Cellular function depends on protein homeostasis, also known as proteostasis1. Proteostasis requires the efficient folding of nascent proteins, as well as the maintenance of mature protein folding. Overseeing proteostasis is a quality control process involving the surveillance of protein-folding status, i.e. protein quality analysis, and the subsequent degradation of proteins that do not pass quality analysis. Imbalanced proteostasis resulting from the accumulation of misfolded and unfolded proteins can drive proteotoxicity, cell death and ultimately organ failure. Many diseases, including neurodegenerative, hepatic, endocrine, and cardiovascular disorders, are thought to be associated with, if not caused by the organ failure resulting from impaired protein folding2. For example, in the heart impaired protein folding is associated with hypertrophic and dilated cardiomyopathies, as well as ischemic heart disease3. Therefore, a better appreciation of the mechanisms governing the recognition and degradation of misfolded proteins will improve our understanding of cardiac physiology and pathology.
Cells have evolved an elaborate protein quality control system that is designed to adjust the molecular machinery required to enhance protein-folding capacity, and/or to degrade misfolded proteins via the ubiquitin proteasome system (UPS). If the UPS is sufficient to remove misfolded proteins, proteotoxicity can be averted; however, UPS insufficiency can result in proteotoxicity and eventual organ dysfunction. Accordingly, the UPS aspect of protein quality control is essential for normal cell and organ function4.
Most UPS-mediated protein degradation is an ATP-dependent process that involves ubiquitin-specific E1 activating, E2 conjugating and E3 ubiquitin ligases, the latter of which function in concert with chaperones to identify and ubiquitylate appropriate target proteins, such as misfolded proteins that contribute to impaired heart function (Figure). The resulting ubiquitylated proteins are then transferred to the proteasome, where they are degraded. The E3 ubiquitin ligases confer substrate specificity; therefore, they constitute the rate-limiting step of misfolded protein ubiquitylation5. Since the UPS plays an important role in proteostasis in the heart, and since E3 ubiquitin ligases catalyze the rate-limiting step of the UPS, the E3 ubiquitin ligases, such as the muscle specific ubiquitin ligases, atrogin-1, MuRF1, MuRF3, CHIP, Mdm2, and Trim 32 have attracted interest as potential targets for the design of novel therapies for heart disease6.
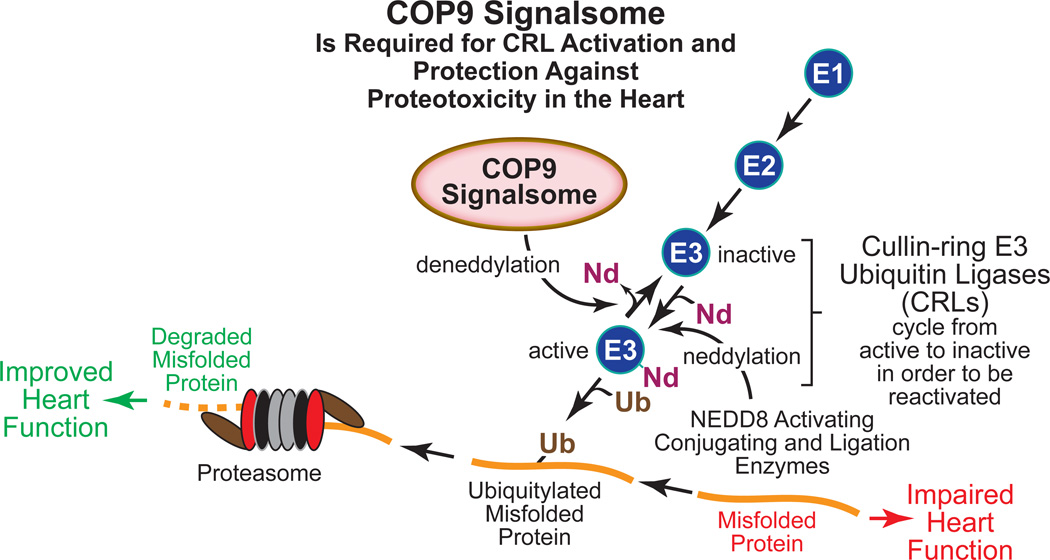
Figure. The COP9 Signalsome Activates Cullin-ring E3 Ubiquitin Ligases and Protects the Heart from Proteotoxicity.
Various cardiac pathologies are associated with accumulation of misfolded protein in cardiac myocytes, which due to proteotoxicity, impairs heart function. The cullin-ring E3 ubiquitin ligases (CRLs) constitute the largest group of the E3 ubiquitin ligases in all cell types; thus, CRLs are responsible for recognizing a large proportion of misfolded toxic proteins and targeting them for proteasome-mediated degradation by ubiquitylation. This removal of misfolded toxic proteins can improve heart function. CRL activation requires neddylation, i.e. covalent addition of the ubiquitin-like 81 amino acid protein, NEDD8 (Nd) by NEDD8-specific activating, conjugation and ligation enzymes, much like those used by the ubiquitylation machinery. But in order to regain the ability to bind a new substrate for ubiquitylation, CRLs must undergo COP9 signalsome (CSN)-mediated deneddylation. The study by Su et al in this issue of Circulation Research has shown that in a mouse model of proteinopathy in the heart, the CSN supports increased ubiquitylation of misfolded proteins and improves cardiac function.
The E3 ubiquitin ligases, of which there are hundreds, fall into two main categories, depending on the existence of either a HECT (homologous to E6-AP carboxy-terminus) or a RING (really interesting new gene) finger domains in the catalytic sites. In the human genome there are about 40 genes encoding HECT domain E3 ubiquitin ligases and nearly 400 genes encoding RING finger domain E3 ubiquitin ligases. Thus, the RING finger domain proteins constitute the vast majority of E3 ubiquitin ligases6. Amongst the RING finger domain E3 ubiquitin ligases are the cullin-RING family of ubiquitin ligases (CRLs), which are by far the most abundant gene products in the RING finger protein family. In fact, CRLs are responsible for the ubiquitylation of as much as 20% of proteins that are targeted for proteasome-mediated degradation7. Therefore, it is important to understand the mechanisms by which the activities of the CRLs are regulated, because CRL-mediated ubiquitylation of misfolded proteins in cardiac myocytes may play an important role in maintaining proteostasis and thus, proper heart function.
The constitutive photomorphogenic 9 signalsome (COP9 signalsome, or CSN) is a critical regulator of CRL activity but until recently, it had not been studied in the heart. The CSN was identified first in Arabidopsis thaliana as a complex protein comprised of 8 subunits. In Arabidopsis the CSN was shown to regulate light-dependent development. Subsequently, the CSN has been found in many other species and is evolutionarily conserved from yeast to humans. The CSN plays critical roles in numerous processes including development, DNA repair and cytokine signaling. The main biochemical activity of the CSN is deneddylation, which is the removal of NEDD8 (neural precursor cell expressed developmentally down-regulated protein 8) from neddylated proteins. NEDD8 is an 81 amino acid ubiquitin-like protein that shares ~60% amino acid sequence identity with ubiquitin. Like ubiquitylation, neddylation of proteins, which requires NEDD8 specific E1 actviating, E2 conjugating and E3 ligases, has been shown to regulate many processes, including transcription, signal transduction, autophagy and cell death. Dysregulation of neddylation has been linked to a variety of diseases, including heart failure. Even though CRLs must be neddylated to be active, paradoxically it is by removing NEDD8 from CRL ubiquitin ligases that the CSN increases CRL-mediated protein ubiquitylation (Figure). This paradox can be explained by evidence suggesting that CRLs cannot engage a new substrate for ubiquitylation unless they recycle through the deneddylation/reneddylation process; the CSN is required for that process8, 9.
Since proteotoxicity from misfolded proteins can cause cardiomyopathy and impaired heart function, and since the CRLs, and their major regulator, the CSN were shown in other cell types to enhance the degradation of misfolded proteins, the effects of CSN deletion in the heart was examined initially in the lab of Xuejun (XJ) Wang. Germline ablation of any of the CSN subunits had previously been shown to be embryonic lethal; thus, in the Wang lab, Su et al studied roles for the CSN in the mouse heart using a conditional gene targeting approach in which a critical subunit of the CSN, CSN8, was deleted in mouse hearts soon after birth. However, these mice prematurely died of dilated cardiomyopathy and heart failure at ~30d of age10. While these findings showed that CSN8 and thus CSN activity are required for normal postnatal cardiac development and function, the premature death of the mice precluded an examination of roles for the CSN in the ubiquitylation and degradation of misfolded proteins in the adult mouse heart. Accordingly, to address this question, in this issue of Circulation Research, Su et al engineered a mouse model in which CSN activity was attenuated, but not completely inactive; this involved the preparation of a new line of mice in which one allele of CSN8 was deleted in a cardiac-specific manner11. Initial characterization showed that these CSN8 hypopmorphic mice exhibited an approximate 80% reduction of CSN8 in the heart and that they survived into adulthood. Moreover, CRL neddylation was increased in the hearts of the CSN8 hypopmorphic mice, consistent with roles for the CSN in CRL deneddylation.
Su et al went on to examine the effects of reduced CSN activity on cardiac pathology in a previously studied12 mouse model of proteinopathy induced by overexpression of a mutant form of the small heat shock protein CryAB, i.e. CryABR120G12. When CSN8 hypomorphic mice were crossed with transgenic mice that overexpress CryABR120G, their hearts exhibited increased CryAB-containing aggregates, which have been associated with cardiomyopathy. The CryABR120G-expressing CSN8 hypomorphic mouse hearts also had decreased levels of ubiquitylated proteins, impaired cardiac function and increased mortality. Moreover, when examined in cultured cardiac myocytes, knockdown of CSN8, or chemical inactivation of CRLs decreased the ubiquitylation and degradation of CryABR120G but not native CryAB, and increased cell death in response to CryABR120G expression. Thus, Su et al concluded that the CSN promotes the ubiquitylation and degradation of misfolded CryABR120G but not properly folded CryAB. Therefore, the inferrence is the CSN protects against proteotoxicity caused by the misfolding of proteins in the heart (Figure).
Su et al focused their studies on the effects of CSN inactivation on the degradation of several model overexpressed misfolded proteins, such as CryABR120G; thus, it remains to be determined how many other proteins are affected by CSN in the heart. However, since the CSN activates CRLs, and since CRLs are responsible for widespread ubiquitylation of misfolded proteins in other cell and tissue types, it is reasonable to assume that via CRL activation, CSN has a similar widespread function in misfolded protein ubiquitylation in the heart. This assumption is supported by the study by Su et al that CSN deletion or hypomorphism reduced the levels of ubiquitylated proteins, in general. Moreover, Su et al showed that CSN is important for nutrient starvation stress-induced autophagy in cultured cardiac myocytes, which is believed to involve the misfolding of a broad spectrum of cardiac proteins. Additionally, this study raises the question of whether the CSN plays a protective role in the ischemic and failing heart, in which function is impaired by protein misfolding. Future examination of the effects of CSN hypomorphism in myocardial infarction, as well as pressure overload models of heart failure will begin to address this question.
In addition to marking proteins that become misfolded for proteasome-mediated degradation, the CSN has been shown to be involved in the selective stabilization and destabilization of key signaling proteins that is important in regulating their cellular functions9. For example, CSN stabilizes some transcription factors (e.g. MYC, p53 and HY5), cell cycle regulators (e.g. Cyclin E and p27), and signaling proteins (e.g. β-catenin and Smad7). CSN destabilizes c-Jun and HIF1-α. Thus, in addition to targeting misfolded proteins for proteasome mediated degradation, it is probable that in the heart CSN is likely to regulate numerous other cellular processes.
The study by Su et al also revealed possible roles for neddylation as a regulator of cardiac myocyte function. While neddylation has been shown to regulate cell proliferation, protein homeostasis, mitochondria turnover, transcription factor activity, receptor signaling, cell death effectors and autophagy in cancer, inflammation, immunodeficiency, and neurodegernerative diseases, little is known about the functions of neddylation in the heart13, 14. Substrates for neddylation, such as p53, Mdm2 and the epidermal growth factor receptor have been examined in various non-cardiac cell types. Now, the study by Su et al provides the initial evidence that neddylation is critical for the function of CRLs in the heart. Moreover, these studies demonstrate that CSN-mediated deneddylation of CRLs is required for maintenance of optimal heart function
Future studies on roles for the COP9 signalsome in the heart will undoubtedly reveal many new functions for this complicated, evolutionarily conserved multi-subunit protein. The elegant experiments on the CNS in the heart that have been reported by Su et al10, 11 have not only provided an important foundation of information, but have also resulted in the generation of novel animal models that will facilitate future studies that are sure to reveal previously unappreciated roles for the COP9 signalsome in proteostasis in the heart.
Acknowledgments
The author wishes to acknowledge Dr. Shirin Doroudgar, Dr. Junkang Jin, Adrian Arrieta, Winston Stauffer, Erik Blackwood, and Amber Pentoney for insightful discussions.
Sources of Funding: National Institutes of Health P01 HL085577, R01 HL121539, R01 HL127439,
Footnotes
DISCLOSURES: None
REFERENCES
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Christians ES, Mustafi SB, Benjamin IJ. Chaperones and cardiac misfolding protein diseases. Curr Protein Pept Sci. 2014;15:189–204. doi: 10.2174/1389203715666140331111518. [DOI] [PubMed] [Google Scholar]
- 3.Sandri M, Robbins J. Proteotoxicity: an underappreciated pathology in cardiac disease. J Mol Cell Cardiol. 2014;71:3–10. doi: 10.1016/j.yjmcc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol. 2014;71:16–24. doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279:532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 6.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. 2008;1782:749–763. doi: 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Martin DS. The COP9 signalosome and cullin-RING ligases in the heart. Am J Cardiovasc Dis. 2015;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 9.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su H, Li J, Zhang H, Ma W, Wei N, Liu J, Wang X. COP9 Signalosome Controls the Degradation of Cytosolic Misfolded Proteins and Protects Against Cardiac Proteotoxicity. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.115.306783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 13.Kandala S, Kim IM, Su H. Neddylation and deneddylation in cardiac biology. Am J Cardiovasc Dis. 2014;4:140–158. [PMC free article] [PubMed] [Google Scholar]
- 14.Stastna M, Van Eyk JE. Posttranslational modifications of lysine and evolving role in heart pathologies-recent developments. Proteomics. 2015;15:1164–1180. doi: 10.1002/pmic.201400312. [DOI] [PubMed] [Google Scholar]