Abstract
Galectin-3 is a multifunctional β-galactoside-binding lectin that is involved in multiple biological functions which are upregulated in malignancies, including cell growth, adhesion, proliferation, progression and metastasis, as well as apoptosis. A previous study has confirmed the roles of galecin-3 overexpression in the biological behavior of Eca109 human esophageal cancer (EC) cells. In the present study, small interfering (si)RNA-mediated galectin-3 silencing was performed to analyze the effects of decreased galectin-3 expression on the biological behavior of EC cells. Western blot and quantitative polymerase chain reaction analyses were utilized to confirm galectin-3 knockdown at the protein and mRNA level (P<0.05 vs. siRNA-control and untransfected groups). Cell proliferation was assessed using the Cell Counting Kit-8 assay. At 72 and 96 h after transfection, the proliferation of Eca109 cells in the siRNA-Gal-3 group was decreased compared with that in the siRNA-Control and untransfected groups (P<0.001 and P=0.004, respectively). Furthermore, Transwell assays demonstrated that inhibition of galecin-3 significantly reduced the migration and invasion of Eca109 cells compared with that in the other groups (P<0.05). Finally, apoptosis of Eca109 cells was detected using Annexin V/7-amino-actinomycin double-staining and flow cytometric analysis. Galectin-3 knockdown significantly enhanced the apoptotic rate of Eca109 cells compared with that in the siRNA-control and untreated groups (P=0.031 and P=0.047, respectively). In conclusion, following successful knockdown of galecin-3 expression in Eca109 cells, the cell proliferation, migration and invasion were reduced, while the apoptosis was enhanced, which indicates that galectin silencing may represent a therapeutic strategy for EC.
Keywords: galectin-3, RNA interference, biological function, esophageal carcinoma
Introduction
Galectin-3, a member of the carbohydrate-binding protein family, shows high affinity for β-galactoside-containing glycoconjugates (1). Galectin-3 is a unique chimera-type galectin containing three distinct structural domains, including a short NH2-terminal domain of 12 amino acids, which regulates its cellular targeting, a C-terminal carbohydrate recognition domain (CRD) with an amino-terminal tandem repeat, the subcellular localization of which determines the wide distribution and numerous biological functions of galectin-3, and a repetitive collagen-like sequence rich in glycine, tyrosine and proline that acts as a substrate for matrix metalloproteinases (2–4). Galectin-3, dependent on its sub-cellular location, has been shown to participate in multiple biological functions, including cell growth, cell adhesion, migration, metastasis, cell-cycle regulation, malignant transformation, cell differentiation and inhibition of apoptosis (5–8). For example, cell-surface galectin-3 is involved in cell adhesion, migration and cell-cell as well as cell-matrix interaction (9). Cytoplasmic galectin-3 mainly acts as an anti-apoptotic factor (10,11), whereas nuclear galectin-3 participates in pre-mRNA splicing (12) and may regulate gene expression at the transcriptional level (11,13).
Studies have demonstrated that galectin-3 is highly expressed during neoplastic progression and metastasis in numerous types of human malignant tumor. In melanoma, upregulation of galectin-3 expression was implicated in tumor growth and metastasis (14). Lotan et al (15) observed that galectin-3 expression in certain primary and metastatic carcinomas was elevated compared with that in adjacent normal mucosa. A study by Inohara et al (16) indicated that galectin-3 is consistently overexpressed in thyroid carcinomas of follicular cell origin, whereas no expression of galectin-3 is found in normal thyroid tissues, nodular goiters and follicular adenomas. Galectin-3 expression in clear-cell renal cell carcinoma with distant metastasis was significantly higher than in those without distant metastasis (6).
Esophageal cancer (EC) is a type of invasive malignant cancer with high mortality due to its early metastasis and post-operative recurrence. In 2014, an estimated 18,170 people were diagnosed with esophageal cancer and 15,450 people succumbed to the disease in the United States (17). Based on the report by the Esophageal Cancer Collaboration (WECC), patient survival decreased with increased tumor invasion, as well as presence of regional lymph node metastases and distant metastases (18). Numerous studies have been performed to explore the mechanisms of tumor progression and metastasis in EC (19–24); however, the exact mechanism underlying the aggressiveness of this cancer type has largely remained elusive. A previous study by our group (19) demonstrated that overexpression of galectin-3 exerts important effects on the biological behavior of Eca109 human EC cells, resulting in enhanced proliferation, migration and invasion, as well as decreased apoptosis. Thus, the present study examined the effects of galectin-3 knockdown using small interfering RNA (siRNA) on multiple biological functions, including proliferation, migration, invasion and apoptosis, in Eca109 cells. The present study indicated that siRNA-mediated knockdown of galectin-3 maybe represent a promising targeted therapy approach for EC.
Materials and methods
Cell culture
The Eca109 human esophageal cancer cell line was obtained from the Shandong Academy of Medical Sciences (Jinan, China). All cells were cultured at 37°C in tissue culture flasks (Corning-Costar, Corning, NY, USA) containing Dulbecco's modified Eagle's medium (DMEM)-F12 (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (HyClone; GE Healthcare, Little Chalfont, UK) in a humidified incubator containing 95% air and 5% CO2.
siRNA transfection
The siRNAs (galectin-3 siRNA and non-silencing siRNA) were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The siRNA-Gal-3 sequences were as follows: Gal-3-homo-422 (siGal3-1), 5′-GCCACUGAUUGUGCCUUAUTT-3′; Gal-3-homo-746 (siGal3-2), 5′-GUACAAUCAUCGGGUUAAATT-3′. As the non-silencing siRNA (siRNA-control), a scrambled sequence (5′-UUCUUCGAACGUGUCACGUTT-3′), which does not target any known mammalian gene, was used as a negative control. The transfection efficiency of siRNA was evaluated by using negative control FAM-siRNA (Shanghai GenePharma Co., Ltd.), which emitted faint green fluorescence. The lyophilized siRNAs were dissolved in diethylpyrocarbonate-treated water according to the manufacturer's instructions. Transfection of Eca109 cells with galectin-3 siRNA was performed using HiperFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Eca109 cells (10×104 cells in 2 ml complete culture medium) were seeded into a six-well plate at 24 h prior to transfection. The cells treated with siRNA were harvested within 48–72 h after transfection for further experiments.
Western blot analysis
Four groups of Eca109 cells (untreated, siRNA-control, siGal3-1 and siGal3-2) were harvested within 72 h after transfection. Total protein was extracted using radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. The protein concentration was determined using a Bicinchoninic Acid Protein Assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). 10% SDS-PAGE was used to isolate the total protein extracts (30 µg/well), which were transferred onto a polyvinylidene membrane (Millipore, Billerica, MA, USA). The band representing galectin-3 was determined using Protein Marker (Beyotime Institute of Biotechnology, Haimen, China). The membrane was subsequently blocked with 5% non-fat milk at 37°C for 1 h. Galectin-3 was immunodetected with monoclonal anti-galectin-3 antibody (cat. no. ab2785; 1:1,000; Abcam, Cambridge, UK) containing a ~30 kDa recombinant fragment of human galectin-3 full-length protein, overnight at 4°C. After washing with Tris-buffered saline containing Tween-20 three times, the membranes were incubated with sheep anti-rat immunoglobulin G conjugated to peroxidase (1:500; Sigma-Aldrich). Based on the manufacturer's instructions, immunosignals were visualized using the Protein Detector 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium western blotting kit (Beyotime Institute of Biotechnology). A FluorChem E instrument (Cell Biosciences, Santa Clara, CA, USA) was utilized to capture enhanced chemiluminescence images. Quantification of the galectin-3 band intensity was performed using ImageJ software (version 1.62; National Institutes of Health, Bethesda, MD, USA). Another membrane that was incubated with anti-β-actin (1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and green fluorescence protein-conjugated (1:1,000; CoWin Biotech Co., Ltd., Beijing, China) antibodies was prepared in an identical manner.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Four groups of Eca109 cells (untreated, siRNA-control, siGal3-1 and siGal3-2) were harvested 48 h post-transfection. Total RNA was separated using RNA-solv reagent (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer's instructions. After detection of the total RNA concentration by spectrophotometry at a wavelength of 260 nm (SpectraMax 190; Molecular Devices, Sunnyvale, CA, USA), total RNA incubated at 65°C for 5 min was used to synthesize cDNA using a Rever Tra Ace® qPCR-RT kit (Toyobo, Osaka, Japan) in a 10 µl reaction system containing 2 µl 5X RT buffer, 0.5 µl Enzyme Mix, 0.5 µl Primer Mix, 2 µl total RNA and 5 µl nuclease-free water, with the following reaction conditions: 5 min at 95°C and 15 min at 37°C. An ABI ViiA7 Dx instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to conduct PCR reactions with a 10 µl reaction mixture containing 5 µl SYBR® Green (Toyobo), 2 µl nuclease-free water, 1 µl forward primer, 1 µl reverse primer and 1 µl cDNA. The thermocycling conditions were as follows: 35 cycles of denaturation at 95°C for 15 sec, annealing at 61°C for 15 sec and extension at 72°C for 45 sec. The human galectin-3 mRNA sequences in GenBank (accession no. NM_02306) were used to design the gene-specific primers (BioSune Co., Ltd., Jinan, China) as follows: Forward, 5′-GGTGAAGCCCAATGCAAACA-3′ and reverse, 5′-TGCAACCTTGAAGTGGTCAG-3′. Amplification of human β-actin mRNA was performed as a reference to normalize sample loading using the following primers (Boshang Co., Ltd.): Forward, 5′-TGGCACCCAGCACAATGAA-3′ and reverse, 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. PCR results were quantified using the ΔCt method according to the following formula: Expression ratio = 2−ΔCt, where ΔCt = ΔCt target gene − ΔCt endogenous control gene (β-actin) (25).
Cell proliferation assay
Eca-109 cells (3×103 cells/well) transfected with either control or galectin-3 siRNA were dispensed into 96-well round-bottomed microtiter plates in a total volume of 100 µl/well and incubated for 24–96 h at 37°C in 5% CO2. Subsequently, 10 µl Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) solution in 100 µl complete DMEM was added to each well, followed by incubation for 4 h. The absorbance was measured at 450 nm using a spectrophotometer (Spectramax 190; Molecular Devices).
Transwell migration and invasion assay
Transwell polycarbonate filters (8.0-µm pore size; Costar®; Sigma-Aldrich) in 24-well plate were used to assess the migration and invasion of Eca109 cells. For the migration assay, transfected or untransfected Eca109 cells at 4×104/200 µl in serum-free DMEM were seeded into the upper chambers, while 600 µl DMEM containing 2.5% FBS was added to the lower chamber. After incubation at 37°C in 5% CO2 for 48 h, cells on the upper surface of the filter were removed using a cotton swab. The cells migrated to the lower side of the filter were fixed in methanol for 15 min at room temperature, stained with 0.25% crystal violet (Beyotime Institute of Biotechnology) for 10 min at room temperature and washed with phosphate-buffered saline following staining. Images of the cells migrated to the lower side of the filter were captured (magnification, ×40) and counted (magnification, ×100) under an inverted fluorescence microscope by an independent investigator.
For the invasion assay, transfected or untransfected Eca109 cells at 8×104/200 µl in serum-free DMEM were seeded into the upper Transwell chambers, which were pre-coated with 100 µl 1:5 diluted Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), while 600 µl DMEM containing 10% FBS was added to the lower chambers. Following incubation at 37°C in 5% CO2 for 48 h, the cells and Matrigel on the upper surface of the membrane were removed using a cotton swab. The number of invaded cells on the lower side of the membrane was determined in an identical manner to that stated for the migration assay.
Cell apoptosis assay
Apoptotic cells were analyzed by using an Annexin V-Phycoerythrin (PE) Apoptosis Detection kit (BD Biosciences) and flow cytometric analysis (FACSAria II; BD Biosciences). Untransfected Eca109 cells or Eca109 cells transfected with siRNA or control-siRNA for 48 h were re-suspended in 1X working solution at 1×106 cells/ml. Following addition of 5 µl Annexin V-PE and 5 µl 7-amino-actinomycin stain (7-AAD; eBioscience, San Diego, CA, USA), cells were incubated at 25°C in the dark for 15 min. Subsequently, 400 µl binding buffer was added and cells were analyzed by flow cytometry within 1 h of staining.
Statistical analysis
All statistical analyses were performed using SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). Values are expressed as the mean ± standard deviation. Unpaired Student's t-tests were utilized for comparisons between values. P<0.05 was considered to indicate a statistically significant difference.
Results
Confirmation of galectin-3 silencing
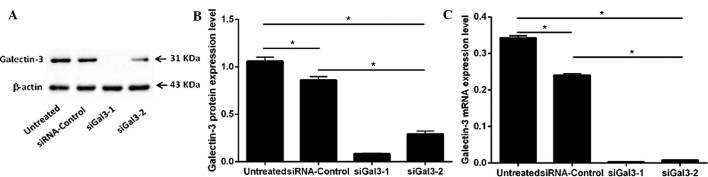
The protein expression of galectin-3 in the four groups of Eca109 cells (siGal3-1, siGal3-2, siRNA-control and untreated) was examined by western blot analysis (Fig. 1A and B). Variant expression was detected in the 31-kDa band representing galectin-3. The expression of galectin-3 in the two siGal3-transfected groups was significantly decreased compared with that in the siRNA-control and untreated groups (siGal3-1, P=0.001 and P=0.001; siGal3-2, P=0.002 and P=0.003, respectively). The galectin-3 suppression rates of siGal3-1 and −2 reached 90.2 and 67.9%, respectively.
Figure 1.
Protein and mRNA expression levels of galectin-3 in Eca109 human esophageal cancer cells. (A and B) Protein expression of galectin-3 in siGal3-1, siGal3-2, siRNA-Control and untreated cells was determined by western blot analysis 72 h post-transfection. A 31-kDa band representing the location of galectin-3 was visualized in four groups. Galectin-3 expression was significantly reduced in the siGal-3 groups compared with that in the in siRNA-Control and untreated groups (P<0.05). (C) mRNA expression of galectin-3 in the four groups 48 h post-transfection. Compared with the siRNA-Control and untreated groups, galectin-3 was significantly reduced in the siGal-3 groups (P<0.001). Values are expressed as the mean ± standard deviation. *P≤0.05. siRNA, small interfering RNA; Gal, galectin.
Furthermore, the mRNA levels of galectin-3 in the four groups (siGal3-1, siGal3-2, siRNA-control and untreated) were determined by RT-qPCR (Fig. 1C). Compared with the siRNA-control cells or untreated cells, galectin-3 mRNA was significantly reduced in the siGal3-1- and siGal3-2-treated cells (P<0.05) with galectin-3 suppression rates of 97.6 and 95.2%, respectively.
While a significant decrease in galectin-3 expression in the siRNA-control group compared with that in the untreated group was observed at the protein and mRNA level (P<0.05), this difference was minor compared to the knockdown effect of siGal3-1 and −2.
The results suggested that Gal-3-homo-422 was more effective at galectin-3 silencing than Gal-3-homo-746. Therefore, Gal-3-homo-422 was used for gene knockdown in the subsequent functional experiments.
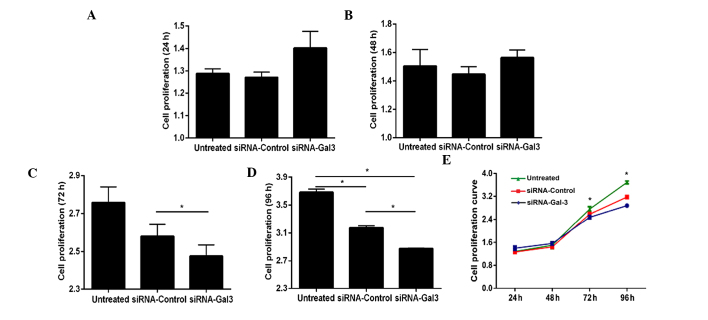
Galectin-3 silencing inhibits EC-cell proliferation
The effects of galectin-3 silencing on the proliferation of Eca109 cells were assessed using a CCK-8 assay (Fig. 2). While transfection with siRNA for 24 and 48 h did not significantly affect the proliferation of Eca109 cells (P>0.05) (Fig. 2A, B and E), the proliferative rate was significantly decreased following 72 h of transfection (P<0.001), and it was further decreased at 96 h after transfection (P=0.004 and P=0.001 vs. siRNA-control and untreated groups, respectively) (Fig. 2C–E).
Figure 2.
Proliferation capacity of siRNA-Gal-3- and siRNA-control-transfected as well as untreated Eca109 cells. At (A) 24 h and (B) 48 h after transfection with siRNA, no significant differences were observed among three groups (P>0.05). (C) At 72 h after transfection, the proliferation of Eca109 cells in the siRNA-Gal-3 group was lower than that in the siRNA-control group. (D) At 96 h after transfection, the siRNA-Gal-3 treated group showed a significantly decreased proliferation compared with that in the siRNA-control and untreated groups (P=0.004 and P=0.001, respectively). (E) Cell proliferation curve of Eca109 cells in the three groups. Values are expressed as the mean ± standard deviation. *P≤0.05. siRNA, small interfering RNA; Gal, galectin.
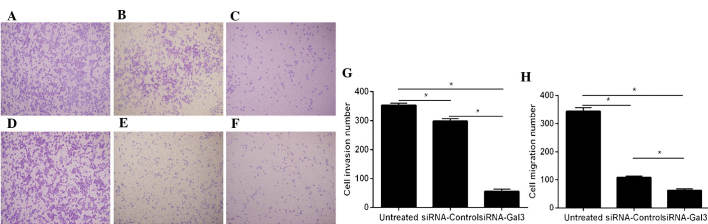
Galectin-3 silencing inhibits the migration and invasion of EC cells
The migratory and invasive capacities of Eca109 cells were detected using Transwell assays (Fig. 3). A significantly decrease of cell invasion was observed in the siRNA-Gal-3 group (55.3±7.37) compared with that in the siRNA-control (298.3±8.33; P=0.001) and untreated (354±7; P<0.001) groups.
Figure 3.
Gal-3 knockdown inhibits the invasive and migratory abilities of Eca109 cells. Microscopy images of crystal violet-stained cells in the siRNA-Gal-3, siRNA-Control and untreated groups, respectively, on the lower side of Transwell membranes (magnification, ×4.2) in (A–C) the invasion and (D–F) the migration assay. (G) Quantification of invaded cells revealed significantly decreased invasion in the siRNA-Gal-3 group compared with that in the siRNA- Control and untreated groups (P=0.002 and P=0.001, respectively). (H) Quantification of migrated cells revealed reduced migration of siRNA-Gal-3-treated Eca109 cells compared with that in the siRNA-Control and untreated groups (P=0.001 and P<0.001, respectively). Values are expressed as the mean ± standard deviation. *P≤0.05. siRNA, small interfering RNA; Gal, galectin.
Furthermore, the migration of Eca109 cells in the siRNA-Gal-3 group (62.3±5.51) was obviously decreased compared with that in the siRNA-control (109±4.59; P=0.002) and untreated (344.3±13.01, P=0.001) groups.
In the invasion as well as in the migration assay, significant differences were present between the siRNA-control and untreated groups (P<0.05); however, these effects were minor compared with those of siRNA-Gal-3.
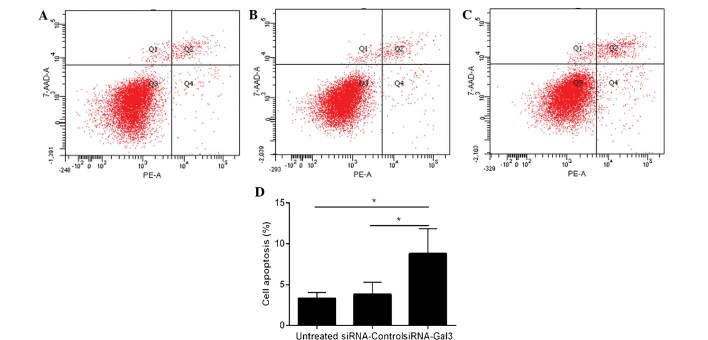
Galectin-3 silencing enhances apoptosis of EC cells
Flow cytometry was utilized to determine the apoptotic rate of Eca109 cells (Fig. 4). The cells treated with siRNA-Gal-3 (8.8±3.08%) exhibited a higher apoptotic rate than those in the siRNA-control (3.8±1.51%; P=0.031) and untreated (3.33±0.72%; P=0.047) groups.
Figure 4.
Gal-3 knockdown enhances apoptosis of Eca109 cells. Flow cytometric analysis of apoptotic cells in the (A) siRNA-Gal-3, (B) siRNA-Control and (C) untreated groups. (D) The siRNA-Gal-3 group exhibited a significantly higher apoptotic rate than the siRNA-Control and the untreated group (P=0.031 and P=0.047, respectively). Values are expressed as the mean ± standard deviation. *P≤0.05. siRNA, small interfering RNA; Gal, galectin; 7-AAD, 7-amino-actinomycin; PE, phycoerythrin.
Discussion
Esophageal cancer is an aggressive digestive tract malignancy and patients with increased tumor invasion or metastasis generally exhibit reduced survival rates (18). At present, the process of discovering effective treatment methods for EC is challenging due to the biological behavior of EC cells, including enhanced migration and invasion as well as evasion of apoptosis. Studies have demonstrated that galectin-3 is highly expressed in various tumor types and linked with tumor progression and metastasis (6,14–16). A previous study by our group revealed that overexpression of galectin-3 enhanced the aggressive behavior of human esophageal Eca109 cells, including increased proliferation, migration and invasion, as well as reduced apoptosis (19). The present study successfully performed siRNA-mediated silencing of galectin-3 in order to assess its effects on multiple biological functions of the Eca109 human EC cell line.
The migration and invasion of cancer cells has been reported to be linked with adhesion and proteolysis of extracellular matrix (ECM) components, which can be regulated by integrins. Integrins are a family of 24 ubiquitous heterodimeric transmembrane receptors that are expressed in multiple cell types and are widely involved as key regulators of inflammation, apoptosis and metastasis (26). In numerous malignancies, overexpressed galectin-3 has been shown to mediate cell migration and invasion by binding to integrins or regulating their expression (27). The present study performed knockdown of galectin-3 using siRNA-Gal-3, which was shown to significantly reduce the migration and invasion abilities of Eca109 cells compared with those of untreated cells or those transfected with siRNA-control. This result indicated a possible link between galectin-3 expression and cell motility, which was consistent with studies on other malignant tumor types. Kobayashi et al (28) showed that transient silencing of galectin-3 reduced the migration and invasion of three human pancreatic cancer cell lines. Honjo et al (29) revealed that in human breast carcinoma cells, the suppression of galectin-3 expression blocked tumorigenicity and led to a significant inhibition of tumor growth in nude mice. Bresalier et al (30) discovered that a decrease of galectin-3 expression was linked with a significant reduction of liver colonization and spontaneous metastasis of human colon cancer cells, whereas overexpression of galectin-3 led to an enhanced metastastic potential. It has been reported that inhibition of galectin-3 expression reduced the motility of human colon cancer and glioblastoma cells (31,32). Galectin-3 was shown to regulate metastasis of breast and prostate cancer cells by binding to cell adhesion-associated molecules and suppressing cell-cell and cell-ECM interactions in vitro as well as in vivo (33). Upregulation of galectin-3 expression in lung carcinoma cells was reported to promote cell motility and invasion (34). However, a study by Junking et al (35) indicated that galectin-3 silencing enhanced cell motility; downregulation of galectin-3 expression was associated with lymphatic invasion, metastasis potential and poor differentiation of cholangiocarcinoma (CCA). Of note, several studies indicated that galectin-3 was overexpressed or downregulated in the same type of malignant tumor. For example, in human colorectal carcinoma, upregulation of galectin-3 expression was observed, which was linked with cancer progression and metastasis (36,37), and Zaia Povegliano et al (38) discovered that galectin-3 was highest in the most advanced stage of colorectal cancer. By contrast, downregulation of galectin-3 expression has also been observed in colorectal carcinoma (39). These contradictory findings indicated that differential galectin-3 expression leading to increased or decreased cell invasion and metastasis may be associated with the cancer type or stage and their associated properties (35); however, the exact mechanisms require further exploration. In the present study, a significant difference was also observed between siRNA-control and untreated groups in terms of galectin-3 expression as well as cell migration and invasion, probably owing to the influences of transfection reagent and negative control siRNA on Eca109 cells.
The present study also analyzed the effects of galectin-3 silencing on Eca109-cell proliferation. At 72 and 96 h after transfection, decreased cell proliferation in the siRNA-Gal-3 treated group was observed compared with that in the siRNA-control and untreated groups, whereas at 24 and 48 h after transfection, no significant difference in the proliferative rate was found among the three groups. Galectin-3 was therefore shown to be associated with cell proliferation in Eca109 cells, and inhibiting tumor cell proliferation by downregulation of galectin-3 expression may contribute to the interference with tumor progression.
Anti-apoptotic activity, the most distinct function of galectin-3, has been shown to rely on its sub-cellular localization with cytoplasmic galectin-3 acting as a main anti-apoptotic factor. Galectin-3 has been demonstrated to exert critical roles in the evasion of apoptosis by interacting with the anti-apoptotic nuclear factor (NF)-κB signaling pathway and inducing the activation of anti-apoptotic transcription factors (40). In turn, NF-κB was shown to be involved in the regulation of galectin-3 (41) and the expression of genes associated with the immune response, inflammatory processes and apoptosis (42,43). Furthermore, synexin was observed to mediate the translocation of galectin-3 to perinuclear mitochondrial membranes to suppress changes in the mitochondrial membrane potential and to thereby inhibit apoptosis (10). In the present study, Annexin V/7-AAD double-staining was utilized to assess the effects of galectin-3 on the apoptotic rate of EC cells. Silencing of galectin-3 was shown to promote apoptosis of Eca109 cells, which was concordant with the results of previous studies and confirmed the anti-apoptotic activity of galectin-3 in tumor cells. Shi et al (37) revealed that silencing of galectin-3 with siRNA contributed to the stimulation of apoptosis in human colorectal cancer cells. Furthermore, accumulating evidence implied that galectin-3-mediated resistance to apoptosis is linked with resistance to chemotherapy. Lin et al (44) demonstrated that siRNA-mediated galectin-3 knockdown induced apoptosis, while increased expression of galectin-3 enhanced chemoresistance to doxorubicin with reduced doxo-rubicin-induced apoptosis and expression of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). In bladder carcinoma cells, upregulation of galectin-3 expression was also shown to inhibit TRAIL-induced apoptosis (45). In addition, Choi et al (46) reported that breast carcinoma cells with galectin-3 overexpression exhibited enhanced resistance to N-(4-hydroxyphenyl) retinamide-induced apoptosis. Tepsiri et al (47) analyzed five human intrahepatic CCA cell lines, revealing that the KKU-100 CCA cell line, in which galectin-3 was most highly expressed, was resistant to all chemotherapeutic drugs tested, whereas the KKU-M055 cell line, which did not express galectin-3, was most sensitive to the chemotherapeutic drugs. These studies indicated that targeting of galectin-3 may represent a potential clinical application for the treatment of malignant tumors. Although the molecular mechanisms by which galectin-3 affects biological functions of cancer cells remain to be fully elucidated, galectin-3 represents a promising target for the treatment of EC, which warrants further development.
In conclusion, the present study revealed that siRNA-mediated galecin-3 silencing in the Eca109 EC cell line resulted in the inhibition of cell proliferation, migration and invasion, as well as an increase of cell apoptosis. While the exact mechanism of the effects of galectin-3 on human EC cells and other cancer types require further investigation to, galectin-3 silencing was indicated to be a promising treatment strategy for EC.
Acknowledgments
The present study was supported by a grant from the Natural Science Foundation of Shandong Province (no. ZR2012HM095).
References
- 1.Dumic J, Dabelic S, Flögel M. Galectin-3: An open-ended story. Biochim Biophys Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K, et al. Galectins: A family of animal β-galactoside-binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 3.Ochieng J, Fridman R, Nangia-Makker P, Kleiner DE, Liotta LA, Stetler-Stevenson WG, Raz A. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 1994;33:14109–14114. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- 4.Gong HC, Honjo Y, Nangia-Makker P, Hogan V, Mazurak N, Bresalier RS, Raz A. The NH2 terminus of galectin-3 governs cellular compartmentalization and functions in cancer cells. Cancer Res. 1999;59:6239–6245. [PubMed] [Google Scholar]
- 5.Zhang HY, Jin L, Stilling GA, Ruebel KH, Coonse K, Tanizaki Y, Raz A, Lloyd RV. RUNX1 and RUNX2 upregulate Galectin-3 expression in human pituitary tumors. Endocrine. 2009;35:101–111. doi: 10.1007/s12020-008-9129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakaki M, Fukumori T, Fukawa T, Elsamman E, Shiirevnyamba A, Nakatsuji H, Kanayama HO. Clinical significance of Galectin-3 in clear cell renal cell carcinoma. J Med Invest. 2010;57:152–157. doi: 10.2152/jmi.57.152. [DOI] [PubMed] [Google Scholar]
- 7.Jia W, Kidoya H, Yamakawa D, Naito H, Takakura N. Galectin-3 accelerates M2 macrophage infiltration and angio-genesis in tumors. Am J Pathol. 2013;182:1821–1831. doi: 10.1016/j.ajpath.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Wu SW, Yu L, Zhou L, Cheng ZN, Tao YS. Expression of Gal-3 and CD82/KAI1 proteins in non-small cell lung cancer and their clinical significance. Chinese Journal of Oncology. 2013;35:124–128. doi: 10.3760/cma.j.issn.0253-3766.2013.02.010. In Chinese. [DOI] [PubMed] [Google Scholar]
- 9.Ochieng J, Furtak V, Lukyanov P. Extracellular functions of galectin-3. Glycoconj J. 2004;19:527–535. doi: 10.1023/B:GLYC.0000014082.99675.2f. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, Finley RL, Jr, Raz A, Kim HR. Galectin-3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. A role for synexin in galectin-3 translocation. J Biol Chem. 2002;277:15819–15827. doi: 10.1074/jbc.M200154200. [DOI] [PubMed] [Google Scholar]
- 11.Takenaka Y, Fukumori T, Yoshii T, Oka N, Inohara H, Kim HR, Bresalier RS, Raz A. Nuclear export of phosphorylated galectin-3 regulates its antiapoptotic activity in response to chemotherapeutic drugs. Mol Cell Biol. 2004;24:4395–4406. doi: 10.1128/MCB.24.10.4395-4406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson RJ, Wang W, Wang JL. Understanding the biochemical activities of galectin-1 and galectin-3 in the nucleus. Glycoconj J. 2004;19:499–506. doi: 10.1023/B:GLYC.0000014079.87862.c7. [DOI] [PubMed] [Google Scholar]
- 13.Shimura T, Takenaka Y, Tsutsumi S, Hogan V, Kikuchi A, Raz A. Galectin-3, a novel binding partner of beta-catenin. Cancer Res. 2004;64:6363–6367. doi: 10.1158/0008-5472.CAN-04-1816. [DOI] [PubMed] [Google Scholar]
- 14.Braeuer RR, Shoshan E, Kamiya T, Bar-Eli M. The sweet and bitter sides of galectins in melanoma progression. Pigment Cell Melanoma Res. 2012;25:592–601. doi: 10.1111/j.1755-148X.2012.01026.x. [DOI] [PubMed] [Google Scholar]
- 15.Lotan R, Ito H, Yasui W, Yokozaki H, Lotan D, Tahara E. Expression of a 31-kDa lactose-binding lectin in normal human gastric mucosa and in primary and metastatic gastric carcinomas. Int J Cancer. 1994;56:474–480. doi: 10.1002/ijc.2910560404. [DOI] [PubMed] [Google Scholar]
- 16.Inohara H, Honjo Y, Yoshii T, Akahani S, Yoshida J, Hattori K, Okamoto S, Sawada T, Raz A, Kubo T. Expression of galectin-3 in fine-needle aspirates as a diagnostic marker differentiating benign from malignant thyroid neoplasms. Cancer. 1999;85:2475–2484. doi: 10.1002/(SICI)1097-0142(19990601)85:11<2475::AID-CNCR25>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 18.Rice TW, Rusch VW, Apperson-Hansen C, Allen MS, Chen LQ, Hunter JG, Kesler KA, Law S, Lerut TE, Reed CE, et al. Worldwide esophageal cancer collaboration. Dis Esophagus. 2009;22:1–8. doi: 10.1111/j.1442-2050.2008.00901.x. [DOI] [PubMed] [Google Scholar]
- 19.Liang N, Song X, Xie J, Xu D, Liu F, Yu X, Tian Y, Liu Z, Qiao L, Zhang J. Effect of galectin-3 on the behavior of Eca-109 human esophageal cancer cells. Mol Med Rep. 2015;11:896–902. doi: 10.3892/mmr.2014.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma M, Liu JF. The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2015 doi: 10.1111/jgh.13180. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Qin X, Li Y, Zhang X, Niu R, Zhang H, Cui A, An W, Wang X. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. Am J Transl Res. 2015;7:1390–1403. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Yue H, Li C, Chen H, Yuan Y. Protein arginine methyltransferase 1 promoted the growth and migration of cancer cells in esophageal squamous cell carcinoma. Tumour Biol. 2015 doi: 10.1007/s13277-015-4098-3. [DOI] [PubMed] [Google Scholar]
- 23.Pei Y, Wang P, Liu H, He F, Ming L. FOXQ1 promotes esophageal cancer proliferation and metastasis by negatively modulating CDH1. Biomed Pharmacother. 2015;74:89–94. doi: 10.1016/j.biopha.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Zhu SC, Li SG, Zhao Y, Xu JR, Song CY. TKTL1 promotes cell proliferation and metastasis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2015;74:71–76. doi: 10.1016/j.biopha.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 27.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T, Shimura T, Yajima T, Kubo N, Araki K, Tsutsumi S, Suzuki H, Kuwano H, Raz A. Transient gene silencing of galectin-3 suppresses pancreatic cancer cell migration and invasion through degradation of β-catenin. Int J Cancer. 2011;129:2775–2786. doi: 10.1002/ijc.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honjo Y, Nangia-Makker P, Inohara H, Raz A. Down-regulation of Galectin-3 suppresses tumorigenicity of human breast carcinoma cells. Clin Cancer Res. 2001;7:661–668. [PubMed] [Google Scholar]
- 30.Bresalier RS, Mazurek N, Sternberg LR, Byrd JC, Yunker CK, Nangia-Makker P, Raz A. Metastasis of human colon cancer is altered by modifying expression of the beta-galactoside-binding protein galectin-3. Gastroenterology. 1998;115:287–296. doi: 10.1016/S0016-5085(98)70195-7. [DOI] [PubMed] [Google Scholar]
- 31.Hittelet A, Camby I, Nagy N, Legendre H, Bronckart Y, Decaestecker C, Kaltner H, Nifant'ev NE, Bovin NV, Pector JC, et al. Binding sites for lewis antigens are expressed by human colon cancer cells and negatively affect their migration. Lab Invest. 2003;83:777–787. doi: 10.1097/01.LAB.0000073129.62433.39. [DOI] [PubMed] [Google Scholar]
- 32.Debray C, Vereecken P, Belot N, Teillard P, Brion JP, Pandolfo M, Pochet R. Multifaceted role of galectin-3 on human glioblastoma cell motility. Biochem Biophys Res Commun. 2004;325:1393–1398. doi: 10.1016/j.bbrc.2004.10.181. [DOI] [PubMed] [Google Scholar]
- 33.Glinsky VV, Glinsky GV, Glinskii OV, Huxley VH, Turk JR, Mossine VV, Deutscher SL, Pienta KJ, Quinn TP. Intravascular metastatic cancer cell homotypic aggregation at the sites of primary attachment to the endothelium. Cancer Res. 2003;63:3805–3811. [PubMed] [Google Scholar]
- 34.O'Driscoll L, Linehan R, Liang YH, Joyce H, Oglesby I, Clynes M. Galectin-3 expression alters adhesion, motility and invasion in a lung cell line (DLKP), in vitro. Anticancer Res. 2002;22:3117–3125. [PubMed] [Google Scholar]
- 35.Junking M, Wongkham C, Sripa B, Sawanyawisuth K, Araki N, Wongkham S. Decreased expression of galectin-3 is associated with metastatic potential of liver fluke-associated cholangiocarcinoma. Eur J Cancer. 2008;44:619–626. doi: 10.1016/j.ejca.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Endo K, Kohnoe S, Tsujita E, Watanabe A, Nakashima H, Baba H, Maehara Y. Galectin-3 expression is a potent prognostic marker in colorectal cancer. Anticancer Res. 2005;25:3117–3121. [PubMed] [Google Scholar]
- 37.Shi Y, He B, Kuchenbecker KM, You L, Xu Z, Mikami I, Yagui-Beltran A, Clement G, Lin YC, Okamoto J, et al. Inhibition of Wnt-2 and galectin-3 synergistically destabilizes beta-catenin and induces apoptosis in human colorectal cancer cells. Int J Cancer. 2007;121:1175–1181. doi: 10.1002/ijc.22848. [DOI] [PubMed] [Google Scholar]
- 38.Zaia Povegliano L, Oshima CT, de Oliveira Lima F, Andrade Scherholz PL, Manoukian Forones N. Immunoexpression of galectin-3 in colorectal cancer and its relationship with survival. J Gastrointest Cancer. 2011;42:217–221. doi: 10.1007/s12029-010-9189-1. [DOI] [PubMed] [Google Scholar]
- 39.Lotz MM, Andrews CW, Jr, Korzelius CA, Lee EC, Steele GD, Jr, Clarke A, Mercurio AM. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci USA. 1993;90:3466–3470. doi: 10.1073/pnas.90.8.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Sakai T, Sano N, Fukui K. Nucling mediates apoptosis by inhibiting expression of galectin-3 through interference with nuclear factor kappaB signaling. Biochem J. 2004;380:31–41. doi: 10.1042/bj20031300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumic J, Lauc G, Flögel M. Expression of galectin-3 in cells exposed to stress-roles of jun and NF-kappaB. Cell Physiol Biochem. 2000;10:149–158. doi: 10.1159/000016345. [DOI] [PubMed] [Google Scholar]
- 42.Baichwal VR, Baeuerle PA. Activate NF-kappaB or die? Curr Biol. 1997;7:R94–R96. doi: 10.1016/S0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 43.Baeuerle PA, Henkel T. Function and activation of NF-kappaB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 44.Lin CI, Whang EE, Abramson MA, Donner DB, Bertagnolli MM, Moore FD, Jr, Ruan DT. Galectin-3 regulates apoptosis and doxorubicin chemoresistance in papillary thyroid cancer cells. Biochem Biophys Res Commun. 2009;379:626–631. doi: 10.1016/j.bbrc.2008.12.153. [DOI] [PubMed] [Google Scholar]
- 45.Oka N, Nakahara S, Takenaka Y, Fukumori T, Hogan V, Kanayama HO, Yanagawa T, Raz A. Galectin-3 inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by activating Akt in human bladder carcinoma cells. Cancer Res. 2005;65:7546–7553. doi: 10.1158/0008-5472.CAN-05-1197. [DOI] [PubMed] [Google Scholar]
- 46.Choi JH, Chun KH, Raz A, Lotan R. Inhibition of N-(4-hydroxyphenyl) retinamide-induced apoptosis in breast cancer cells by galectin-3. Cancer Biol Ther. 2004;3:447–452. doi: 10.4161/cbt.3.5.808. [DOI] [PubMed] [Google Scholar]
- 47.Tepsiri N, Chaturat L, Sripa B, Namwat W, Wongkham S, Bhudhisawasdi V, Tassaneeyakul W. Drug sensitivity and drug resistance profiles of human intrahepatic cholangiocar-cinoma cell lines. World J Gastroenterol. 2005;11:2748–2753. doi: 10.3748/wjg.v11.i18.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]