribosome biogenesis pathway was the most significantly enriched pathway. A previous study investigating dyskeratosis congenita demonstrated that the failure of ribosome biogenesis and the induction of DNA damage activates the p53 signaling pathway and induces cell senescence in X-DC or AD-DC fibroblasts (80). In the turquoise module in the present study, protein processing in the ER was the most highly enriched signaling pathway. The ER is the principal organelle for several cellular functions, including protein folding, maturation and the maintenance of cellular homeostasis (81). Wiel et al (82) reported that the calcium channel, inositol 1,4,5-trisphosphate receptor type 2 and mitochondrial calcium uniporter led to calcium release from the ER and mitochondrial, respectively, resulting in the induction of cellular senescence. This pathway is independent of the retinoblastoma and p53 pathways (82). In addition, a previous study reported the repression of ER stress and the promotion of oncogene-induced cellular senescence by peroxisome proliferator-activated receptor β/δ in mouse keratinocytes (83). The linoleic acid metabolism signaling pathway was enriched in the yellow module. Linoleic acid is a polyunsaturated essential fatty acid, which maintains normal physiological functions and is involved in disease prevention (84). It has been demonstrated that linoleic acid affects the cell cycle and proliferation. For example, in mouse embryonic stem cells, Kim et al (85) reported that linoleic acid induces the cell cycle through multiple signaling pathways, including Ca2+/protein kinase C, phosphoinositide 3 kinase and MAPKs. In addition, in mouse pancreatic β-cells, others have demonstrated that linoleic acid elevates the expression levels of cell cycle inhibitors, p16 and p18, in vivo and in vitro.
The results of the present study enabled the identification of the gene modules and hub genes which exhibit crucial biological activity in pneumocyte senescence induced by thoracic irradiation. The modules with different activities were involved in the progress of senescence, including immune system responses, post-transcriptional modifications, post-translational modifications, cell signal transduction pathways, cellular motility, hormone regulation and biomolecule metabolism. The analyses demonstrated that various significant responses arose during irradiation-induced pneumocyte senescence. Hub genes associated with cellular senescence may offer an important source of novel hypotheses, experimental directions and novel therapeutic targets. The results of the present study not only further current understanding of the mechanism of senescence, but also provide considerable information for understanding RILI and for generating innovative therapies to treat this life-threatening disease.
It is increasingly evident that biological functions arise from complex interactions among macromolecules, including proteins, DNA and RNA. Microarray techniques and gene co-expression profile network analyses are becoming necessary to examine these complex interactions. The novel WGCNA method is a popular microarray data analysis approach due to its lower rate of false positive connections and its impressive module detection ability. From a systemic perspective, the results of the present study provide a comprehensive summary of irradiation-induced pneumocyte senescence analyzed using the WGCNA method.
Acknowledgments
The present study was supported by grants from the National Program on Key Basic Research Project (Program 973; grant no. 2011CB964800-G) and the National Natural Science Foundation of China (grant nos. 81129020 and 81372928).
References
- 1.Wang J, Feng J, Jia W, Chang S, Li S, Li Y. Lignin engineering through laccase modification: A promising field for energy plant improvement. Biotechnol Biofuels. 2015;8:145. doi: 10.1186/s13068-015-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Citrin DE, Shankavaram U, Horton JA, Shield W, III, Zhao S, Asano H, White A, Sowers A, Thetford A, Chung EJ. Role of type II pneumocyte senescence in radiation-induced lung fibrosis. J Natl Cancer Inst. 2013;105:1474–1484. doi: 10.1093/jnci/djt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Schabath MB, Nguyen A, Wilson P, Sommerer KR, Thompson ZJ, Chiappori AA. Temporal trends from 1986 to 2008 in overall survival of small cell lung cancer patients. Lung Cancer. 2014;86:14–21. doi: 10.1016/j.lungcan.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedicini P, Strigari L, Benassi M, Caivano R, Fiorentino A, Nappi A, Salvatore M, Storto G. Critical dose and toxicity index of organs at risk in radiotherapy: analyzing the calculated effects of modified dose fractionation in non-small cell lung cancer. Med Dosim. 2014;39:23–30. doi: 10.1016/j.meddos.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linton PJ, Gurney M, Sengstock D, Mentzer RM, Jr, Gottlieb RA. This old heart: Cardiac aging and autophagy. J Mol Cell Cardiol. 2015;83:44–54. doi: 10.1016/j.yjmcc.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park YH. Stem cell therapy for sensorineural hearing loss, still alive? J Audiol Otol. 2015;19:63–67. doi: 10.7874/jao.2015.19.2.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer. 2015;90:121–127. doi: 10.1016/j.lungcan.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farup J, Madaro L, Puri PL, Mikkelsen UR. Interactions between muscle stem cells, mesenchymal-derived cells and immune cells in muscle homeostasis, regeneration and disease. Cell Death Dis. 2015;6:e1830. doi: 10.1038/cddis.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikhed Y, Daiber A, Steven S. Mitochondrial oxidative stress, mitochondrial DNA damage and their role in age-related vascular dysfunction. Int J Mol Sci. 2015;16:15918–15953. doi: 10.3390/ijms160715918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill RP. Radiation effects on the respiratory system. Brit J Radiol Suppl. 2005;27:75–81. doi: 10.1259/bjr/34124307. [DOI] [Google Scholar]
- 13.Ley B, Brown KK, Collard HR. Molecular biomarkers in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;307:L681–L691. doi: 10.1152/ajplung.00014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez Bort JA, Hackl M, Höflmayer H, Jadhav V, Harreither E, Kumar N, Ernst W, Grillari J, Borth N. Dynamic mRNA and miRNA profiling of CHO-K1 suspension cell cultures. Biotechnol J. 2012;7:500–515. doi: 10.1002/biot.201100143. [DOI] [PubMed] [Google Scholar]
- 15.Xie L, Zhou J, Zhang S, Chen Q, Lai R, Ding W, Song C, Meng X, Wu J. Integrating microRNA and mRNA expression profiles in response to radiation-induced injury in rat lung. Radiat Oncol. 2014;9:111. doi: 10.1186/1748-717X-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan V, Howland M. Gene expression responses in human lung fibroblasts exposed to alpha particle radiation. Toxicol In Vitro. 2014;28:1222–1229. doi: 10.1016/j.tiv.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 18.Chou WC, Cheng AL, Brotto M, Chuang CY. Visual gene-network analysis reveals the cancer gene co-expression in human endometrial cancer. BMC Genomics. 2014;15:300. doi: 10.1186/1471-2164-15-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong S, Boks MP, Fuller TF, Strengman E, Janson E, de Kovel CG, Ori AP, Vi N, Mulder F, Blom JD, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS One. 2012;7:e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown J, et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R, et al. NCBI GEO: Mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tejera E, Bernardes J, Rebelo I. Co-expression network analysis and genetic algorithms for gene prioritization in preeclampsia. BMC Med Genomics. 2013;6:51. doi: 10.1186/1755-8794-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecca P, Re A. Detecting modules in biological networks by edge weight clustering and entropy significance. Front Genet. 2015;6:265. doi: 10.3389/fgene.2015.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic tree cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan N, Chung MK, Smith JD, Hsu J, Serre D, Newton DW, Castel L, Soltesz E, Pettersson G, Gillinov AM, et al. Weighted gene coexpression network analysis of human left atrial tissue identifies gene modules associated with atrial fibrillation. Circ Cardiovasc Genet. 2013;6:362–371. doi: 10.1161/CIRCGENETICS.113.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peña-Castillo L, Mercer RG, Gurinovich A, Callister SJ, Wright AT, Westbye AB, Beatty JT, Lang AS. Gene co-expression network analysis in Rhodobacter capsulatus and application to comparative expression analysis of Rhodobacter sphaeroides. BMC Genomics. 2014;15:730. doi: 10.1186/1471-2164-15-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filteau M, Pavey SA, St-Cyr J, Bernatchez L. Gene coexpression networks reveal key drivers of phenotypic divergence in lake whitefish. Mol Biol Evol. 2013;30:1384–1396. doi: 10.1093/molbev/mst053. [DOI] [PubMed] [Google Scholar]
- 33.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Yang XY, Shi WJ. Identifying differentially expressed genes and pathways in two types of non-small cell lung cancer: Adenocarcinoma and squamous cell carcinoma. Genet Mol Res. 2014;13:95–102. doi: 10.4238/2014.January.8.8. [DOI] [PubMed] [Google Scholar]
- 35.Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, Kurosaki T. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 37.Rueda B, Gourh P, Broen J, Agarwal SK, Simeon C, Ortego-Centeno N, Vonk MC, Coenen M, Riemekasten G, Hunzelmann N, et al. BANK1 functional variants are associated with susceptibility to diffuse systemic sclerosis in Caucasians. Ann Rheum Dis. 2010;69:700–705. doi: 10.1136/ard.2009.118174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, Liu Y, Di X, Kang H, Zeng H, Zhao Y, Cai K, Pang T, Wang S, Yao Y, Hu X. EIF4E over-expresses and enhances cell proliferation and cell cycle progression in nasopharyngeal carcinoma. Med Oncol. 2013;30:400. doi: 10.1007/s12032-012-0400-z. [DOI] [PubMed] [Google Scholar]
- 39.Blesa JR, Hernández JM, Hernández-Yago J. NRF-2 transcription factor is essential in promoting human Tomm70 gene expression. Mitochondrion. 2004;3:251–259. doi: 10.1016/j.mito.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y, Kim HD, Kim J. Cytoplasmic ribosomal protein S3 (rpS3) plays a pivotal role in mitochondrial DNA damage surveillance. Biochim Biophys Acta. 2013;1833:2943–2952. doi: 10.1016/j.bbamcr.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-kB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20:994–1011. doi: 10.1038/cr.2010.103. [DOI] [PubMed] [Google Scholar]
- 43.Kasama Y, Saito M, Takano T, Nishimura T, Satoh M, Wang Z, Ali SN, Harada S, Kohara M, Tsukiyama-Kohara K. Translocase of outer mitochondrial membrane 70 induces interferon response and is impaired by hepatitis C virus NS3. Virus Res. 2012;163:405–409. doi: 10.1016/j.virusres.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Resnitzky D, Yarden A, Zipori D, Kimchi A. Autocrine beta-related interferon controls c-myc suppression and growth arrest during hematopoietic cell differentiation. Cell. 1986;46:31–40. doi: 10.1016/0092-8674(86)90857-3. [DOI] [PubMed] [Google Scholar]
- 45.Bertolin G, Ferrando-Miguel R, Jacoupy M, Traver S, Grenier K, Greene AW, Dauphin A, Waharte F, Bayot A, Salamero J, et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy. 2013;9:1801–1817. doi: 10.4161/auto.25884. [DOI] [PubMed] [Google Scholar]
- 46.Kang HT, Lee KB, Kim SY, Choi HR, Park SC. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao H, Han B, Lodyga M, Bai XH, Wang Y, Liu M. The actin-binding domain of actin filament-associated protein (AFAP) is involved in the regulation of cytoskeletal structure. Cell Mol Life Sci. 2012;69:1137–1151. doi: 10.1007/s00018-011-0812-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder BN1, Cho Y, Qian Y, Coad JE, Flynn DC, Cunnick JM. AFAP1L1 is a novel adaptor protein of the AFAP family that interacts with cortactin and localizes to invadosomes. Eur J Cell Biol. 2011;90:376–389. doi: 10.1016/j.ejcb.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frame MC. Newest findings on the oldest oncogene; how activated src does it. J Cell Sci. 2004;117:989–998. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Park SI, Artime MC, Summy JM, Shah AN, Bomser JA, Dorfleutner A, Flynn DC, Gallick GE. AFAP-110 is overex-pressed in prostate cancer and contributes to tumorigenic growth by regulating focal contacts. J Clin Invest. 2007;117:2962–2973. doi: 10.1172/JCI30710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han B, Xiao H, Xu J, Lodyga M, Bai XH, Jin T, Liu M. Actin filament associated protein mediates c-Src related SRE/AP-1 transcriptional activation. FEBS Lett. 2011;585:471–477. doi: 10.1016/j.febslet.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Ye N, Ding Y, Wild C, Shen Q, Zhou J. Small molecule inhibitors targeting activator protein 1 (AP-1) J Med Chem. 2014;57:6930–6948. doi: 10.1021/jm5004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann S, Vögtle T, Bender M, Rose-John S, Nieswandt B. The SLAM family member CD84 is regulated by ADAM10 and calpain in platelets. J Thromb Haemost. 2012;10:2581–2592. doi: 10.1111/jth.12013. [DOI] [PubMed] [Google Scholar]
- 54.Wang A, Batteux F, Wakeland EK. The role of SLAM/CD2 polymorphisms in systemic autoimmunity. Curr Opin Immunol. 2010;22:706–714. doi: 10.1016/j.coi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Binsky-Ehrenreich I, Marom A, Sobotta MC, Shvidel L, Berrebi A, Hazan-Halevy I, Kay S, Aloshin A, Sagi I, Goldenberg DM. CD84 is a survival receptor for CLL cells. Oncogene. 2014;33:1006–1016. doi: 10.1038/onc.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tangye SG, Nichols KE, Hare NJ, van de Weerdt BC. Functional requirements for interactions between CD84 and Src homology 2 domain-containing proteins and their contribution to human T cell activation. J Immunol. 2003;171:2485–2495. doi: 10.4049/jimmunol.171.5.2485. [DOI] [PubMed] [Google Scholar]
- 57.Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell. 2005;16:2325–2338. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brobeil A, Graf M, Eiber M, Wimmer M. Interaction of PTPIP51 with Tubulin, CGI-99 and Nuf2 During cell cycle progression. Biomolecules. 2012;2:122–142. doi: 10.3390/biom2010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaneko N, Miura K, Gu Z, Karasawa H, Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390:1235–1240. doi: 10.1016/j.bbrc.2009.10.127. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki H, Fukuhara M, Yamaura T, et al. Multiple therapeutic peptide vaccines consisting of combined novel cancer testis antigens and anti-angiogenic peptides for patients with non-small cell lung cancer. J Transl Med. 2013;11:97. doi: 10.1186/1479-5876-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung KA, Kwak MK. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules. 2010;15:7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujita R, Takayama-Tsujimoto M, Satoh H, Gutiérrez L, Aburatani H, Fujii S, Sarai A, Bresnick EH, Yamamoto M, Motohashi H. NF-E2 p45 is important for establishing normal function of platelets. Mol Cell Biol. 2013;33:2659–2670. doi: 10.1128/MCB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasiorek JJ, Nouhi Z, Blank V. Abnormal differentiation of erythroid precursors in p45 NF-E2(−/−) mice. Exp Hematol. 2012;40:393–400. doi: 10.1016/j.exphem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 66.Yigit N, Covey S, Barouk-Fox S, Turker T, Geyer JT, Orazi A. Nuclear factor-erythroid 2, nerve growth factor receptor, and CD34-microvessel density are differentially expressed in primary myelofibrosis, polycythemia vera, and essential thrombocythemia. Hum Pathol. 2015;46:1217–1225. doi: 10.1016/j.humpath.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Chan JY, Kwong M, Lo M, Emerson R, Kuypers FA. Reduced oxidative-stress response in red blood cells from p45NFE2-deficient mice. Blood. 2001;97:2151–2158. doi: 10.1182/blood.V97.7.2151. [DOI] [PubMed] [Google Scholar]
- 68.Leontieva OV, Blagosklonny MV. Tumor promoter-induced cellular senescence: Cell cycle arrest followed by geroconversion. Oncotarget. 2014;5:12715–12727. doi: 10.18632/oncotarget.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 70.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 71.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao EC, Hsu YT, Chuah QY, Lee YJ, Hu JY, Huang TC, Yang PM, Chiu SJ. Radiation induces senescence and a bystander effect through metabolic alterations. 2014;5:e1255. doi: 10.1038/cddis.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh AK, Li J. A stereoselective synthesis of (+)-herbox-idiene/GEX1A. Org Lett. 2011;13:66–69. doi: 10.1021/ol102549a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, Mizui Y. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. Febs J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 76.Pawellek A, McElroy S, Samatov T, Mitchell L, Woodland A, Ryder U, Gray D, Lührmann R, Lamond AI. Identification of small molecule inhibitors of pre-mRNA splicing. J Biol Chem. 2014;289:34683–34698. doi: 10.1074/jbc.M114.590976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hegde AN, Haynes KA, Bach SV, Beckelman BC. Local ubiquitin-proteasome-mediated proteolysis and long-term synaptic plasticity. Front Mol Neurosci. 2014;7:96. doi: 10.3389/fnmol.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu Y, Li N, Xiang R, Sun P. Emerging roles of the p38 MAPK and PI3K/AKT/mTOR pathways in oncogene-induced senescence. Trends Biochem Sci. 2014;39:268–276. doi: 10.1016/j.tibs.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muñoz-Espín D, Serrano M. Cellular senescence: From physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 80.Carrillo J, González A, Manguán-García C, Pintado-Berninches L, Perona R. p53 pathway activation by telomere attrition in X-DC primary fibroblasts occurs in the absence of ribosome biogenesis failure and as a consequence of DNA damage. Clin Transl Oncol. 2014;16:529–538. doi: 10.1007/s12094-013-1112-3. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Yu H, Zhang J, Gao J, Ge X, Lou G. Hesperidin inhibits HeLa cell proliferation through apoptosis mediated by endoplasmic reticulum stress pathways and cell cycle arrest. BMC Cancer. 2015;15:682. doi: 10.1186/s12885-015-1706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiel C, Lallet-Daher H, Gitenay D, Gras B, Le Calvé B, Augert A, Ferrand M, Prevarskaya N, Simonnet H, Vindrieux D, Bernard D. Endoplasmic reticulum calcium release through ITPR2 channels leads to mitochondrial calcium accumulation and senescence. Nat Commun. 2014;5:3792. doi: 10.1038/ncomms4792. [DOI] [PubMed] [Google Scholar]
- 83.Zhu B, Ferry CH, Markell LK, Blazanin N, Glick AB, Gonzalez FJ, Peters JM. The nuclear receptor peroxisome proliferator-activated receptor-β/Δ (PPARβ/Δ) promotes oncogene-induced cellular senescence through repression of endoplasmic reticulum stress. J Biol Chem. 2014;289:20102–20119. doi: 10.1074/jbc.M114.551069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choque B, Catheline D, Rioux V, Legrand P. Linoleic acid: Between doubts and certainties. Biochimie. 2014;96:14–21. doi: 10.1016/j.biochi.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 85.Kim MH, Kim MO, Kim YH, Kim JS, Han HJ. Linoleic acid induces mouse embryonic stem cell proliferation via Ca2+/PKC, PI3K/Akt, and MAPKs. Cell Physiol Biochem. 2009;23:53–64. doi: 10.1159/000204090. [DOI] [PubMed] [Google Scholar]