Abstract
Increasing evidence revealed that miRNAs, the vital regulators of gene expression, are involved in various cellular processes, including cell growth, differentiation, apoptosis and progression. In addition, miRNAs act as oncogenes and/or tumor suppressors. The present study aimed to verify the potential roles of miR148b in human renal cancer cells. miR-148b was found to be downregulated in human renal cancel tissues and human renal cancer cell lines. Functional studies demonstrated that plasmid-mediated overexpression of miR-148b promoted cell proliferation, increased the S-phase population of the cell cycle and enhanced apoptosis in the 786-O and OS-RC-2 renal cancer cell lines, while it did not appear to affect the total number of viable cells according to a Cell Counting Kit-8 assay. Subsequently, a luciferase reporter assay verified that miR148b directly targeted mitogen-activated protein kinase (MAPK) kinase kinase 9 (MAP3K9), an upstream activator of MAPK kinase/c-Jun N-terminal kinase (JNK) signaling, suppressing the protein but not the mRNA levels. Furthermore, western blot analysis indicated that overexpression of miR148b in renal cancer cells inhibited MAPK/JNK signaling by decreasing the expression of phosphorylated (p)JNK. In addition, over-expression of MAP3K9 and pJNK was detected in clinical renal cell carcinoma specimens compared with that in their normal adjacent tissues. The present study therefore suggested that miR-148b exerts an oncogenic function by enhancing the proliferation and apoptosis of renal cancer cells by inhibiting the MAPK/JNK pathway.
Keywords: microRNA-148b, renal cancer, mitogen-activated protein kinase kinase kinase 9, c-Jun N-terminal kinase pathway, proliferation, apoptosis, cell cycle, post-transcriptional regulation
Introduction
Renal cell carcinoma is the most common cause of mortality among adult kidney cancers and its global incidence has been increasing by ~3% per year in Ireland (1). Derived from the renal tubular epithelial cells, clear cell renal cell carcinoma is the most common pathology sub-type of renal cancer (2). The prognosis of patients with renal cell carcinoma remains poor due to limited treatment strategies, and prognostic methods require to be refined (3). The discovery of novel diagnostic and prognostic biomarkers for the renal cell carcinoma is required in order to optimize patient selection for specific treatments.
MicroRNAs (miRNAs/miRs), a class of highly conserved non-coding and single-stranded RNAs, possess the ability to regulate gene expression at the post-transcriptional level by binding primarily to the 3′untranslated region (UTR) of targeted mRNA. It has been frequently reported that the genesis and progression of renal cancer is associated with abnormal miRNA expression (4). Furthermore, miRNAs are potential tumor biomarkers and are associated with various signaling pathways, including Von Hippel-Lindau/hypoxia-inducible factor (5,6), phosphoinositide-3 kinase/Akt/mammalian target of rapamycin (7), Wnt/Frizzled (8), Hippo (9), mitogen-activated protein kinase (MAPK) (10,11) and nuclear factor-κB (12), in renal cancer cells. Among them, MAPKs have important roles in signal transduction and multiple biological processes, including cell proliferation, differentiation and survival. The MAPK family comprises three sub-groups: Extracellular signal-regulated kinase 1/2, stress-activated protein kinase (SAPK)/c-Jun N-terminal kinase (JNK) and p38 MAPK (13). Of note, SAPK/JNK signaling has a dual function by exerting pro- as well as anti-apoptotic effects, depending on cell type, duration of its activation, the nature of the death stimulus and the activity of other signaling pathways (14).
miR-148b was shown to be downregulated in several types of human cancer, including breast cancer (15), lung cancer (16), hepatocellular carcinoma (17), pancreatic cancer (18), gastric cancer (19) and colorectal cancer (20). However, the role of miR-148b in renal cell carcinoma as well as the underlying molecular mechanisms have remained elusive. Therefore, the present study evaluated the expression of miR-148b in renal cancer tissues and investigated its roles in the 786-O and OS-RC-2 renal cancer cell lines. The effects of miR-148b on the proliferation, viability, apoptosis and cell cycle distribution, as well as MAPK signaling in renal cancer cells were assessed. Furthermore, as MAP3K9 acts as an upstream activator of the MAPK kinase (MKK)/JNK signaling pathway (21), a luciferase reporter assay was performed in order to investigate whether MAP3K9 is a functional target of miR-148b in human renal cancer cells. The present study revealed that miR-148b exerts an oncogenic function in human renal cancer cells by enhancing apoptosis and proliferation through targeting the MAPK/JNK signaling pathway via MAP3K9.
Materials and methods
Clinical specimens
A total of twelve pairs of renal clear cell carcinoma and adjacent non-cancerous tissues were collected between November 2013 and April 2014 from 12 patients (age, 68.80±9.7 years; eight men and four women) who were histologically diagnosed with renal clear cell carcinoma and who did not suffer from other severe diseases or kidney disease, and who underwent nephrectomy at the West China Hospital of Sichuan University (Chengdu, China). All specimens were immediately frozen in liquid nitrogen, ground into powder and stored at −80°C until RNA and protein extraction. Prior written informed consent was obtained from the patients with regard to experimentation on their tissue specimens and the patients' privacy rights of human subjects were respected. The protocol of the present study was approved by the ethics committee of the College of Laboratory Medicine, Chongqing Medical University (Chongqing, China).
Cell culture and transfection
The 786-O and OS-RC-2 human renal cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific). The 293T normal renal cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (high glucose; Gibco; Thermo Fisher Scientific) supplemented with 10% FBS. All cell lines were cultured at 37°C in a humidified atmosphere containing 5% CO2.
miR-148b-3p (UCA GUG CAU CAC AGA ACU UUG U) and negative control (NC) mimics (scrambled miRNA control) were synthesized by RiboBio Co. (Guangzhou, China). The miR-148b mimics (50 nM) or NC mimics (50 nM) was transfected into 786-O and OS-RC-2 cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific) according to the manufacturer's instructions.
Bioinformatics analysis and plasmid construction
All plasmids and vectors were supplied by the Department of Laboratory Medicine of the Chongqing Medical University unless otherwise stated. Target genes of miR-148b were predicted using the TargetScan prediction software (http://www.targetscan.org/). The 3′UTR of human MAP3K9 was identified to contain a putative target sites for miR-148b (TGC ACTGA). For dual-luciferase assays, the plasmid containing the wild-type (WT) target sequence, pCDNA3.1-lucif-erase-hMAP3K9-3′UTR-WT, was obtained by cloning the 3′-UTR of human MAP3K9 into the BamHI (Thermo Fisher Scientific) and EcoRI (Thermo Fisher Scientific) sites of the pCDNA3.1-luciferase vector. The MAP3K9 3′UTR was amplified from genomic DNA using polymerase chain reaction (PCR) amplification with the following primers: Forward, 5′-CGC GGA TCC ACA GCC AGC GGA GAT GAGG-3′ and reverse, 5′-CCG GAA TTC CCA AGT CCC AAT GTC CAGG-3′. As a negative control, a sequence with a mutation in the putative miR-148b target site (MUT) was inserted to obtain the plasmid pCDNA3.1-luciferase-hMAP3K9-3′UTR-MUT. The target site sequence (TGC ACTGA) was replaced with the MUT sequence (GAT CAGTG). For this, the mutation method of homologous recombination was employed by using the following primers: Forward, 5′-TGAGATC-CAGCCCTACTTCT GAT CAG TGT AAT GCA CTT-3′ (MUT) and reverse, 5′-CTT CAA AGT GCA TTA CAC TGA TCAG AAG TAG GGC TGGA-3′ (MUT).
Dual luciferase reporter assay
To verify the predicted target genes and the associated signaling pathways of miR-148b, dual luciferase assays were performed in 293T cells. One day prior to transfection, 293T cells were cultured in 24-well plates (Corning-Costar, Corning, NY, USA). As the cells reached 70% confluence, they were co-transfected with 1.25 μl 50 nM miR-148b mimics/NC mimics together with 0.5 μg target gene plasmids (pCDNA3.1-luciferase-hMAP3K9-3′UTR-WT or pCDNA3.1-luciferase-hMAP3K9-3′UTR-MUT) or the following Wnt/transforming growth factor (TGF)-β/MAPK signaling pathway reporter plasmids (Agilent Technologies, Inc., Santa Clara, CA, USA): 0.5 μg TopFlash/0.5 μg (CAGA) 12-LUC/30 ng pFA-Elk1 (activator plasmid) + 0.5 μg pFR-Luc (reporter plasmid) with 2.5 μl Lipofectamine® 3000 as the transfection agent. Normalization was achieved by co-transfection of 20 ng pRL-SV40 plasmid per well. Following 48 h of incubation, Renilla and Firefly luciferase activities were measured using a Dual-Luciferase Reporter Assay kit (cat. no. E1910; Promega Corp., Madison, WI, USA) according to the manufacturer's instructions.
Reverse-transcription quantitative (RT-q)PCR
Following tissue homogenization, total RNA was extracted from transfected or untransfected cells using TRIzol reagent (Thermo Fisher Scientific). For the detection of MAP3K9, RT was performed using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The mRNA levels were detected using the SYBR-Green qRT-PCR kit (Takara Bio Inc., Otsu, Japan) with the following primers: MAP3K9 sense, 5′-TTC CCC AGC AAC TAC GTG AC-3′ and anti-sense, 5′-TCT AAC AAC TGA ATG GGC GGG-3′; miR-148b sense, 5′-CAC GTC TCA GTG CAT CAC AGA-3′ and anti-sense, 5′-GTG CAG GGT CCG AGG T-3′. The levels of MAP3K9 mRNA were normalized to 18S (sense, GTAACCCGTTGAACCCCATT; antisense, CCATCCAATCGGTAGTAGCG; Invitrogen; Thermo Fisher Scientific) and the expression of miR-148b was normalized to U6 (sense, CTCGCTTCGGCAGCACA; antisense, AACGCTTCACGAATTTGCGT; Invitrogen; Thermo Fisher Scientific) using the ΔΔCt method (22). PCR was conducted using a CFX96™ Real-Time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with a PCR mix containing cDNA template primers, SYBR® Premix Ex Taq reagent (Takara Bio Inc.,), and diethylpyrocarbonate-treated water (Takara Bio Inc.). The following thermocycling conditions were used: 94°C for 10 sec followed by 40 cycles of 94°C for 10 sec, 60°C for 20 sec and 72°C for 10 sec.
Western blot analysis
Protein was extracted from 786-O and OS-RC-2 cells using radioimmunoprecipitation assay lysis buffer (Beyotime Biotechnology Company, Haimen, China). Total protein was quantified using NanoDrop 2000 (Thermo Fisher Scientific). Equal amounts (100 μg) of protein from each extract were separated by a 12% SDS-PAGE and then electrotransferred onto a polyvinylidene difluoride membrane for 1 h at 300 mA. After blocking with 5% non-fat milk in Tris-buffered saline containing Tween 20 for 1 h at room temperature, blots were incubated with phosphor (p)-SAPK/JNK (Thr183/Tyr185) rabbit monoclonal antibody (1:1,000 dilution; cat no. 4671; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-MAP3K9 rabbit polyclonal antibody (1:500 dilution; cat no. bs-10424R; Bioss, Shanghai, China) or anti-β-tubulin mouse monoclonal antibody (1:1,000 dilution; cat no. HC101; Transgen, Beijing, China) overnight at 4°C. Following incubation with the corresponding secondary antibody, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Ig)G (1:2,000 dilution; cat. no. SA00001-1; Proteintech, Chicago, IL, USA) or goat anti-rabbit IgG (1:2,000 dilution; cat. no. SA00001-2; Proteintech), respectively, at room temperature for 1 h, the protein bands were visualized using an Immobilon Western Chemiluminescent HRP Substrate kit (cat. no. WBKLS0100; Millipore, Billerica, MA, USA) and a ChemiDoc XRS+ Imaging system (Bio-Rad Laboratories, Inc.). Grey value analysis of protein bands was performed for quantification of the protein levels using ImageJ software version 1 (National Institutes of Health, Bethesda, MD, USA).
Cell counting kit-8 (CCK-8) assay
786-O and OS-RC-2 cells (2×104 cells per well) were seeded onto 96-well plates. After 24 h of incubation, 786-O and OS-RC-2 cells were co-transfected with miR-148b mimics or control mimics. At 48 h after transfection, the proliferation of 786-O and OS-RC-2 cells was determined using the CCK-8 (Beyotime Institute of Biotechnology) with 10 μl CCK-8 stain added per well according to the manufacturer's instructions. The apparatus used for colorimetric analysis was the Bio-Tek Synergy 2 microplate reader (Bio-Tek Instruments, Inc., Winooski, VT, USA), at a wavelength of 450 nm.
5-Ethynyl-2′-deoxyuridine (EdU) assay
The EdU assay, which is based on the determination of DNA replication activity (23), was used to quantify cell proliferation in the present study. 786-O and OS-RC-2 cells (1×105 cells per well) were seeded onto 24-well plates. After 24 h of incubation, 786-O and OS-RC-2 cells were co-transfected with miR-148b mimics or control mimics. At 48 h after transfection, the proliferation of 786-O and OS-RC-2 cells was determined using the EdU DNA Proliferation Detection kit (RiboBio) according to the manufacturer's instructions and DAPI reagent (Sangon Biotech Co., Ltd., Shanghai, China). The nuclei were photographed using fluorescence microscopy (Eclipse TE300; Nikon, Melville, NY, USA).
Cell cycle analysis
786-O and OS-RC-2 cells (5×105 cells per well) were seeded into six-well plates. Following complete attachment, cells were transfected with miR-148b mimics or NC mimics. After 48 h, cells were harvested, washed three times with cold phosphate-buffered saline and fixed in cold 75% ethanol for at least 8 h at 4°C. Cells were then treated with RNase A (Sangon Biotech Co., Ltd.) and stained with propidium iodide (PI; Sangon Biotech Co., Ltd.) followed by flow cytometric analysis using a FACSVantage SE flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Apoptosis assay
The FITC Annexin V Apoptosis Detection kit I (BD Pharmingen, Franklin Lakes, NJ, USA) was used for determination of apoptotic rates. 786-O and OS-RC-2 cells (5×105 cells per well) were seeded into six-well plates. Following complete attachment, cells were transfected with miR-148b mimics (50 nM) or NC mimics (50 nM). After 48 h of incubation, cells were harvested, re-suspended in 1X binding buffer and incubated with 5 μl of fluorescein isothiocyanate-conjugated Annexin V and 5 μl PI for 15 min at 25°C in the dark. Flow cytometric analysis was performed within 1 h.
Statistical analysis
All statistical analyses were performed using Graphpad Prism 5 software (GraphPad Inc., La Jolla, CA, USA). All quantitative values are expressed as the mean ± standard deviation. One-way analysis of variance was performed for comparisons between groups. All other results are representative of three independent experiments. P<0.05 was considered to indicate a statistically significant difference between values.
Results
miR-148b is downregulated in renal cancer tissues and renal cancer cell lines
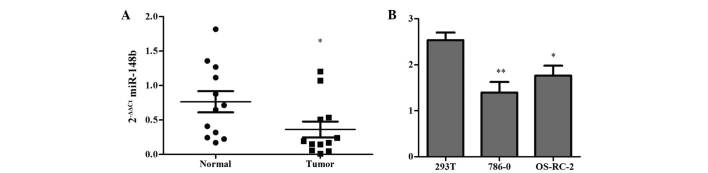
To determine the potential significance of miR-148b in renal cancer, the present study determined the expression levels of miR-148b in renal cancer tissues and cell lines. RT-qPCR analysis revealed that miR-148b was down-regulated in 12 renal cancer tissues compared with that in their paired adjacent non-tumorous tissues (Fig. 1A), as well as in the 786-O and OS-RC-2 renal cancer cell lines compared with that in the 293T normal renal cell line (Fig. 1B). These results suggested that miR148b was downregulated in renal cancer tissues and renal cancer cell lines.
Figure 1.
miR-148b expression in clinical samples and cell lines. (A) miR-148b expression levels in normal vs. tumor tissues from 12 patients with renal clear cell carcinoma. Each data-point represents the result for one patient, horizontal lines indicate the mean value and error bars represent the SD. *P<0.05 vs. normal tissues. (B) miR-148b expression levels in the 293T normal renal cell line and the 786-0 and OS-RC-2 renal cancer cell lines. Values are expressed as the mean ± SD. *P<0.05, **P<0.01 vs. 293T cells. miR, microRNA; SD, standard deviation; Ct, cycle threshold.
Overexpression of miR-148b does not affect the total number of viable renal cancer cells
To explore the effects of miR-148b in renal cell carcinoma, 786-O and OS-RC-2 cells were transfected with miR-148b mimics or negative control mimics and following 48 h, cell viability was assessed using the CCK-8 kit. As shown in Fig. 2A, miR-148b expression was significantly increased at 48 h post-transfection of miR-148b mimics However, no significant effect of miR-148b transfection on the number of viable cells was observed (Fig. 2B). These results suggested that overexpression of miR-148b had no effect on the total number of viable renal cancer cells.
Figure 2.
Overexpression of miR-148b does not significantly affect the total number of viable renal cancer cells. (A) miR-148b expression levels in 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b-3p mimics (miR-148b) or NC mimics (miR-NC). miR-148b expression was normalized to U6 expression. (B) The total number of viable 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or NC mimics was assessed using the Cell Counting Kit-8 assay. Values are expressed as the mean ± standard deviation. NC, negative control; miR, microRNA.
Overexpression of miR-148b promotes proliferation of renal cancer cells
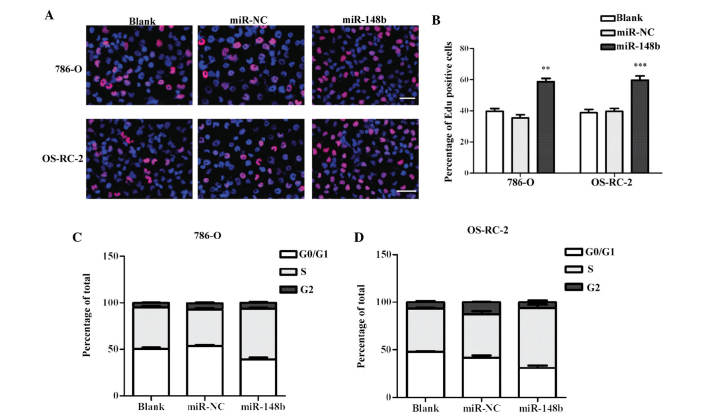
To examine effect of miR-148b on renal cancer cell proliferation, cells were stained with by EdU and DAPI at 48 h post-transfection with miR-148b and evaluated by fluorescence microscopy. As demonstrated in Fig. 3A, overexpression of miR-148b significantly promoted DNA replication activity in 786-O and OS-RC-2 cells. Furthermore, cell cycle analysis illustrated that the cell population in G0/G1 phase cells was decreased, that in S-phase was significantly increased in 786-O and OS-RC-2 cells at 48 h after transfection with miR-148b mimics (Fig. 3B). These results indicated that the DNA replication activity of renal cancer cells was enhanced by miR148b, and that the percentage of cells undergoing DNA synthesis in S-phase was increased. These results led to the conclusion that overexpression of miR-148b promoted the proliferation of 786-O and OS-RC-2 cells.
Figure 3.
Ectopic expression of miR-148b induces proliferation and increases the S-phase population of the cell cycle in renal cancer cell lines. (A) Proliferation of 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or NC mimics was analyzed using the EdU assay with visualization by fluorescent microscopy (magnification, ×400; scale bar, 50 μm; red, EdU; blue, DAPI). Images are representative of three independent experiments. (B) The percentage of EdU-positive cells was quantified. **P<0.01, ***P<0.001 vs blank/miR-NC groups. (C and D) Cell cycle distribution of 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or NC mimics. The blank group was treated with transfection regent and distilled water at the same volume. Values are expressed as the mean ± standard deviation. EdU, 5-ethynyl-2′-deoxyuridine; NC, negative control; miR, microRNA.
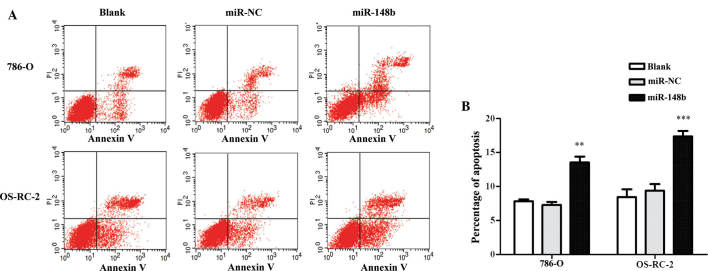
miR148b enhances the apoptotic rate of renal cancer cells
To estimate whether miR148b can affect the apoptosis of renal cancer cells, Annexin V/PI double staining and flow cytometric analysis were performed. It was revealed that miR148b enhanced the apoptotic rate of renal cancer cells (Fig. 4A and B). While miR-148b promoted cell proliferation, it also enhanced the apoptotic rate of 786-O and OS-RC-2, while the total number of viable cells was retained.
Figure 4.
Ectopic expression of miR-148b induces apoptosis in renal cancer cell lines. (A) Analysis of apoptosis of 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or NC mimics by PI and Annexin V double staining and flow cytometric detection. Dot plots shown are representatives of three independent experiments. (B) Early apoptotic rates obtained by quantification of A. Values are expressed as the mean ± standard deviation. **P<0.01; ***P<0.001 vs. blank/NC groups. NC, negative control; miR, microRNA; PI, propidium iodide.
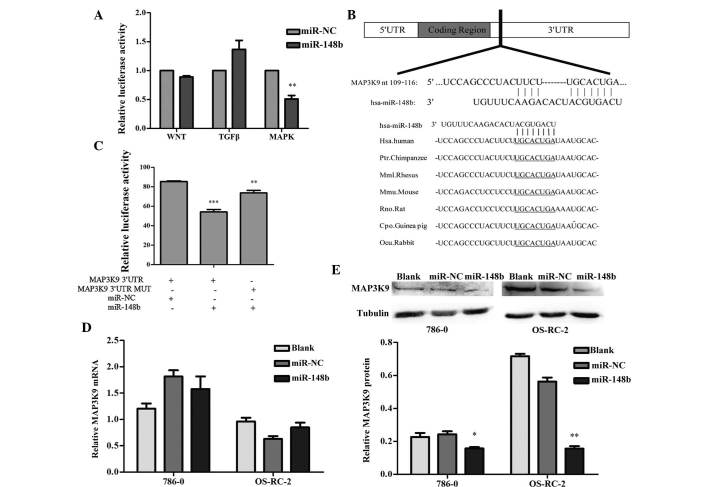
miR-148b inhibits MAPK/JNK signaling by directly targeting MAP3K9 in renal cancer cells
To identify the signaling pathway via which miR-148b exerts its effects in renal cell carcinoma, the present study performed a luciferase reporter assay, which included reporter plasmids for Wnt, TGF-β and MAPK. The results indicated that the MAPK signaling pathway was regulated by miR-148b (Fig. 5A). To identify which gene upstream of the MAPK signaling pathway was regulated by miR-148b, TargetScan, which is a bioinformatics tool for miRNA target screening, was used to. The 3′UTR of human MAP3K9, which is an upstream activator of the MAPK/JNK signaling pathway, was shown to contain a putative target site for miR-148b, which is highly conserved across species (Fig. 5B). To experimentally verify that MAP3K9 is a direct target of miR-148b, a luciferase assay was performed in 293T cells, which revealed a significant decrease in luciferase expression in the group co-transfected with the wild-type 3′UTR-containing reporter plasmid and miR-148b mimics, while the mutation in the putative binding site in the 3′UTR of MAP3K9 prevented the inhibitory effects of miR-148b on luciferase expression (Fig. 5C). This result demonstrated that miR-148b directly bound to the 3′UTR of MAP3K9. Furthermore, western blot analysis revealed that overexpression of miR-148b markedly decreased the protein expression of MAP3K9 in renal cancer cells (Fig. 5D), while the mRNA levels were not affected according to RT-qPCR (Fig. 5E).
Figure 5.
miR-148b reduces MAPK/JNK signaling by targeting MAP3K9 in renal cancer cell lines. (A) Dual luciferase assays were performed in 293T cells at 48 h after transfection with miR-148b/control mimics and signaling-pathway reporter plasmids [TopFlash/(CAGA)12-LUC/pFA-Elk1, pFR-Luc]. **P<0.01 vs. miR-NC group. (B) Schematic representation illustrating that the 3′UTR of MAP3K9 contains a putative target site for miR-148b, which is highly conserved across species. (C) 293T cells were transiently transfected with luciferase reporter plasmids containing the wild-type MAP3K9 3′UTR or the MAP3K9 3′UTR with a mutation in the predicted miR-148b target sequence, together with miR-148b or NC mimics. pRL-SV40 was included as an internal control. Following 48 h of incubation, cells were collected and luciferase activity was assayed. **P<0.01, ***P<0.001 vs. NC- and MAP3K9 3′UTR-transfected group. (D) Reverse-transcription polymerase chain reaction analysis of MAP3K9 mRNA expression in 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or NC mimics, which was quantified using the 2−ΔΔCT method. (E) Western blot analysis of MAP3K9 protein expression in 786-0 and OS-RC-2 cells at 48 h after transfection with miR-148b or control mimics. A western blot representative of three independent experiments is shown. Protein bands were quantified by densitometric grey value analysis. *P<0.05, **P<0.01 vs. blank/NC groups. The blank group was treated with transfection regent and distilled water at the same volume. Values are expressed as the mean ± standard deviation. NC, negative control; miR, microRNA; UTR, untranslated region; MAPK9, mitogen-activated protein kinase kinase kinase 9; hsa, Homo sapiens; MUT mutated.
To further elucidate the mechanism by which miR-148 affects the MAPK pathway, the present study examined the protein expression of p-SAPK/JNK, which is a downstream signaling protein of MAP3K9, at 48 h after transfection with miR-148b mimics. As shown in Fig. 6A, the phosphorylation level of JNK was significantly decreased by miR-148b. These results indicated that miR-148b regulates MAPK/JNK signaling via MAP3K9 post-transcriptionally.
Figure 6.
Expression of MAP3K9 and pJNK in renal cancer cell lines and tissues. (A) Effects of miR-148b overexpression on the expression of MAP3K9 and pJNK in the 786-0 and OS-RC-2 renal cancer cell lines at 48 h post-transfection. **P<0.01; ***P<0.001 vs. blank/NC groups. The blank group was treated with transfection regent and distilled water at the same volume. (B) Expression of MAP3K9 and pJNK in five pairs of renal cancer tissues and their corresponding adjacent non-tumorous tissues. Representative western blots of three independent experiments and quantified protein levels obtained by densitometric grey value analysis are shown. Values are expressed as the mean ± standard deviation. NC, negative control; pJNK, phosphorylated c-Jun N-terminal kinase; miR, microRNA; MAPK9, mitogen-activated protein kinase kinase kinase 9; N, non-tumorous tissue; T, tumorous tissue.
miR-148b is upregulated, while MAP3K9 and pJNK are down- regulated in renal cancer tissues
The present study evaluated the expression of miR148b, MAP3K9 and pJNK in tissues from five patients with renal cancer, revealing that miR148b was upregulated in these renal cancer tissues compared with that in their paired adjacent non-tumorous tissues, while MAP3K9 and pJNK were downregulated (Fig. 6B). These results indicated an inverse correlation between miR-148b and MAP3K9 and a positive correlation between the expression of MAP3K9 and JNK.
Discussion
Tumorigenesis is a complex process which is frequently accompanied with the aberrant expression of miRNAs. miRNAs may serve as diagnostic tumor biomarkers, indicators of tumor prognosis and targets for therapy (24). Downregulation of miR-148b has been identified in various types of human cancer and is therefore considered to be a tumor-suppressive miRNA (15–20). miR-148b was demonstrated to inhibit cell proliferation and invasion, suppress the progression of cancer and to induce apoptosis. However, the potential function of miR-148b in human renal cell carcinoma has remained elusive. The present study detected miR-148b in renal cancer tissues and cell lines and assessed the effects of miR-148b in the 786-O and OS-RC-2 renal cancer cell lines as well as the underlying mechanisms.
The present study revealed that miR-148b was significantly downregulated in renal cell carcinoma; furthermore, overexpression of miR-148b enhanced the proliferation and apoptosis of renal cancer cells, while not affecting the total number of viable cells. It was therefore indicated that following 48 h of transfection, miR-148b mimics enhanced the proliferation and apoptosis of renal cancer cell growth to a similar extent. Furthermore, the present study confirmed that miR-148b exerts effects via inhibiting the MAPK signaling pathway in renal cancer cells with the upstream MAP3K9 being the direct target of miR-148b as indicated by a luciferase reporter assay, RT-qPCR and western blot analysis. MAP3K9, also known as MLK1, is an essential component of the MAPK/JNK signal transduction pathway (25); once activated, MAP3K9 acts as an upstream activator of the MKK/JNK signal transduction cascade through phosphorylating MAP2K4/MKK4 and MAP2K7/MKK7, which in turn activates the JNKs (26). It was previously reported that MLK1/MLK2 deficiency did not impair kidney development and function, and extensive functional redundancy between MLK1/MLK2 and MLK3 was present (27). The present study indicated that miR-148b inhibited JNK activation by targeting MLK1. JNK signaling represents a double-edged sword in the malignant transformation and tumorigenesis of cells, as it exerts either pro- or anti-apoptotic effects (28). The present study demonstrated that miR-148b targeted MAP3K9 and simultaneously increases apoptosis as well as proliferation and associated DNA synthesis, thereby indicating that miR-148b has an oncogenic function by upregulating a variety of cellular processes, while keeping the number of viable cells in a balance. This result may enhance the current understanding of miRNA-dependent regulation of gene expression in regulating proliferation and apoptosis in renal cancer. However, the present study only assessed the effects of miR-148b upregulation in renal cancer cells at 48 h following transfection; further evaluation is required in order to reveal the time-dependent effects of miR-148b on proliferation and apoptosis as well as MAPK/JNK signaling in renal cancer cells. In human renal cancer cells, JNK may have complex roles and crosstalk with other pathways may exist, and the exact mechanisms require further elucidation.
In addition, the present study revealed that detected that miR-148b was overexpressed in renal cancer tissues, while MAP3K9 and pJNK levels were downregulated compared to those in normal adjacent tissues. An inverse correlation between the expression of miR-148b and MAP3K9, and a positive correlation between the expression of MAP3K9 and JNK activation was indicated.
In conclusion, the present study revealed that miR-148b was upregulated in renal cancer cells and upregulates proliferation and apoptosis. miR-148b significantly decreased the G0/G1-phase population and increased the S-phase population and enhanced DNA synthesis in renal cancer cells. miR-148b was revealed to directly target the 3′UTR of MAP3K9, which inhibits the JNK pathway. The present study demonstrated that miR-148b exerts an oncogenic function in human renal cancer and revealed the underlying mechanisms of its stimulatory effects on proliferation and apoptosis by inhibiting the MAPK/JNK pathway. miR-148b may be involved in the progression of renal cancer and represents a potential biomarker and target for the treatment of renal cancer. To assess the prognostic value of miR-148b, future studies will focus on detecting the variety of changes occurring to cellular processes following knockdown of miR-148b expression, or on examining the JNK signaling pathway using an inhibitor to investigate the biological mechanisms underlying renal cancer cell function.
Acknowledgments
The authors sincerely acknowledge the help of their laboratory colleagues and thank all members of the Core Facility of Genetically Engineered Mice, West China Hospital, West China Medical School, Sichuan University (Chengdu, China) for their assistance in this study. This work was supported by the National Basic Research Program of China (grant no. 2011CB944002 to QZ) and the National Natural Science Foundation of China (grant no. 31271563 to QZ).
References
- 1.Falebita OA, Mancini S, Kiely E, Comber H. Rising incidence of renal cell carcinoma in Ireland. Int Urol Nephrol. 2009;41:7–12. doi: 10.1007/s11255-008-9413-0. [DOI] [PubMed] [Google Scholar]
- 2.Störkel S, van den Berg E. Morphological classification of renal cancer. World J Urol. 1995;13:153–158. doi: 10.1007/BF00184870. [DOI] [PubMed] [Google Scholar]
- 3.Bukowski RM, Negrier S, Elson P. Prognostic factors in patients with advanced renal cell carcinoma: Development of an international kidney cancer working group. Clin Cancer Res. 2004;10:S6310–S6314. doi: 10.1158/1078-0432.CCR-050000. [DOI] [PubMed] [Google Scholar]
- 4.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 5.An J, Liu H, Magyar CE, Guo Y, Veena MS, Srivatsan ES, Huang J, Rettig MB. Hyperactivated JNK is a therapeutic target in pVHL-deficient renal cell carcinoma. Cancer Res. 2013;73:1374–1385. doi: 10.1158/0008-5472.CAN-12-2362. [DOI] [PubMed] [Google Scholar]
- 6.Zeng L, Bai M, Mittal AK, El-Jouni W, Zhou J, Cohen DM, Zhou MI, Cohen HT. Candidate tumor suppressor and pVHL partner Jade-1 binds and inhibits AKT in renal cell carcinoma. Cancer Res. 2013;73:5371–5380. doi: 10.1158/0008-5472.CAN-12-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pal SK, He M, Tong T, Wu H, Liu X, Lau C, Wang JH, Warden C, Wu X, Signoretti S, et al. RNA-seq reveals aurora kinase driven-mTOR pathway activation in patients with sarcomatoid metastatic renal cell carcinoma. Mol Cancer Res. 2015;13:130–137. doi: 10.1158/1541-7786.MCR-14-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Von Schulz-Hausmann SA, Schmeel LC, Schmeel FC, Schmidt-Wolf IG. Targeting the Wnt/beta-catenin pathway in renal cell carcinoma. Anticancer Res. 2014;34:4101–4108. [PubMed] [Google Scholar]
- 9.Schütte U, Bisht S, Heukamp LC, Kebschull M, Florin A, Haarmann J, Hoffmann P, Bendas G, Buettner R, Brossart P, Feldmann G. Hippo signaling mediates proliferation, invasiveness, and metastatic potential of clear cell renal cell carcinoma. Transl Oncol. 2014;7:309–321. doi: 10.1016/j.tranon.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang D, Ding Y, Luo WM, Bender S, Qian CN, Kort E, Zhang ZF, VandenBeldt K, Duesbery NS, Resau JH, Teh BT. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res. 2008;68:81–88. doi: 10.1158/0008-5472.CAN-07-5311. [DOI] [PubMed] [Google Scholar]
- 11.Zhan YH, Liu J, Qu XJ, Hou KZ, Wang KF, Liu YP, Wu B. β-Elemene induces apoptosis in human renal-cell carcinoma 786-0 cells through inhibition of MAPK/ERK and PI3K/Akt/mTOR signalling pathways. Asian Pac J Cancer Prev. 2012;13:2739–2744. doi: 10.7314/APJCP.2012.13.6.2739. [DOI] [PubMed] [Google Scholar]
- 12.Du HF, Ou LP, Song XD, Fan YR, Yang X, Tan B, Quan Z, Luo CL, Wu XH. Nuclear factor-kB signaling pathway is involved in phospholipase Cε-regulated proliferation in human renal cell carcinoma cells. Mol Cell Biochem. 2014;389:265–275. doi: 10.1007/s11010-013-1948-4. [DOI] [PubMed] [Google Scholar]
- 13.Munshi A, Ramesh R. Mitogen-activated protein kinases and their role in radiation response. Genes Cancer. 2013;4:401–408. doi: 10.1177/1947601913485414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Lin A. Role of JNK activation in apoptosis: A double-edged sword. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- 15.Cimino D, De Pittà C, Orso F, Zampini M, Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 16.Liu GL, Liu X, Lv XB, Wang XP, Fang XS, Sang Y. miR-148b functions as a tumor suppressor in non-small cell lung cancer by targeting carcinoembryonic antigen (CEA) Int J Clin Exp Med. 2014;7:1990–1999. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Zheng W, Hai J. MicroRNA-148b expression is decreased in hepatocellular carcinoma and associated with prognosis. Med Oncol. 2014;31:984. doi: 10.1007/s12032-014-0984-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, Wang CY. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther. 2013;12:83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 19.Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Y, Xu Y, Wang Z, Chen Y, Yue Z, Gao P, Xing C, Xu H. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int J Cancer. 2012;131:1042–1051. doi: 10.1002/ijc.26485. [DOI] [PubMed] [Google Scholar]
- 21.Fawdar S, Trotter EW, Li Y, Stephenson NL, Hanke F, Marusiak AA, Edwards ZC, Ientile S, Waszkowycz B, Miller CJ, Brognard J. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc Natl Acad Sci USA. 2013;110:12426–12431. doi: 10.1073/pnas.1305207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro J, Marinho-Dias J, Monteiro P, Loureiro J, Baldaque I, Medeiros R, Sousa H. miR-34a and miR-125b expression in HPV infection and cervical cancer development. Biomed Res Int. 2015;2015:304584. doi: 10.1155/2015/304584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu Z, Mao Z, Wang H, Fang Y, Chen T, Wan Q, Wang M, Wang N, Xiao J, Wei H, et al. MiR-181b targets Six2 and inhibits the proliferation of metanephric mesenchymal cells in vitro. Biochem Biophys Res Commun. 2013;440:495–501. doi: 10.1016/j.bbrc.2013.09.059. [DOI] [PubMed] [Google Scholar]
- 24.Grange C, Collino F, Tapparo M, Camussi G. Oncogenic micro-RNAs and renal cell carcinoma. Front Oncol. 2014;4:49. doi: 10.3389/fonc.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durkin JT, Holskin BP, Kopec KK, Reed MS, Spais CM, Steffy BM, Gessner G, Angeles TS, Pohl J, Ator MA, Meyer SL. Phosphoregulation of mixed-lineage kinase 1 activity by multiple phosphorylation in the activation loop. Biochemistry. 2004;43:16348–16355. doi: 10.1021/bi049866y. [DOI] [PubMed] [Google Scholar]
- 26.Stronach B, Lennox AL, Garlena RA. Domain specificity of MAP3K family members, MLK and Tak1, for JNK signaling in drosophila. Genetics. 2014;197:497–513. doi: 10.1534/genetics.113.160937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisson N, Tremblay M, Robinson F, Kaplan DR, Trusko SP, Moss T. Mice lacking both mixed-lineage kinase genes Mlk1 and Mlk2 retain a wild type phenotype. Cell Cycle. 2008;7:909–916. doi: 10.4161/cc.7.7.5610. [DOI] [PubMed] [Google Scholar]
- 28.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]