Abstract
How the immune system remembers a previous encounter with a pathogen and responds more efficiently to a subsequent encounter has been one of the central enigmas for immunologists for over a century. The identification of pathogen-specific memory lymphocytes that arise after an infection provided a cellular basis for immunological memory. But the molecular mechanisms of immunological memory remain only partially understood. The emerging evidence suggests that epigenetic changes have a key role in controlling the distinct transcriptional profiles of memory lymphocytes and thus in shaping their function. In this Review, we summarize the recent progress that has been made in assessing the differential gene expression and chromatin modifications in memory CD4+ and CD8+ T cells, and we present our current understanding of the molecular basis of memory T cell function.
Immunological memory is one of the defining features of the adaptive immune response and its induction is the basis for immunization and vaccination1–2. The cellular foundation of immunological memory resides in the existence of memory lymphocytes, which carry the ’memory’ of a previous exposure to an antigen, along with an altered and enhanced functional capacity. Although immunological memory has been intensively studied in the past few decades, the mechanisms underlying the generation and maintenance of memory lymphocytes during and after an immune response remain only partially understood. Nevertheless, the key features of memory lymphocytes that make these cells distinct from naive lymphocytes are known; specifically, memory lymphocytes are longer lived and have a reduced activation threshold and enhanced effector functions.
The distinct features of a cell are determined principally by its transcriptional profile. Accordingly, the underlying basis of the acquired functions of memory lymphocytes is, primarily, their distinct patterns of gene expression. How memory lymphocytes acquire their gene expression patterns is not fully known, but recent advances in identifying unique patterns of gene expression and epigenetic regulation define a chromatin state that underpins memory T cell gene expression and function. In this Review, we summarize the current knowledge of gene expression profiles and kinetics in memory T cells during activation and differentiation, and we discuss the epigenetic features that are associated with the unique transcriptional profiles of memory T cells.
Gene expression in memory T cells
Memory T cells differentiate from naive T cells after antigenic stimulation and exhibit a sustained and enhanced response to the primary stimulus (antigen) during a subsequent encounter. Memory T cell populations are heterogeneous and can be divided into two main subsets: central memory T cells (TCM cells) and effector memory T cells (TEM cells)3,4. TCM cells express CD62L (also known as L-selectin) and CC-chemokine receptor 7 (CCR7), circulate in lymphoid organs and have the stem cell-like ability to differentiate and proliferate after receiving proper signals (from antigens or cytokines). TEM cells do not express CD62L or CCR7, and they circulate in non-lymphoid tissues. In addition, TEM cells express high levels of effector molecules, such as cytokines (in the case of CD4+ TEM cells) or granzyme B and perform (in the case of CD8+ TEM cells), enabling them to perform rapid effector functions following activation.
Most human studies have defined memory T cells by their selective expression of CD45RA, CD45RO, CD62L and/or CCR7, as well as other cell-surface markers, whereas memory T cells in mice are often studied using antigen-specific approaches. Despite differences in the approaches used to study human and mouse memory T cell responses, the core transcriptional features of memory T cells are shared.
The development of microarray technology has revolutionized gene expression analysis and provided a powerful tool for the genome-wide evaluation of cellular gene expression profiles5,6. Thus, the transcriptional identity of a cell can be defined by a set of signature genes and can be used for comparison between different types of cells, as well as between the same types of cells at different stages of differentiation. Analyses of gene expression profiles of memory T cells have been reported for both CD4+ and CD8+ T cells from humans and mice7–17. Because of the differences in experimental platforms and analysis tools (as well as in specific details such as the mouse strains and the criteria used to identify memory T cells) across various microarray experiments, considerable variations exist in the details of the gene expression data. Here, we analyse published findings on CD4+ and CD8+ memory T cells from both humans and mice using the deposited microarray datasets from the Gene Expression Omnibus (GEO) database hosted by the National Center for Biotechnology Information (NCBI) (dataset numbers GSE24759, GSE22880, GSE14422, GSE23663, GSE24151, GSE26928, GSE32596, GSE21360 and GSE13743)13–16,18–21. We discuss transcriptional features that are shared between all memory T cells and compare them with those shared between CD4+ and CD8+ memory T cells and between TCM and TEM cells.
Global transcriptional profiles of memory T cells
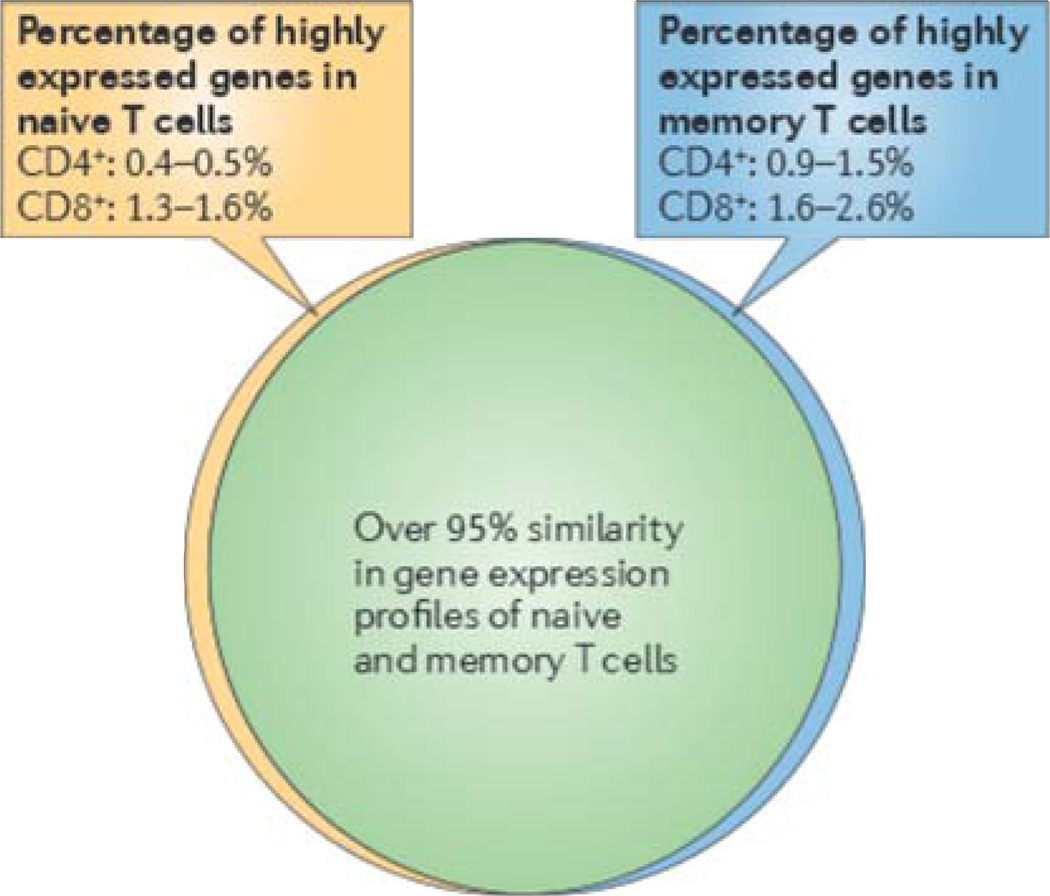
The analysis of gene expression characteristics in memory T cells is often carried out by comparing these characteristics with the gene expression characteristics of precursor naive T cells. Despite their substantial functional differences, naive and memory T cells share a great degree of similarity (~95%) in their overall gene expression profiles (FIG. 1). Indeed, only a few percent of their expressed genes (equivalent to a few hundred genes) are expressed at significantly different levels between naive and memory T cells13–16,18–21. Although genes that are highly expressed only in naive T cells are likely to be important in understanding the function of memory T cells, the roles of most of these genes are poorly characterized and their precise contributions to memory T cell function are not all clear. Therefore, we focus here on well-characterized genes that are highly expressed in memory T cells. Broadly speaking, these genes can be divided into different functional classes: genes with known immune function; genes that promote T cell survival and homeostasis; and genes with multiple or undefined functions (TABLE 1).
Figure 1. Comparison of overall gene expression in naive and memory T cells.
The numbers in this figure are based on the analysis of nine published or deposited datasets from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (dataset numbers GSE24759, GSE22880, GSE14422, GSE23663, GSE24151, GSE26928, GSE32596, GSE21360 and GSE13743). These datasets describe the differences in gene expression between naive and memory CD4+ and CD8+ T cells from humans and mice. Because different array analyses have different numbers of annotated genes, the genes that are expressed significantly more highly (expression difference fold > 2.0 and P< 0.05) in naive or memory T cells are presented as a percentage of the total number of genes analysed. The ranges reflect the lowest to the highest percentages in the nine datasets analysed here. In general, fewer genes are differentially expressed between naive and memory CD4+ T cells than between naive and memory CD8+ T cells.
Table 1.
Chromatin states of selected genes that are highly expressed by human memory T cells*
| Gene symbol | Gene name | Chromatin state‡ | Memory T cell subset§ | Functional subgroup |
|---|---|---|---|---|
| Immune function | ||||
| CD58 | CD58 molecule | Open | CD4+ ≈ CD8+ | Activation |
| CLECL1 | C-type lectin-like 1 | Open | CD4+ ≈ CD8+ | Activation |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | Open | CD4+ ≈ CD8+ | Activation |
| HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 | Open | CD4+ ≈ CD8+ | Activation |
| SLAMF1 | Signalling lymphocytic activation molecule family member 1 | Open | CD4+ ≈ CD8+ | Activation |
| HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | Open | CD8+ > CD4+ | Activation |
| HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | Open | CD8+ | Activation |
| CD63 | CD63 molecule | Open | CD4+ ≈ CD8+ | Adhesion or migration |
| CXCR3 | CXC-chemokine receptor 3 | Open | CD4+ ≈ CD8+ | Adhesion or migration |
| CXCR5 | CXC-chemokine receptor 5 | Open | CD4+ ≈ CD8+ | Adhesion or migration |
| CCR5 | CC-chemokine receptor 5 | Open | CD8+ > CD4+ | Adhesion or migration |
| CCR6 | CC-chemokine receptor 6 | ND | CD4+ | Adhesion or migration |
| CX3CR1 | CX3C-chemokine receptor 1 | Open | CD8+ | Adhesion or migration |
| DUSP4 | Dual specificity phosphatase 4 | Open | CD4+ ≈ CD8+ | Signalling |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 | Open | CD4+ ≈ CD8+ | Signalling |
| PTGER2 | Prostaglandin E receptor 2 | Open | CD4+ ≈ CD8+ | Signalling |
| RGS1 | Regulator of G-protein signalling 1 | Open | CD4+ ≈ CD8+ | Signalling |
| S100A4 | S100 calcium-binding protein A4 | Open | CD4+ ≈ CD8+ | Signalling |
| DUSP5 | Dual specificity phosphatase 5 | ND | CD4+ | Signalling |
| LY96 | Lymphocyte antigen 96 | ND | CD4+ | Signalling |
| NOD2 | Nucleotide-binding oligomerization domain-containing 2 | ND | CD4+ | Signalling |
| STAM | Signal transducing adaptor molecule 1 | ND | CD4+ | Signalling |
| CFH | Complement factor H | Open | CD8+ > CD4+ | Effector molecule |
| GZMA | Granzyme A | Open | CD8+ > CD4+ | Effector molecule |
| GZMK | Granzyme K | Open | CD8+ > CD4+ | Effector molecule |
| GZMB | Granzyme B | Open | CD8+ | Effector molecule |
| GZMH | Granzyme H | Open | CD8+ | Effector molecule |
| CTSW | Cathepsin W | Open | CD8+ | Effector molecule |
| PRF1 | Perforin 1 | Open | CD8+ | Effector molecule |
| CD160 | CD160 molecule | Open | CD8+ | Effector molecule |
| CD244 | CD244 molecule | Open | CD8+ | Effector molecule |
| NCR3 | Natural cytotoxicity triggering receptor 3 | Open | CD8+ | Killing-related function |
| KLRC1 | Killer cell lectin-like receptor subfamily C, member 1 | Open | CD8+ | Killing-related function |
| KLRF1 | Killer cell lectin-like receptor subfamily F, member 1 | Open | CD8+ | Killing-related function |
| KLRG1 | Killer cell lectin-like receptor subfamily G, member 1 | Open | CD8+ | Killing-related function |
| Survival and homeostasis | ||||
| IFNG | Interferon gamma | Open | CD4+ ≈ CD8+ | Cytokine or chemokine |
| TNF | Tumor necrosis factor | Open | CD4+ = CD8+ | Cytokine or chemokine |
| CCL5 | CC-chemokine ligand 5 | Open | CD8+ > CD4+ | Cytokine or chemokine |
| IL2RB | Interleukin-2 receptor beta | Open | CD8+ > CD4+ | Receptor |
| IL10RA | Interleukin-10 receptor alpha | Open | CD4+ ≈ CD8+ | Receptor |
| LCALS1 | Lectin, galactoside-binding, soluble, 1 | Open | CD4+ ≈ CD8+ | Receptor |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 | Open | CD4+ ≈ CD8+ | Receptor |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member IB | Open | CD4+ ≈ CD8+ | Receptor |
| CD74 | CD74 molecule (HLA class II histocompatibility antigen gamma chain) | Open | CD4+ ≈ CD8+ | Receptor |
| FAS | FAS (TNF receptor superfamily, member 6) | Open | CD4+ ≈ CD8+ | Receptor |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | Open | CD4+ ≈ CD8+ | Receptor |
| Multiple or undefined functions | ||||
| MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homologue (avian) | Open | CD4+ ≈ CD8+ | Transcriptional regulator |
| TOX | Thymocyte selection-associated high mobility group box | Open | CD4+ ≈ CD8+ | Transcriptional regulator |
| TBX21 | T-box 21 (encodes T-bet) | Open | CD8+ > CD4+ | Transcriptional regulator |
ND, not determined (no genome-wide data).
The selection of highly expressed genes in human memory T cells was based on six datasets from the Gene Expression Omnibus (CEO) database (GSE14422, GSE26928, GSE24759, GSE24151, GSE22880 and GSE23663)13,15,16,18,19,21. The comparison between CD4+ and CD8+ memory T cells used the GSE24579 dataset16
Chromatin state was determined based on levels of trimethylation of histone H4 lysine 4 (H4K4me3) in CD8+ memory T cells.
CD4+ ≈ CD8+, similarly highly expressed in both CD4+ and CD8+ memory T cells; CD8+ > CD4+, highly expressed in both CD4+ and CD8+ memory T cells but expressed significantly more highly in CD8+ memory T cells; CD4+, highly expressed only in CD4+ memory T cells; CD8+, highly expressed only in CD8+ memory T cells.
The genes that are highly expressed in memory T cells and that are involved in immune function can be further divided into several subgroups. First, there are genes that regulate the activation of T cells, including MHC class II genes (HLA-DRA, HLA-DRB1, HLA-DPA1 and HLA-DPB1). The second subgroup comprises genes involved in the migration of T cells, including adhesion molecule and chemokine receptor genes, such as CCR5, CCR6, CXC-chemokine receptor 3 (CXCR3) and CXCR5. The third subgroup of genes is involved in intracellular signalling; these genes include mitogen-activated protein kinase kinase kinase 5 (MAP3K5), dual specificity phosphatase 4 (DUSP4), regulator of G-protein signalling 1 (RGS1) and S100 calcium-binding protein A4 (S100A4). Finally, the fourth subgroup includes effector molecule genes, such as granzyme A (GZMA) and GZMK.
The genes involved in memory T cell survival and homeostasis can be further separated into two subgroups. The first subgroup includes cytokine and chemokine genes, such as CC-chemokine ligand 5 (CCL5), interferon-γ (IFNG) and tumour necrosis factor (TNF). Genes in the second subgroup encode receptors, such as interleukin receptors (IL2RB and IL10RA), lectins (LGALS1 and LGALS3), tumour necrosis factor receptor superfamily, member IB (TNFRSF1B), CD74 and FAS.
The third class of genes that are highly expressed in memory T cells includes genes encoding transcriptional regulators, which serve an array of diverse functions. Such genes include MAF, thymocyte selection-associated high mobility group box (TOX) and TBX21 (which encodes T-bet).
Comparing human CD4+ and CD8+ memory T cells, there seem to be more differentially expressed genes in CD8+ memory T cells than in CD4+ memory T cells (FIG. 1). Analysing these differentially expressed genes shows that CD8+ memory T cells highly express genes that encode key effector molecules of cytotoxicity and killing — such as granzymes (GZMB and GZMH), cathepsin W (CTSW), perform 1 (PRF1), CD160, CD244 and killer cell lectin-like receptors (KLRC1, BCLRF1 and KLRG1) — as well as genes that encode chemokines (CCL5), complement factor H (CFH), interleukin receptors (IL2RB) and a master transcription factor (TBX21) (TABLE 1). These highly expressed genes make perfect sense for explaining the enhanced cytotoxic functions of CD8+ memory T cells.
Although most of the highly expressed genes in memory T cells are shared between TCM and TEM cells, some genes are more highly expressed in either TCM or TEM cells. CCR2, LGALS1, LGALS3, the MHC class II genes HLA-DPB1, HLA-DQA1, HLA-DRA, HLA-DRB5 and HLA-DRB6, and integrin αM (ITGAM) are more abundantly expressed in human CD4+ TEM cells than in human CD4+ TCM cells (on the basis of analyses of the GSE24759 and GSE26928 datasets)15,16. Genes that are more abundantly expressed in human CD8+ TEM cells than in human CD8+ TCM cells include genes encoding cell-surface receptors — such as chemokine receptors (CCR6 and CCR9), killer cell lectin-like receptors (KLRC1, KLRD1, KLRF1 and CD244), integrins (ITGA4 and ITGAE), interleukin receptors (IL18R1 and IL23R) and the apoptosis inducer FAS ligand (FASLG) — as well as cytotoxic molecule genes (GZMH) and IFNG (on the basis of analyses of the GSE14422, GSE24759 and GSE23663 datasets)13,16,21. The highly expressed genes in TEM cells explain the effector function and the characteristic migration and circulation of these cells.
Collectively, the identification of highly expressed genes in CD4+ and CD8+ memory T cells, as well as in TCM and TEM cells, at the genome-wide level offers a transcriptional basis for explaining some of the unique features of memory T cells and of individual memory T cell subsets, such as enhanced effector functions, unique circulation and homing patterns, and homeostasis. It is essential to further characterize the roles of the genes with unknown functions that are highly expressed in memory T cells. A better understanding of the function of these genes will certainly reveal new insights into different aspects of memory T cells, and will lead to the elucidation of how the transcriptional state of these genes contributes to the overall function of memory T cells.
Kinetic features of memory T cell gene expression
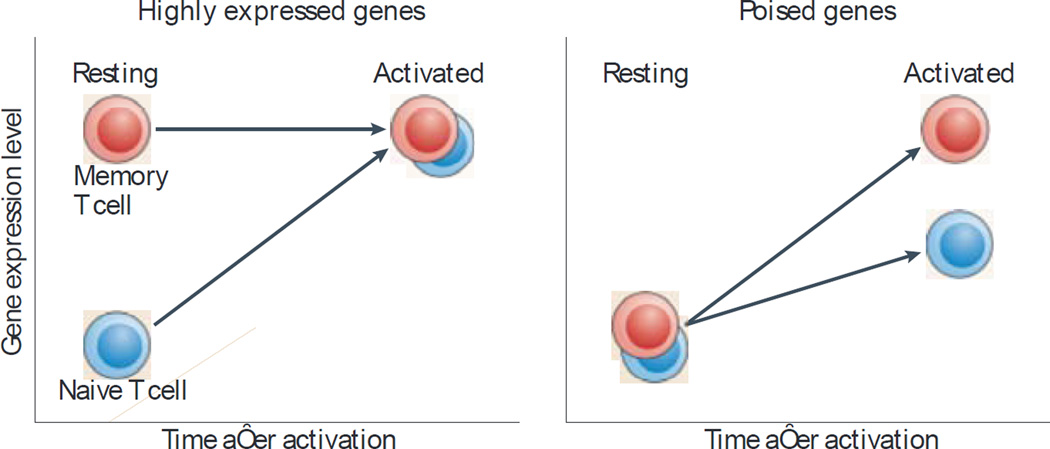
Based on the kinetics of gene expression before and after antigen-mediated T cell activation, genes that are highly expressed in memory T cells compared with naive T cells can be further divided into two main classes (FIG. 2). First, there are genes that are highly expressed by resting memory T cells but not by resting naive T cells7,13–16,18–21. Second, there are activation-induced genes that are upregulated more rapidly in activated memory T cells than in activated naive T cells. Expression of this second group of genes is similar between resting naive and memory T cells, but they are expressed at higher levels by activated memory T cells than by activated naive T cells; these genes are also called ’poised’ genes8,13,22.
Figure 2. Types of genes that are differentially expressed in memory T cells based on their expression kinetics before and after T cell activation.
There are two main kinetic patterns of expression for genes that are expressed at higher levels in memory T cells than in naive T cells. First, there are genes that are highly expressed in resting memory T cells compared with resting naive T cells. These highly expressed genes in resting memory T cells include genes involved in migration, homeostasis and readiness for activation. Second, there are genes that are highly expressed only after the activation of memory T cells; these genes are termed poised genes. Such poised genes are tightly regulated when the T cell is in the resting state but are rapidly induced after T cell activation. It is apparent that the function of these poised genes is not desired in the resting state, and therefore they are minimally expressed. These two patterns of expression for genes that are highly expressed in memory T cells show that the expression of such genes is precisely controlled in a time- and space-dependent manner to fulfil the function of memory T cells.
In human CD8+ memory T cells, the highly expressed genes include killer cell lectin-like receptor genes (KLRC1, KLRF1 and KLRG1) and granzyme genes (GZMA, GZMB, GZMH and GZMK), which mediate the cytotoxic and killing functions of CD8+ T cells13. Activation-induced poised genes in memory T cells include interleukin-3 (IL3), IL5, IL10 and IL21 and these genes provide a fast and robust effector function for memory T cells following activation13.
These two distinct patterns of highly expressed genes in memory T cells based on expression kinetics provide an additional level of regulation to precisely control gene function in a time- and space-dependent manner. Although the signals that maintain the high-level expression of specific genes in resting memory T cells are not completely understood, the functional role of the well-characterized genes discussed in the previous section is obvious for the enhanced effector functions, migration and homeostasis of memory T cells. The expression of the poised genes is tightly regulated in the resting state and after activation, indicating that the activities of these genes are only desired following T cell activation. Further characterization of the kinetic changes in the expression of genes that are highly expressed in memory T cells will offer new insights into how memory T cells are generated and maintained from a transcriptional perspective.
Epigenetic regulation in memory T cells
The sustained and distinct patterns of gene expression in cells are believed to be controlled by epigenetic changes at the chromatin level23,24. The molecular nature and scope of the epigenetic changes during memory T cell development are currently under intensive study25, and chemical modifications of DNA and histones are the most commonly studied epigenetic changes.
Epigenetic markers and their roles in chromatin state and transcription
Chromatin structure is dynamic and differs between one region (or gene) on a chromosome and another26–28. There are two basic states of chromatin: an open chromatin state that is accessible to DNA-binding proteins (such as transcription factors, transcriptional activators or repressors, and RNA polymerase) and thus facilitates active transcription; and a closed chromatin (heterochromatin) state that lacks accessibility for the transcriptional machinery and thus is associated with gene silencing. Several epigenetic markers have been identified in association with specific chromatin states and transcription levels23 (TABLE 2).
Table 2.
Chemical modifications of DNA and histones and their association with chromatin and transcription states
| Target | Modification | Nucleotide or amino acid |
Residue position | Chromatin state | Transcription state |
|---|---|---|---|---|---|
| DNA | Methylation | Cytosine (C) | CpG islands | Closed | Repressed |
| Histones | Acetylation | Lysine (K) | H2AK5, H2BK12, H2BK15, H3K9, H3K14, H3K18, H3K56, H4K5, H4K8, H4K13, H4K16 | Open | Active |
| Methylation | Arginine (R) | H3R17, H3R23, H4R3 | Open | Active | |
| Methylation | Lysine (K) | H3K4, H3K36, H3K79 | Open | Active | |
| H3K9, H3K27, H4K20 | Closed | Repressed | |||
| Phosphorylation | Serine (S) or threonine (T) | H3T3, H3S10, H3S28, H2BS14 | Open | Active | |
| Sumoylation | Lysine (K) | H2AK126, H2BK6, H2BK7 | Closed | Repressed | |
| Ubiquitylation | Lysine (K) | H2AK119 | Closed | Repressed | |
| H2BK120 | Open | Active |
Covalent modifications of DNA, in particular methylation, have been associated with chromatin states and transcriptional activity29. Methylation of cytosines within DNA occurs predominantly at clusters of CpG dinudeotides (known as CpG islands). It has been shown that approximately 70% of annotated gene promoters are associated with a CpG island30. Low-level or no methylation of CpG islands is associated with open chromatin and active gene transcription, whereas a high level of CpG island methylation is linked to heterochromatin and transcriptional silencing. Some CpG islands are located at a distance from transcription start sites (for example, at enhancers), and these sites can also influence transcription31.
Covalent modifications of the amino-terminal tails of histones (namely, histones H2A, H2B, H3 and H4) have also been shown to regulate the chromatin state and transcription26,27. Histone modifications that are associated with open chromatin include: acetylation of H2A, H2B, H3 lysine 9 (H3K9), H3K14, H4K5 and H4K16; methylation of H3K4, H3K36 and H3K79; phosphorylation of H3 threonine 3 (H3T3), H3 serine 10 (H3S10) and H3S28; and ubiquitylation of H2BK120. Histone modifications that are associated with closed chromatin include: methylation of H3K9, H3K27 and H4K20; ubiquitylation of H2AK119; and sumoylation of H2BK6 or H2BK7 and H2AK126.
Conventional chromatin immunoprecipitation (ChIP) and reverse transcription PCR (RT-PCR) assays allow assessment of the chromatin state and transcription of selected genes, yielding valuable information on their chromatin state and transcriptional status during T cell differentiation32,33. Furthermore, the development of the ChIP-seq technique, which is a combination of ChIP and high-throughput sequencing methods, allows the direct assessment of the chromatin state, in terms of histone modifications and DNA-binding proteins, at the whole-genome level34. Combining results from global gene expression analysis methods (that is, microarrays and RNA sequencing) with ChIP-seq data makes it feasible for the first time to produce a genome-wide picture of the chromatin state and transcriptional activity of a population of cells.
DNA methylation and transcription in memory T cells
DNA methylation of CpG islands in genes encoding cytokines and their receptors, effector molecules, and their regulators has been long studied in memory T cells35–41. In CD4+ memory T cells, CCR6, RAR-related orphan receptor C (RORC) and genes encoding ligands for P-selectin and E-selectin have been shown to be hypomethylated, in contrast to the hypermethylated states of these loci in naive CD4+ T cells35–37. Correspondingly, higher expression of CCR6 and RORC is found in memory T cells than in naive T cells. In CD8+ memory T cells, low levels of DNA methylation at the IFNG and IL2 loci and concomitant high protein expression levels have been observed after activation38–40. Furthermore, rapid DNA demethylation in the promoters of these genes occurs in CD8+ memory T cells but not CD8+ naive T cells after activation39. Such dynamic changes in DNA methylation are also found at the programmed cell death 1 (Pdcd1) gene locus (which encodes a key regulator of cell proliferation and exhaustion) during naive to effector to memory CD8+ T cell differentiation following viral infection42.
As the DNA methylation status is stable and passes from a parental cell to its descendants after memory T cell division38, the feature of enhanced or poised gene expression is retained in the descendants of memory T cells. Thus, DNA methylation-related chromatin changes provide a transcriptional basis for the enhanced effector response, a key feature of memory T cells. In addition, the DNA methylation status of a cell death-related gene (Noxa; also known as Pmaip1) has been shown to influence the survival of CD4+ memory T cells41; indeed, the repression of Noxa expression by DNA methylation is necessary for the survival of these cells.
It is evident that DNA methylation has a key role in the regulation of gene expression in memory T cells. However, the full scope of the involvement of DNA methylation in the differential gene expression of memory T cells remains to be determined. A recent genome-wide analysis of changes in DNA methylation during immune cell differentiation observed a robust change in DNA methylation during early lineage differentiation but only a minor change during later stages of differentiation43. It is therefore possible that the changes in DNA methylation status during naive to memory T cell differentiation might be limited to key genes of memory T cell function, such as cytokines and growth-related molecules and their regulators.
Histone modifications and gene expression in memory T cells
Similarly to DNA methylation, histone acetylation has been analysed in cytokine genes during CD4+ memory T cell differentiation32,44. Histone hyperacetylation is associated with an active chromatin conformation and is found at the promoters of IFNG and IL4 in memory T helper 1 (TH1) and TH2 cells, respectively45,46. Similarly, histone hyperacetylation is observed at the promoters of genes encoding cytokines (such as IFNγ), effector molecules (such as granzyme B and perform 1), and their regulators in CD8+ memory T cells40,47–49. More importantly, induced hyperacetylation or hypoacetylation at these gene loci results in an increase or decrease, respectively, in their expression in CD8+ T cells. These findings show that histone acetylation at these effector gene loci is necessary for their enhanced expression in memory T cells, thus providing evidence that histone acetylation state regulates memory T cell function.
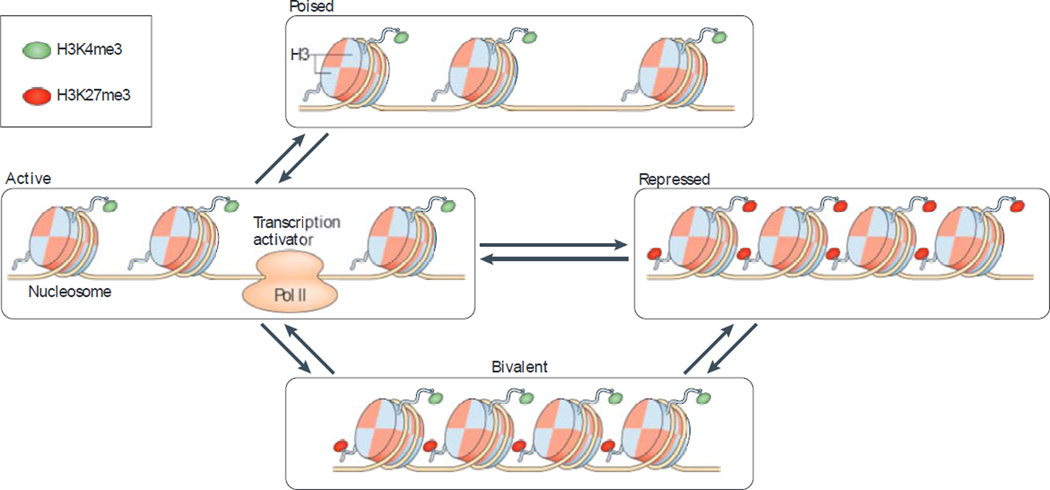
Analyses of genome-wide histone methylation (specifically, trimethylation of H3K4 (H3K4me3) and H3K27me3) and gene expression show a general correlation between gene expression and the distribution of histone methylation. Indeed, H3K4me3 is positively correlated and H3K27me3 is negatively correlated with gene expression13–50. Further analysis reveals four distinct states that are determined by histone methylation and that differentially control gene expression: active, poised, bivalent and repressed (FIG. 3). During the differentiation of a CD4+ naive T cell to an effector T cell, the chromatin state is altered at the gene locus encoding the key transcription factor that controls the differentiation of the particular effector T cell subset (that is, TBX21 for TH1 cells, GATA3 for TH2 cells, RORC for TH17 cells and FOXP3 for regulatory T cells), such that the original closed chromatin state (high levels of H3K27me3 and low levels of H3K4me3) changes to an open chromatin state (high levels of H3K4me3 and low levels of H3K27me3)33,50. In TH2-polarized CD4+ memory T cells, methylation of histones H3 and H4 at the IL4 and IL13 gene loci46 is observed, together with H3K4 methylation at the GATA3 locus (which encodes the master transcriptional regulator of IL-4 expression)51. Furthermore, the histone methyltransferase HRX (which is encoded by MLL and catalyses histone H3 and H4 methylation) is involved in the production of TH2 cell-associated cytokines. Decreased HRX expression results in reduced H3K4me2 levels at the GATA3 and IL4 gene loci, and thus in decreased expression of GATA3 and IL-4, in CD4+ memory T cells52. Collectively, these findings show that histone methylation influences open and closed chromatin states and thereby regulates the differential expression of key genes in CD4+ memory T cells.
Figure 3. The chromatin basis for differential gene expression in memory T cells involves histone methylation.
There are four distinct modes of relationship between histone methylation and gene expression in memory T cells: active, poised, bivalent and repressed. In the active mode, the gene locus has an open chromatin state, which is indicated by high levels of trimethylation at histone H3 lysine 4 (H3K4me3; an activating modification), and there is active gene transcription (indicated by the presence of the transcription activator). In the poised mode, the gene locus has an open chromatin state, similar to that of the active mode, but there is no active gene transcription in resting memory T cells. However, following T cell activation, the transcription of genes in the poised mode can be rapidly initiated. Genes in the bivalent mode contain high levels of both H3K4me3 and the repressive modification H3K27me3 at their loci. Such a chromatin state can change in either direction (to an open or closed state) after T cell activation, and this is followed by the initiation or repression of gene transcription. Genes with a repressed chromatin state contain low levels of H3K4me3 but high levels of H3K27me3 at their loci, and thus their transcription is repressed. The unique chromatin landscape of memory T cells provides a structural basis for the differential gene expression and function of memory T cells. Pol II, RNA polymerase II.
Analyses of histone methylation (specifically, H3K4me3 and H3K27me3) in CD8+ memory T cells in humans13 and mice53 have shown that genes associated with effector functions (such as PRDM1, KLRG1, Ifng and Gzmb) have high levels of H3K4me3 and low levels of H3K27me3. It has also been shown that certain genes — such as inhibitor of DNA binding 2 (ID2), which has a key role in the survival of CD8+ memory T cells following their activation54 — have an open chromatin state but low mRNA levels in resting CD8+ memory T cells (which is termed a poised state)13. This finding reveals a molecular basis for the rapid and enhanced expression of poised genes following the activation of memory T cells. In addition, bivalent chromatin — which contains histone modifications that are associated with both open and closed chromatin (that is, high levels of both H3K4me3 and H3K27me3 at the same region) — is first found in stem cells and is believed to be involved in the differentiation of stem cells after they receive a differentiation signal55,56. Bivalent chromatin has also been observed at several gene loci in memory T cells13. For example, KIAA1804 (mixed lineage kinase 4), which is involved in Toll-like receptor 4 (TLR4) signalling57, has a bivalent chromatin state in resting CD8+ memory T cells but changes to an open chromatin state and is expressed at higher levels after CD8+ memory T cell activation13.
Thus, genes with a bivalent chromatin state in resting memory T cells could more rapidly assume an open chromatin state and initiate transcription in response to T cell activation than genes in a closed chromatin state. It is clear that the four epigenetic states mentioned above (that is, active, poised, bivalent and repressed) provide a means to regulate gene expression to facilitate the function of memory T cells in the resting state and after antigenic activation. Whether the particular states at specific gene loci are fixed properties of memory T cells or can change in memory T cells is currently unclear. It is conceivable that variations in these epigenetic states could be a source of the heterogeneity of memory T cell populations. In the effector phase of CD4+ T cell differentiation, there is increasing evidence of plasticity among the different TH cell subsets58,59. However, it remains to be determined how stable or flexible these specific states of chromatin are at individual gene loci in memory T cells during their homeostasis and after activation.
Complexity of epigenetic changes and their meaning and regulation
In memory T cells, the role of epigenetic changes that involve other post-translational histone modifications (such as ubiquitylation and sumoylation) or the remodelling of nucleosomes within chromatin remains to be elucidated. For the known modifications, it becomes apparent that the same chromatin state can be influenced by several different epigenetic changes; for example, an open chromatin state is influenced by DNA hypomethylation, histone acetylation and certain types of histone methylation and, therefore, is a result of combinatorial changes in multiple modifications60. Intriguingly, not all open chromatin regions with active gene transcription have identical patterns of histone modifications13. It is currently unclear whether the particular combinatorial patterns of epigenetic modifications are unique for specific genes, for specific types of cell and/or for specific stages of differentiation, and whether such differences or redundancies in histone modifications contribute to the heterogeneity of memory T cells. Therefore, further study is needed to characterize different patterns of epigenetic changes and to understand the meanings of particular patterns and their roles in the regulation of differential gene expression in memory T cells. In addition, to facilitate chromatin and gene expression studies with greater resolution, it will be necessary to develop sensitive methods that are capable of analysing the epigenetic changes at the single-cell and single-chromosome levels.
How the epigenetic changes in memory T cells are established and maintained is not known. Current evidence suggests that a series of different steps may be needed to establish an open or closed chromatin conformation. For example, it has been shown that the prior DNA methylation status serves as a basis for recruiting histone-modifying enzymes, such as the H3K4 methyl-transferase SETD1, to regulate transcription61. In addition, enzymes that catalyse histone acetylation and deacetylation — histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively — are both targeted to transcribed regions of active genes (where the function of HDACs is to reset chromatin by removing acetylation62) and are both intricately regulated by histone methyltransferases. The poised genes that are primed by HRX-mediated methylation of histone H3K4 are further modified by a dynamic cycle of acetylation and deacetylation through transient HAT and HDAC binding52. In repressed genes, trimethylation of H3K27 is mediated by the histone methyltransferase EZH2 (enhancer of zeste homologue 2) as part of Polycomb repressive complex 2 (PRC2). The activity of EZH2 in mediating trimethylation of H3K4 is dependent on the activity of HDACs, as limited HDAC activity markedly reduces H3K4me3 levels and gene transcription63. The current challenges are to elucidate the sequences of events that are involved in establishing a particular chromatin state at the loci of differentially expressed genes during memory T cell formation and in its subsequent maintenance, and to determine whether the sequence of epigenetic events that establishes a particular chromatin state is uniform for all gene loci or varies among the different gene loci in memory T cells.
During memory T cell formation, epigenetic changes such as histone acetylation and methylation are thought to be initiated during the effector phase and to persist in the memory phase in the absence of antigenic stimulation64. Therefore, the process of epigenetic modification in memory T cells can be conceptually divided into two stages. First, chromatin changes are established in the loci of genes encoding proteins with effector functions (such as cytokines and cytotoxic molecules) during the differentiation of naive T cells into effector T cells. Second, during the transition of a T cell from an effector to a memory phenotype, the chromatin status is maintained at these gene loci, and this maintenance is accompanied by additional modifications in the chromatin states of genes that have a dominant function in effector T cells and the establishment of new chromatin states in gene loci that are differentially expressed in memory T cells. Elucidation of the precise epigenetic processes at these two stages during memory T cell formation is essential not only for understanding how the epigenetic changes are established and maintained in memory T cells, but also for providing the molecular benchmarks to evaluate the long-lasting protectiveness of an immune response.
Clinical implications
Knowledge of T cell transcription signatures and the epigenetic modifications of these signature genes has significantly enhanced our understanding of the differences between memory T cells and naive T cells at the transcriptional level, and of the chromatin basis for the enhanced transcriptional and functional activities of memory T cells. An effective immune response to a pathogen or a vaccine yields competent effector and memory T cells. The quantity and quality of TEM and TCM cells will determine how well the host is protected from a subsequent encounter with the same pathogen. As the adaptive immune response is highly heterogeneous for different pathogens and in different individuals, the ability to assess the immune function of an individual before and after vaccination is crucial for determining the success of a vaccination. It has been shown that assessing an immune response at the transcriptional level by examining the expression of immune-relevant genes using a systems biology approach provides a comprehensive portrait of an immune response65. By evaluating the expression kinetics of key genes (including those encoding cytokines, effector molecules and their master transcriptional regulators), the outcome of an immune response to a vaccine can be predicted. Thus, it is possible that the level of immune competency can be evaluated based on the transcriptional status and epigenetic features of specific signature genes, in combination with cellular characterization of antigen-specific B and T cells. This information before vaccination could be used to select the type and dosage of the vaccine, and information generated after vaccination would allow for the measurement of the T cell response and would enable us to predict whether a long-lasting memory T cell response is achievable and also to avoid unfruitful or even harmful consequences of vaccination. Furthermore, a better understanding of the immune response of the host could in turn guide the design of vaccines.
Considering the role of epigenetic changes in the gene expression patterns and enhanced function of memory T cells, it is not surprising that there can be detrimental consequences to the body when the epigenetic changes occur at a gene or several genes at an improper time or in an autoreactive T cell. A recent report shows that there are alterations in the DNA methylation and histone acetylation status of T cells from patients with the autoimmune disease systemic lupus erythematosus (SLE), which has a much higher prevalence in women66,67. For example, the gene encoding CD40 ligand (CD40LG), which is involved in B cell-T cell interactions, is demethylated in CD4+ T cells from women with SLE, but not in CD4+ T cells from men with SLE, and this correlates with an increased expression of CD40LG in female patients. Intriguingly, as CD40LG is located on the X chromosome, its demethylated status on the inactive X chromosome is associated with its enhanced expression and therefore may contribute to the striking female predilection of SLE66. Another study described global histone hypoacetylation in the absence of histone H3K4 methylation in CD4+ T cells from patients with SLE compared with CD4+ T cells from healthy controls67. If these autoreactive T cells develop a chromatin landscape of memory T cells, the disease state will be persistent and/or will deteriorate. Clearly, more studies are needed to understand the scope of epigenetic changes in autoreactive T cells. Furthermore, a better understanding of the process of generating epigenetic changes in normal memory T cells may have therapeutic applications in preventing autoreactive T cells from becoming memory T cells.
Conclusion
The identification of differentially expressed genes and their epigenetic regulation provides a chromatin basis for explaining the transcriptional changes and functions of memory T cells. Future studies should further elucidate the functions of many of the highly expressed genes in memory T cells, as well as in naive T cells, to enable us to gain a complete appreciation of memory T cell formation, maintenance and function. In addition, we should focus on how specific epigenetic changes are established and maintained in memory T cells and on how numerous transcription factors and histone-modifying enzymes are coordinated at a specific gene locus and at the correct time during naive to memory T cell differentiation and memory T cell activation. It will also be important to determine whether dysregulation of epigenetic changes contributes to the altered function of the immune system in degenerative processes, such as autoimmunity and ageing.
Acknowledgements
We thank R. Hodes, K. Zhao and the anonymous reviewers for critical reading of the manuscript and helpful suggestions. This research was supported by the Intramural Research Programs of the US National Institute on Aging, National Institutes of Health.
Glossary
- Epigenetic regulation
The modifications on DNA, histones and other targets that collectively determine a stable phenotype without altering the DNA sequence. Epigenetic changes can pass from the parental cells to their offspring and provide a molecular basis for cellular memory.
- Chromatin
The combination of DNA, histones and other proteins that comprises eukaryotic chromosomes. The basic repeating unit of chromatin is the nucleosome, which consists of an octamer of histone proteins around which ~ 146 base pairs of DNA is wound.
- Central memory T cells (TCM cells)
Antigen-experienced T cells that lack immediate effector function but can mediate rapid recall responses. They also rapidly develop the phenotype and function of effector memory T cells after re-stimulation with antigen. TCM cells retain the migratory properties of naive T cells and therefore circulate through the secondary lymphoid organs.
- Effector memory T cells (TEM cells)
Terminally differentiated T cells that lack lymph node-homing receptors but express receptors that enable them to home to inflamed tissues. TEM cells can exert immediate effector functions without the need for further differentiation.
- Microarray
A tool for measuring gene transcription. Its use involves the hybridization of fluorescently labelled cDNA prepared from a cell or tissue of interest with thousands of known oligonucleotides or cDNAs dotted on glass slides or other surfaces. The known DNA ideally represents all of the expressed genes in the species.
- Heterochromatin
High-density regions in the nucleus that are thought to contain compacted chromatin structures associated with silent genes.
- Chromatin immunoprecipitation
A technique that uses antibodies specific for transcription factors or other DNA-binding proteins to precipitate associated DNA sequences from chromatin to study their functional relationship.
- Reverse transcription PCR
A type of PCR in which RNA is converted into complementary DNA (cDNA), which is then amplified.
- ChIP-seq
A technique in which chromatin immunoprecipitation (ChIP) is followed by high-throughput sequencing to generate a genome-wide distribution map of protein–DNA interactions. This technique can be used to measure transcription factor binding and histone modifications.
- Toll-like receptor (TLR)
A member of a family of receptors that are homologous to Drosophila melanogaster Toll. TLRs recognize conserved molecular patterns that are unique to microorganisms. The lipopolysaccharide component of bacterial cell walls is one such ligand. TLRs can also recognize mammalian components and contribute to autoimmunity.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Bevan MJ. Understand memory, design better vaccines. Nature Immunol. 2011;12:463–465. doi: 10.1038/ni.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nature Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sallusto R, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 4.Pepper M, Jenkins MK. Origins of CD4+ effector and central memory T cells. Nature Immunol. 2011;12:467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyatt G, et al. Gene expression microarrays: glimpses of the immunological genome. Nature Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 6.Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 7. Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. This pioneering study showed the changes in global gene expression that occur as CD8+ naive T cells differentiate into effector and then memory T cells after viral infection and suggested that antigen-specific CD8+ T Bells progressively differentiate into memory T cells following viral infection.
- 8.Liu K, et al. Augmentation in expression of activation-induced genes differentiates memory from naive CD4+ T cells and is a molecular mechanism for enhanced cellular response of memory CD4+ T cells. J. Immunol. 2001;166:7335–7344. doi: 10.4049/jimmunol.166.12.7335. [DOI] [PubMed] [Google Scholar]
- 9.Holmes S, He M, Xu I, Lee PP. Memory T cells have gene expression patterns intermediate between naive and effector. Proc. Natl Acad. Sci. USA. 2005;102:5519–5523. doi: 10.1073/pnas.0501437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willinger I, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J. Immunol. 2005;175:5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 11.Luckey CJ, et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haining WN, et al. Identification of an evolutionary conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells. J. Immunol. 2008;181:1859–1868. doi: 10.4049/jimmunol.181.3.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Araki Y, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. This study identifies four different modes of association between histone methylation and differential gene expression in CD8+ memory T cells. providing a chromatin basis for the differential gene expression and function of memory CD8+ T cells.
- 14. Wirth JC, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. This study reveals signature genes of CD8+ memory T cells and their complex regulation following repeated antigenic challenges.
- 15.Chevalier N, et al. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J. Immunol. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 16.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall HD, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi RL, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nature Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 19.Hertoghs KM, et al. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J. Clin. Invest. 2010;120:4077–4090. doi: 10.1172/JCI42758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K, et al. Identification of stem cell transcriptional programs normally expressed in embryonic and neural stem cells in alloreactive CD8+ T cells mediating graft-versus-host disease. Biol. Blood Marrow Transplant. 2010;16:751–771. doi: 10.1016/j.bbmt.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turtle CJ, et al. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161hi CD8α+ semi-invariant T cells. Blood. 2011;118:2752–2762. doi: 10.1182/blood-2011-02-334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai W, et al. Transcriptional control of rapid recall by memory CD4 T cells. J. Immunol. 2011;187:133–140. doi: 10.4049/jimmunol.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 25.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinudeotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avni O, et al. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nature Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 33.Chang S, Aune TM. Dynamic changes in histone-methylation ’marks’ across the locus encoding interferon-γ during the differentiation of T helper type 2 cells. Nature Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 34.Northrup DL, Zhao K. Application of ChIP-Seq and related techniques to the study of immune function. Immunity. 2011;34:830–842. doi: 10.1016/j.immuni.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syrbe U, et al. Differential regulation of P-selectin ligand expression in naive versus memory CD4+ T cells: evidence for epigenetic regulation of involved glycosyltransferase genes. Blood. 2004;104:3243–3248. doi: 10.1182/blood-2003-09-3047. [DOI] [PubMed] [Google Scholar]
- 36.Schmidl C, et al. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naive Treg. Eur. J. Immunol. 2011;41:1491–1498. doi: 10.1002/eji.201041067. [DOI] [PubMed] [Google Scholar]
- 37.Steinfelder S, et al. Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood. 2011;117:2839–2846. doi: 10.1182/blood-2010-06-293027. [DOI] [PubMed] [Google Scholar]
- 38.Fitzpatrick DR, Shirley KM, Kelso A. Stable epigenetic inheritance of regional IFN-γ promoter demethylation in CD44highCD8+ T lymphocytes. J. Immunol. 1999;162:5053–5057. [PubMed] [Google Scholar]
- 39.Kersh EN, et al. Rapid demethylation of the IFN-γ gene occurs in memory but not naive CD8 T cells. J. Immunol. 2006;176:4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 40.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-γ loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 2006;177:1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita M, et al. Bmi 1 regulates memory CD4 T cell survival via repression of the Noxa gene. J. Exp. Med. 2008;205:1109–1120. doi: 10.1084/jem.20072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8+ T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deaton AM, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fields PE, Kim ST, Flavell RA. Changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 45.Messi M, et al. Memory and flexibility of cytokine gene expression as separable properties of human TH1 and TH2 lymphocytes. Nature Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 46.Yamashita M, et al. Interleukin (IL)-4-independent maintenance of histone modification of the IL-4 gene loci in memory Th2 cells. J. Biol. Chem. 2004;279:39454–39464. doi: 10.1074/jbc.M405989200. [DOI] [PubMed] [Google Scholar]
- 47.Fann M, et al. Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8+ T cell response. Blood. 2006;108:3363–3370. doi: 10.1182/blood-2006-02-005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J. Immunol. 2008;180:8102–8108. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Northrop JK, Wells AD, Shen H. Chromatin remodeling as a molecular basis for the enhanced functionality of memory CD8 T cells. J. Immunol. 2008;181:865–868. doi: 10.4049/jimmunol.181.2.865. [DOI] [PubMed] [Google Scholar]
- 50. Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. This study identifies the chromatin states of key transcription factor genes and their target genes in naive CD4+ T cells differentiating into distinct lineages (including TH1, TH2, TH17 and inducible regulatory T cells) and suggests that an epigenetic mechanism underlies the specificity and plasticity of effector and regulatory T cells.
- 51.Nakata Y, et al. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood. 2010;116:1280–1290. doi: 10.1182/blood-2009-05-223255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamashita M, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. This study demonstrates that MLL regulates histone modifications and expression at the GATA3 locus, indicating that MLL has a crucial role in the maintenance of memory TH2 cells.
- 53.Zediak VP, Johnnidis JB, Wherry EJ, Berger SL. Persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J. Immunol. 2011;186:2705–2709. doi: 10.4049/jimmunol.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cannarile MA, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nature Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 55.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 56.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seit-Nebi A, Cheng W, Xu H, Han J. MLK4 has negative effect on TLR4 signaling. Cell. Mol. Immunol. 2012;9:27–33. doi: 10.1038/cmi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nature Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu KT, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Narure. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Narure Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 64.Zediak VP, Wherry EJ, Berger SL. The contribution of epigenetic memory to immunologic memory. Curr. Opin. Genet. Dev. 2011;21:154–159. doi: 10.1016/j.gde.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 65.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nature Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Q, et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 67.Hu N, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 2008;35:804–810. [PubMed] [Google Scholar]