Abstract
Membrane trafficking is essential for plant growth and responses to external signals. The plant unique FYVE domain-containing protein FREE1 is a component of the ESCRT complex (endosomal sorting complex required for transport). FREE1 plays multiple roles in regulating protein trafficking and organelle biogenesis including the formation of intraluminal vesicles of multivesicular body (MVB), vacuolar protein transport and vacuole biogenesis, and autophagic degradation. FREE1 knockout plants show defective MVB formation, abnormal vacuolar transport, fragmented vacuoles, accumulated autophagosomes, and seedling lethality. To further uncover the underlying mechanisms of FREE1 function in plants, we performed a forward genetic screen for mutants that suppressed the seedling lethal phenotype of FREE1-RNAi transgenic plants. The obtained mutants are termed as suppressors of free1 (sof). To date, 229 putative sof mutants have been identified. Barely detecting of FREE1 protein with M3 plants further identified 84 FREE1-related suppressors. Also 145 mutants showing no reduction of FREE1 protein were termed as RNAi-related mutants. Through next-generation sequencing (NGS) of bulked DNA from F2 mapping population of two RNAi-related sof mutants, FREE1-RNAi T-DNA inserted on chromosome 1 was identified and the causal mutation of putative sof mutant is being identified similarly. These FREE1- and RNAi-related sof mutants will be useful tools and resources for illustrating the underlying mechanisms of FREE1 function in intracellular trafficking and organelle biogenesis, as well as for uncovering the new components involved in the regulation of silencing pathways in plants.
Keywords: Suppressors, FREE1, Endomembrane trafficking, Arabidopsis, NGS
INTRODUCTION
The plant endomembrane system contains several functionally distinct membrane-bound organelles including endoplasmic reticulum (ER), golgi apparatus, trans-golgi network (TGN) or early endosome, prevacuolar compartment (PVC) or multivesicular body (MVB) or late endosome, and vacuoles (Lam et al., 2007; Reyes et al., 2011; Gao et al., 2014a). Membrane trafficking within the endomembrane system is essential for protein and lipid material transport and exchange within and in-between cells. In plants, the trafficking system functions for both fundamental cellular activities and also responds to environmental stresses (Samaj et al., 2006; Bar and Avni, 2014; Teh and Hofius, 2014).
Plant cells have highly regulated membrane trafficking pathways that are conserved among eukaryotic cells, but also seem to have evolved additional features with some of the components present in multiple copies (Cvrckova et al., 2012). The plant endomembrane machinery was largely established from homology-based studies (Bassham et al., 2008). For example, the endosomal sorting somplex required for transport (ESCRT) machinery regulates the homeostasis of plasma membrane protein by sorting them into the MVB which eventually fuse with the lytic vacuole to degrade the ubiquitinated plasma membrane (PM) cargo molecules (Henne et al., 2011; Cai et al., 2014). Most of our current knowledge on ESCRT in plants comes mainly by homology with studies performed on yeast and humans. However, orthologs of the subunits of ESCRT-0 have not been identified in plants (Leung et al., 2008).
A recent study of the plant unique FYVE domain containing protein FREE1 (FYVE domain protein required for endosomal sorting 1) showed that FREE1 localizes to the PVC, and interacts with the ESCRT-I component Vps23 to regulate the formation of intraluminal vesicles (ILVs) in PVCs/MVBs thereby facilitating PM protein degradation in the vacuole (Gao et al., 2014b). Mutant plants without functional FREE1 (T-DNA and RNAi mutants) are seedling lethal, defective in MVB ILV biogenesis and vacuolar sorting of membrane proteins. They also show abnormalities in vacuolar trafficking and accumulate small fragmented vacuoles and autophagosomes (Gao et al., 2014b, 2015). FREE1 was found to directly interact with SH3P2, a unique regulator of plant autophagy (Zhuang et al., 2013; Zhuang and Jiang 2014), thus controlling autophagosome-vacuole fusion and finally autophagic degradation in plants (Gao et al., 2015). These studies have unveiled a direct link between the ESCRT machinery and autophagy process, and demonstrate the multiple functional roles of FREE1 in regulating MVB formation, vacuolar protein transport, vacuole biogenesis and autophagy pathway. Because of the multiple functions of FREE1 in Arabidopsis, it is likely that plants have evolved unique mechanisms and components responsible for the FREE1-containing ESCRT pathway. It is now clear that FREE1 has dual roles in regulating membrane trafficking: 1) through FREE1 N-terminal direct interaction with ESCRT1 components VPS23, FREE1 regulates MVB biogenesis and PM protein vacuolar degradation; 2) through FREE1 C-terminal direct interaction with the autophagosome regulator SH3P2 (Zhuang et al., 2013; Zhuang and Jiang 2014), FREE1 regulates autophagosome formation and autophagic degradation (Gao et al., 2015). In addition, the abnormalities in free1 mutant including the dysfunction of vacuolar protein transport and autophagic degradation point to a defect in membrane fusion caused by FREE1 depletion. However, it is still not understood why and how FREE1 depleted cells contain fragmented small vacuoles and how FREE1 loss-of-function leads to seedling death. It is likely that the functions of proteins required for docking and/or fusion with vacuoles might be disrupted in the free1 mutant. Alternatively, FREE1 might work together with other proteins to regulate membrane fusion and vacuole biogenesis through as yet undefined mechanisms.
To address how FREE1 fulfills its multiple roles in regulating membrane trafficking and organelle biogenesis in plants, we performed a suppressor screen using a FREE1 loss-of-function mutant as the starting material. With the emerging technology of next-generation sequencing (NGS), the mutated gene can be identified through an NGS-mapping approach in a short time (Manavella et al., 2012). Here, we report on the NGS-based high-throughput suppressor identification method to find mutants that can rescue the free1 seedling lethal phenotype using inducible FREE1-RNAi plants. Upon dexamethasone (DEX) induction, the FREE1-RNAi transgenic plants are seedling lethal with the FREE1 protein barely detectable. Seeds from FREE1-RNAi plants were treated with EMS for suppressor screening. M2 seeds were sprayed on plates with DEX to identify surviving plants, which we name sof (suppressors of free1. Because some of the isolated mutants could be caused by a disruption of the RNAi process, we also performed a second screen through Western blot analysis using FREE1 antibodies. We selected mutants with significantly reduced FREE1 proteins as putative sof mutants (termed as FREE1-related sof mutants) for further characterization. Using this method, we have successfully isolated 84 FREE1-related sof mutants. These SOF probably encode proteins involved in controlling vacuole biogenesis and vacuolar trafficking. All of these FREE1-related sof mutants and RNAi-related mutants will be useful resources for members of the international research community working on novel mechanisms of regulating organelle and RNA biogenesis in plants.
RESULTS
Establishment of the mutant pool and screening process
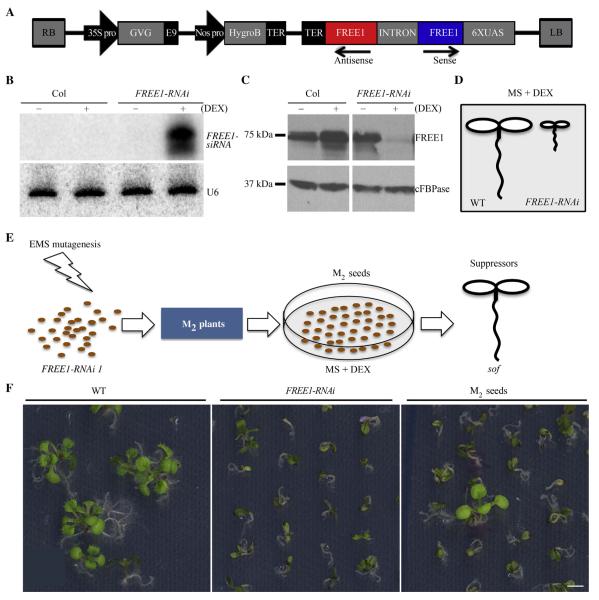
Using the Columbia ecotype (Col) as the wild type, FREE1-RNAi M0 seeds were successfully generated with the wild type transformed with pTA7002 construct containing the hairpin FREE1-RNAi (Fig. 1A). T3 seeds from individual hygromycin resistant T2 lines were screened on both DEX and hygromycin conditions. Line 11# was chosen as mutagenesis material because 3/4 of its T3 showed hygromycin resistance and lethal phenotype on DEX, supporting the single copy insertion of pTA7002-FREE1-RNAi. Homozygous Line 11# was used for following EMS treatment and RNA and protein analysis. The successful induction of the FREE1 silencing system in FREE1-RNAi line was confirmed by RNA blot analysis with significant FREE1-siRNA accumulation and by Western blot analysis with little detection of FREE1 (Fig. 1B and C), and the silencing of FREE1 resulted in seedling lethality (Fig. 1D). For sof mutant screening, M2 seeds were planted on MS plates supplied with 10 μmol/L DEX, and surviving plants were selected (Fig. 1E and F).
Fig. 1. Illustration of sof mutants screening.
A: Schematic diagram of the T-DNA constructs in the binary vector pTA7002, which confers hygromycin resistance in plants. B: RNA blot analysis with FREE1 probe detected accumulated FREE1-siRNA with DEX application in FREE1-RNAi line. Detection of U6 served as loading control. C: Immunoblot analysis with anti-FREE1 antibody. FREE1 protein level was barely detected in FREE1-RNAi plants with DEX treatment. Antibodies against the protein cFBPase served as the loading control. D: Diagram showing the seedling lethal phenotype caused by FREE1 loss of function. E: Schematic illustration of screening procedure. Single insertion Dex-inducible FREE1-RNAi seeds were mutated with EMS, and with DEX induction survived M2 seedlings were selected as putative sof mutants. F: Example of primary screening with M2 seeds plated on MS plates supplied with 10 μmol/L DEX. Scale bar, 5 mm.
Identification of sof mutants
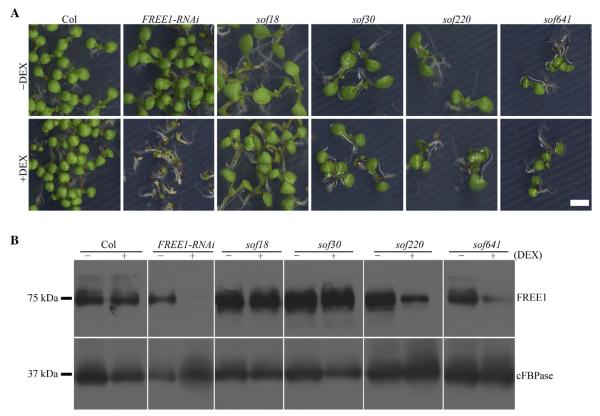
To confirm whether the screened M2 mutations were heritable, M3 seeds collected from individual M2 plants were screened on MS plates with 10 μmol/L DEX. Seedlings that showed recovered phenotype similar to the wild type (as those in Fig. 2A) were further analyzed as putative sof mutants. To classify whether the putative sof mutants were FREE1-related or RNAi related, FREE1 protein levels were determined by Western blot using cFBPase as the loading control. As summarized in Table 1, we screened out 84 FREE1-related and 145 RNAi-related sof mutants. The 84 candidates showed reduced expressions of FREE1 including sof220 and sof641 in Fig. 2B, which were categorized into the FREE1-related mutants (Table S1); 145 candidates (Table S2) showed an unchanged FREE1 protein level including sof18 and sof30, which were grouped as the RNAi-related mutants. Representatives of both types of mutants with altered phenotypes are shown in Figs. S1 and S2, respectively.
Fig. 2. Representative sof mutants characterization with M3 seeds.
A: Seedling survival phenotype of putative sof mutants with M3 seeds plated on MS plates and MS plates supplied with 10 μmol/L DEX. Scale bar, 5 mm. B: Immunoblot analysis with anti-FREE1 antibody. FREE1 protein level was not changed in sof18 and sof30 with DEX, showing that they are RNAi-related mutants. Reduced FREE1 protein level was detected in sof220 and sof641 mutants, showing that they are FREE1-related mutants. Antibodies against the protein cFBPase served as loading control.
Table 1.
Summary of sof screening.
| Generation | Number |
|---|---|
| M1 | 10,000 |
| M2 | 1500 from ~40,000 |
| M3 | 84 FREE1-related |
| M3 | 145 RNAi-related |
For sof screening, 10,000 FREE1-RNAi M0 seeds were mutagenized with EMS, and 1500 sof mutants were screened out from about 40,000 M2 seeds. In total, about 230 mutants were identified for restoring the lethality phenotype in M3 generation. Western blot analysis using anti-FREE1 antibody with M3 generation plants further identified 84 FREE1-related suppressor mutants and 145 RNAi-related mutants.
Fragmented vacuole morphology is restored in FREE1-related sof mutants
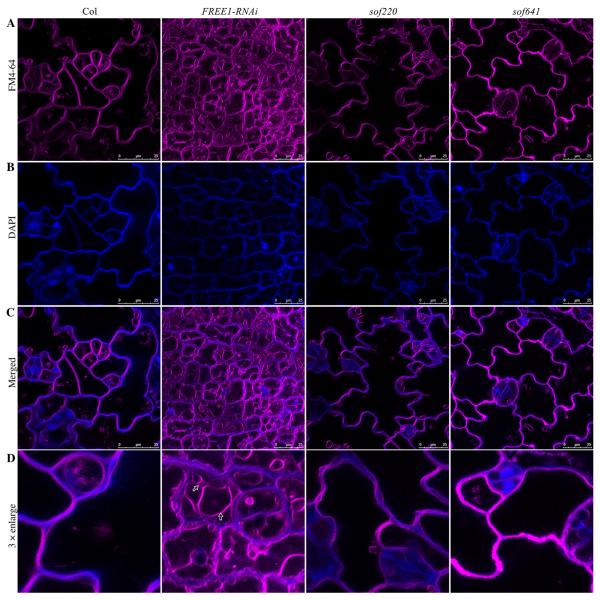
To test whether FREE1-related sof mutants were able to restore the fragmented vacuole phenotype of FREE1-RNAi plants, FM4-64 dye staining was performed to reflect the vacuole morphology. As shown in Fig. 3, upon DEX induction, the FREE1-RNAi plants showed numerous fragmented vacuoles (as indicated by arrows) while the wild type had a central large vacuole. It is possible that down-regulation of endogenous FREE1 may affect cell division or cell shape in the cotyledon epidermal cells compared to the wild-type cells. However, in the two FREE1-related sof mutants sof220 and sof641, vacuole morphology may have been restored to a large central vacuole. Similar patterns were observed in other FREE1-related candidates. Rapid DAPI staining was also applied for visualizing the cell shape (Spitzer et al., 2009).
Fig. 3. Vacuole morphology analysis of representative FREE1-related sof mutants.
A: Vacuole morphology was analyzed using FM 4-64 dye staining. Compared to the wild type plants, fragmented vacuoles were observed in FREE1-RNAi plants. FREE1-related sof mutants sof220 and sof641 showed large central vacuoles. B: The short-time DAPI staining was used to visualize the cell shape. C: Merged result of FM 4-64 dye staining and DAPI staining. Scale bar, 25 μm. D: 3 × enlargement of merged images. Arrows indicated the fragmented vacuoles in FREE1-RNAi plants.
NGS-mapping workflow
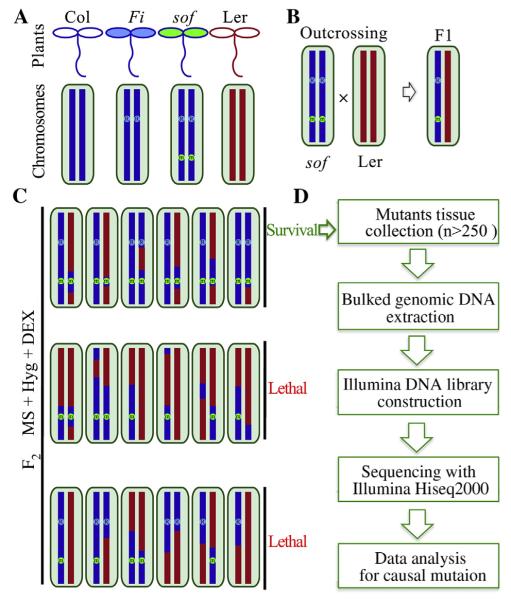
To identify the mutated SOF gene, an outcross-mapping population was established by crossing sof mutants to Landsberg erecta (Ler) wild type. The genome (representing all chromosomes) of Ler is marked in red and Col is marked in blue (Fig. 4A). Based on genetic analysis, FREE1-RNAi transgenic plants are confirmed to harbor a single-insertion (as indicated by R in blue, representing its insertion in certain chromosome). All sof mutants are assumed to harbor a mutation (as indicated by m in green, representing the possible location in any chromosomes) caused by EMS. The putative mutants were crossed with Ler and marked as F1 seeds, and F1 selfing generates F2 populations (Fig. 4B). The F2 seeds were screened on MS plates supplemented with DEX and Hyg. The F2 seeds with small scale (~100 seeds) were first analyzed to see whether that 3/16 of total seedlings was able to survive (for detailed genetic analysis, see Tables S3 and S4). With a 3/16 segregation ratio, we screened the F2 in large scale use a more strict selection to reduce the mis-scored individuals, then more than 250 seedlings showing a convincing surviving phenotype were selected from more than 2500 F2 seeds. The genomic DNA was used for library preparation and DNA sequencing (Fig. 4C and D).
Fig. 4. NGS-mapping workflow.
A: The genome of Ler is in red and Col is in blue. From genetic analysis, we know that FREE1-RNAi transgenic plants harbor a single-insertion that we marked as R in blue. All sof mutants are assumed to harbor a mutation (m in green) caused by EMS. B: The putative mutant was first crossed to the Ler wild type. F1 hybrids inbreeded and gave rise to a segregating F2 mapping population. C: The causal mutation was identified using segregating populations from F2 via outcrossing with Ler ecotype wild type. Survived seedlings from F2 Seeds were pooled together for genomic DNA on medium containing both hygromycin antibiotics and DEX. In the case of recessive sof mutations, this will include only homozygous mutants, which should be around 3/16 of all seeds. D: Illumina sequencing library was constructed using obtained bulked DNA. Whole genome sequencing was performed using Hiseq2000 platform. Further sequencing data analysis would uncover the causal mutation.
NGS-mapping analysis strategy and simulated results
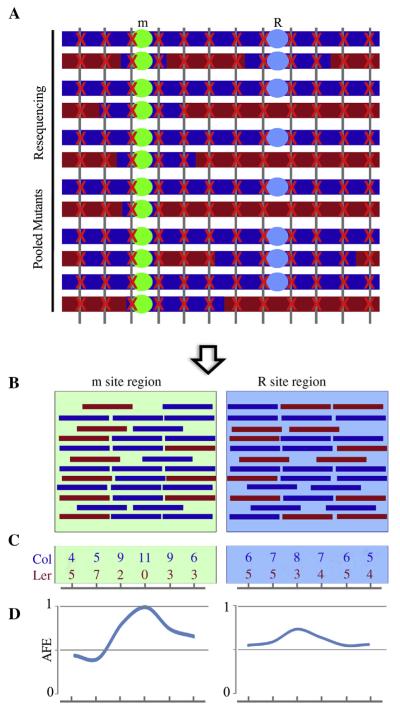
Instead of using traditional PCR-based mapping methods, we used NGS-mapping methods for gene identification. As shown in Fig. 5A, it is clear that the m site (sof suppressor mutation site) was enriched during the sample pooling process, and close to the m site, there will be an over-representation of Col background. Parental genotypes Col and Ler are known, then mapping-by-sequencing can be carried out on the basis of allele frequency estimations (AFE). The mutant AFE was calculated as the percentage of alleles from Col divided by all alleles aligned to the respective chromosome region (both Col and Ler). The Col allele frequency peak should correspond to the causal mutation m site. When the simulated reads (Fig. 5B and C) were plotted, the significant peak representing the enriched m site appeared (Fig. 5D). The fine mapping interval resides within the peak region that showed a high Col allele frequency, and the causal mutation will be identified within the peak regions.
Fig. 5. Analysis strategy and simulative results for mapping-by-sequencing.
A: Schematic of mapping mutants and whole genome sequencing. The m site was selected during the mapping sample pooling process. They are colored after the parental genomes from which they were sampled. The red cross marks reads hit along with the genome. B: Simulative reads aligned to m site region and R site region. Reads in red stand for Ler genome and reads in blue stand for Col genome. C: Examples of reads counts from simulate reads alignment to parental genomes. D: Allele frequency estimations of both m site region and R site region. One Col allele frequency peak should correspond to the causal m site region, while a mild Col allele frequency peak may appear at R site region.
Comparing to the recessive m site, other sites that unlinked with m site should mostly show a Col allele frequency of around 0.5 due to recombination events. However, the dominant R site (FREE1-RNAi T-DNA insertion site) is a little different among all sites unlinked with m site. As illustrated before (Fig. 4C), theoretically the selected F2 should contain mutants that are homozygous at m site (all), homozygous at R site (1/3) and heterozygous mutants at R site (2/3), which means probably there is a mild peak corresponding to the R site (Fig. 5C and D).
Sequencing results of two sof mutants
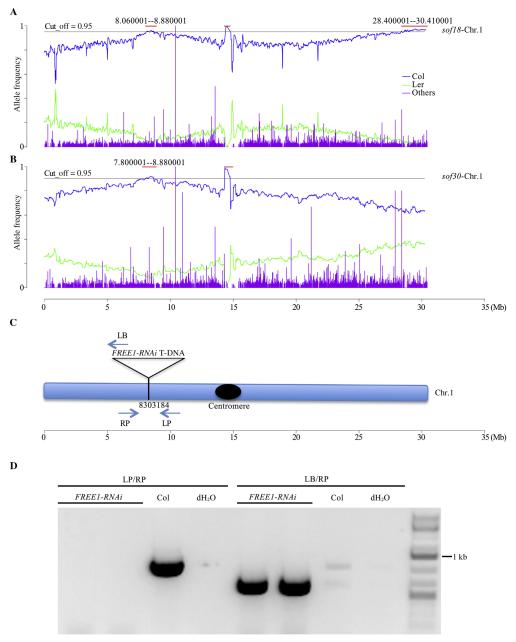
As proof-of-principle, two RNAi-related sof mutants sof18 and sof30 were outcrossed to Ler, and the F2 seeds were selected and sequenced following the previously described workflow (Fig. 4 and Table S4). With the whole-genome sequencing sample, the NGS-Mapping were carried out based on 461,070 SNP markers (Galvao et al., 2012). As expected, a peak specific to sof18 was found located on the far right arm of chromosome 1 (Fig. S3), while the sof30 specific peak located to chromosome 5 (Fig. S4). For each mutant, the fine mapping identified the candidate genes (with none of them published before) bearing meaningful mutations caused by EMS. As to these two novel mutants, further characterizations are in need to illustrate how they are involved in interrupting the FREE1-RNAi process. Meanwhile, for both sof18 and sof30, shared peaks appeared on the left arm of chromosome 1, which indicates the R site of FREE1-RNAi T-DNA (Fig. 6A and B).
Fig. 6. Sequencing results of sof18 & sof30 identified the FREE1-RNAi T-DNA insertion site.
A: Allele frequency analysis result of sof18 on chromosome 1. Two Col-allele peaks located on chromosome 1, the peak appeared in the middle region due to centromere, and the far right peak is specific to sof18. B: Allele frequency analysis result of sof30 chromosome 1. One Col-allele peak located on chromosome 1, and shared by sof18 with the left peak. C: Shared peak on chromosome 1 between sof18 and sof30 corresponded to the R site (FREE1-RNAi T-DNA insertion site), which was the insertion site of FREE1-RNAi T-DNA. D: Genomic DNA PCR confirmed the FREE1-RNAi T-DNA insertion site.
Identification of the FREE1-RNAi T-DNA insertion site by chimeric reads from whole genome sequencing
To confirm that the shared peaks correspond to the R site, we identified the chimeric reads containing sequence of both FREE1-RNAi T-DNA and Col genomic DNA from NGS data of sof30. Chimeric reads alignment to genome found that the chimeric reads located on chromosome 1 at position 8,303,184, which supports the expected R site at this shared peak region (Fig. 6C). The PCR amplified product was further subjected to Sanger sequencing using flanking primers LB and RP, and verified the FREE1-RNAi T-DNA insertion site at position 8,303,184 (Fig. 6D).
DISCUSSION
The significance of FREE1-related sof mutants
In Arabidopsis, forward genetic screening has identified mutants in development and signaling transduction pathways, and has also been applied to membrane trafficking studies (Page and Grossniklaus, 2002). For example, BEN1 was identified through screening for mutants with defects in constitutive endocytosis of the PIN1 auxin transporter (Tanaka et al., 2009). As always, enhancer and suppressor screening is helpful in discovering important players in the same pathway. For example, the identification of NPY1 as yuc1yuc4 enhancer defined its critical role in auxin pathway (Cheng et al., 2007); the identification of the E3 ligase SP1 as suppressor of the Arabidopsis plastid protein import mutation ppi1 led to the discovery of the ubiquitin-proteasome system in regulation of plastid development (Ling et al., 2012). However, it is hard to apply suppressor screening to lethal mutant plants. Here by using inducible FREE1-RNAi plants, we carried out a suppressor screening to identify new components involved in FREE1-mediated membrane trafficking pathways.
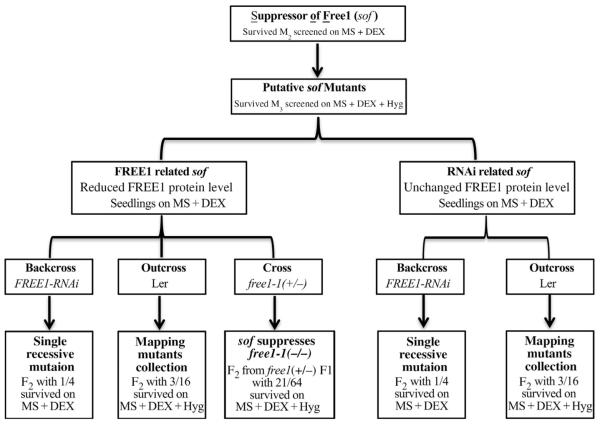
As summarized in Fig. 7, based on the FREE1 protein levels after DEX induction, putative sof mutants were categorized into FREE1-related and RNAi-related groups. Confirmed mutants were backcrossed with FREE1-RNAi to ascertain whether it was a single recessive or dominant mutation (Table S3), and outcrossed with Ler for gene identification (Table S4). For FREE1-related sof mutants, a cross with free1 (T-DNA heterozygous mutants) mutants further verified the rescue of free1 by certain sof mutants (Table S5). For screening of FREE1-related sof mutants via Western blot, we also screened out the mutants that were disrupted in the FREE1-RNAi process as we named RNAi-related sof mutants. To these mutants, detection of FREE1-siRNA with DEX will classify them into two groups, one group with no FREE1-siRNA detected while the other group showed no defects in FREE1-siRNA production. Further identification of these mutants will surely extend our current knowledge about silencing pathways including transgene suppression, siRNA biogenesis and post-transcriptional gene silencing.
Fig. 7. Flowchart summarizing the whole process of screening.
To further uncover the functional mechanism of FREE1, suppressors of free1 (sof for short) were screened using EMS-treated M2 seeds of a single insertion Dexinducible FREE1-RNAi line. Putative M3 seeds were classified into FREE1-related and RNAi-related groups through FREE1 protein level detection. Mutants with reduced FREE1 protein level were termed as FREE1-related sof mutants while those with no changes in FREE1 protein levels were termed RNAi-related sof mutants. For both FREE1-related and RNAi-related sof mutants, a backcross with FREE1-RNAi line was used to detect whether the mutation was a single recessive mutation. To map the mutated gene, outcrossing with Ler was used to establish the mapping mutants pool for both FREE1-related and RNAi-related sof mutants. For FREE1-related sof mutants, a cross with free1-1 T-DNA knockout line was used to verify that the sof mutation was able to suppress the lethal phenotype of free1-1.
Successful isolation of the salt-tolerance mutant sltA in Aspergillus nidulans for the rescue of null mutations in ESCRT-0, I, II and III genes has linked sodium homeostasis with the ESCRT pathway, although the mechanism remains unclear (Calcagno-Pizarelli et al., 2011). As Arabidopsis lacks an SLTA ortholog, it is significant to identify new plant-specific components to help us in understanding the mechanisms for the roles of FREE1 in the plant ESCRT machinery and also in the biogenesis of MVBs. At the whole plant level, we expect to understand the reasons why FREE1 deficient plants are lethal through further studies on the newly identified genes. This will surely allow us to have a better understanding of the role of FREE1 mediated membrane trafficking in early seedling development.
The advantages and problems of this suppressor screen
The severe DEX-inducible lethal phenotype of the FREE1-RNAi seedling provides an excellent basis to genetically dissect the FREE1-mediated membrane trafficking pathway through suppressor screening. Accordingly, we screened the ethyl methanesulfonate (EMS) mutagenized inducible FREE1-RNAi plants for suppressors.
The advantages of using inducible FREE1-RNAi plants include the followings: 1) Since the lethality caused by FREE1-RNAi is dominant, outcrosses of these candidate mutants into the Ler wild-type have allowed us to collect true mutants for gene identification (Table S2); 2) Since the lethal phenotype is inducible, crosses of putative FREE1-related sof mutants with free1 T-DNA insertion mutants have enabled us to identify true sof suppressors in FREE1 knock out mutants (Table S5); 3) We have used next-generation sequencing technology to facilitate the identification of the causal gene(s) in a timesaving and reliable manner.
On the other hand, the screening had the following problems. First, the classification of FREE1-related and RNAi-related mutants was mainly based on FREE1 protein detection with DEX. In our experience, it is hard to differentiate RNAi-related mutants from FREE1-related mutants when the reduction of FREE1 was not significant. Second, some of the sof mutants were not able to reach a flowering stage, making it difficult to identify the gene.
Future work
Our on-going and future work will mainly focus on gene identification and a functional analysis of the putative FREE1-related sof mutants as well as some interesting RNAi-related sof mutants. These mutants are currently being processed for gene identification. Characterizing these mutants will help us gain valuable insights into how FREE1 functions in coordinating normal membrane trafficking pathways and organelle biogenesis in relation to plant growth and development.
In addition, the application of this fast suppressor mutant screening approach can also be applied to other mutants involved in membrane trafficking and organelle biogenesis in plants. Indeed, we have recently extended such approach to screen for the mon1 repressor mutants for new components in regulating vacuole biogenesis (Cui et al., 2014); and suppressor screening using SH3P2-RNAi line for new modules in the autophagic pathway (Zhuang et al., 2013; Zhuang and Jiang 2014); as well as suppressor screening using EXPO-or exocyst-related mutants for the EXPO-mediated unconventional protein secretion pathways in plants (Wang et al., 2010; Ding et al., 2012, 2014a, 2014b).
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis wild type (Col) plants were transformed with a pTA7002 construct (Aoyama and Chua, 1997) harboring a hairpin FREE1-RNAi cassette (Wesley et al., 2001), and hygromycin resistant single insertion FREE1-RNAi transgenic plants were used for mutagenesis. The previously described free1 mutant was designated free1-1 (Gao et al., 2014b). Seeds were surface sterilized and grown on plates with full Murashige and Skoog (MS) salts (pH adjusted to 5.7 with 0.1 mol/L KOH) plus 3% sucrose and 0.8% agar at 22°C under a long-day (16 h light/8 h dark) photoperiod. For induction of FREE1 RNAi, 10 μmol/L DEX (10 mmol/L stock dissolved in ethanol) was added in the medium.
Mutants pool establishment
The mutant pool was established following a previous report with mild modification (An et al., 2010, Zhang et al., 2015). About 10,000 high quality seeds from single insertion FREE1-RNAi transgenic plants were soaked at 4°C in 40 mL dH2O for 60 h, and then washed twice with 50 mL dH2O. Washed seeds were treated with 0.3% EMS for 8 h with gentle shaking (~140 r/min) at room temperature. The seeds were then washed ten times with dH2O (50 mL × 10). M1 seeds were sprayed into 20 large square ports. For M2 seeds collection, M1 plants were divided into 283 subfamilies.
Phenotype screening for mutant isolation
M2 seeds were surface-sterilized and sprayed on MS medium plus with 10 μmol/L DEX. Plates were kept at 4°C in dark for two days before placing in growth chambers. 5-day-old seedlings were screened for survived phenotypes. Selected M2 seedlings were planted into soil for individual M3 seeds collection. Individual M3 seeds were screened on MS medium plus with 10 μmol/L DEX and hygromycin, and 5-day-old seedlings M3 seedlings were screened for survived phenotype.
Protein extraction and Western blot analysis
For total protein extraction from plants, whole seedlings tissues were ground in liquid nitrogen and extracted with a lysis buffer containing 25 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 1 × Complete Protease Inhibitor Cocktail (Roche, USA) and 1% Triton X-100. The protein blot was hybridized with rabbit FREE1 antibodies (Gao et al., 2014b) and rabbit cFBPase antibodies (Agrisera, Cat No. AS04 043, SWEDEN).
FM-DYE staining and confocal microscopy analysis
FM-DYE staining was performed as described (Lam et al., 2008). Stock solutions of FM4-64 (12 mmol/L in DMSO; Invitrogen, USA) were prepared and stored at −20°C. Seedlings were first washed with MS liquid medium and stained for 3 h with FM4-64 which was diluted to working solution at 12 μmol/L with MS liquid medium just before use. After staining, the seedlings were washed twice with fresh medium before confocal imaging. Rapid DAPI staining was carried as described (Spitzer et al., 2009). Images were collected using a 63 × objective water lens in the Leica SP8 Confocal system.
RNA extraction and RNA Northern blot
RNA isolation and northern blotting detection of FREE1-siRNAs were performed following a standard protocol as previously described (Park et al., 2002, Li et al., 2013). In brief, total RNAs of 10-day light-grown seedlings from DEX and MS conditions were extracted using TRIzol reagent. Probes against FREE1 siRNAs were labeled using 5′-end-labeling (32P) to detect siRNAs from total RNAs.
DNA extraction, library preparation and Illumina sequencing
Genomic DNA was prepared using DNeasy Plant Mini Kit (Qiagen, Cat No. 69104, USA) following the manufacturer’s instruction. These sheared DNAs were sequentially ligated with the 3′ and 5′ adapters using the DNA library Preparation Kit (Illumina, USA) according to the manufacturer’s instructions. The libraries were barcoded and sequenced on an Illumina Hiseq2000. The whole sequencing process was carried out following previous reported procedures (Zhong et al., 2013).
Sequence data analysis
For reference genome, the Col-0 (TAIR10) sequence was used. Both sof18 and sof30 are in Col background. Sequencing reads of sof18 and sof30 were aligned against the reference genome sequence (TAIR10) using SOAP2 (http://soap.genomics.org.cn/soapaligner.html), and consensus was called using SAM-TOOLS program (Li et al., 2009). NGS-Mapping was carried out based on 461,070 SNP markers using SHOREmap outcross function (Schneeberger et al., 2009; Galvao et al., 2012). Relatively reliable loci were filtered as below: consensus quality >20 (error rate, 1%), total depth >5. Only EMS induced C/G to T/A SNP markers were further considered as candidates.
Identification of the FREE1-RNAi T-DNA insertion site from whole genome sequencing
The site of the FREE1-RNAi T-DNA insertion was determined using the genome sequencing data from the bulk F2 mapping population of sof30. The strategy was to identify chimeric reads with both T-DNA and genome DNA sequences. The reads from the sof30 population were mapped to the Arabidopsis genome using BWA (Li and Durbin 2009) without mismatches. The unmapped reads were mapped back to the T-DNA insertion sequence using Blast (Zhang and Madden 1997). The reads containing perfect matches to the T-DNA sequence longer than 25 nt and shorter than 76 nt were retained. The unmapped parts of the retained reads were obtained and remapped to the Arabidopsis genome. In this way, the reads containing partial matches to the Arabidopsis genome were obtained, which are chimeric reads containing both T-DNA and flanking genomic DNA sequences.
Sequencing of FREE1-RNAi T-DNA insertion site
Primers (LP_5′-GCTTCTCGAAACCCATTTCATC-3′ and RP_5′-GAGCTTCAGGCTTCATAGGT-3′) flanking the T-DNA insertion site were used to amplify wild type fragment. Primers (LB_5′-ATTTCGGAACCACCATCAAACAG-3′ and RP_5′-GAGCTTCAGGCTTCATAGGT-3′) were used to amplify the mosaic fragment. The mosaic fragment was sequenced using standard Sanger sequencing methods.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH GM114660 to Y. Zhao, the Research Grants Council of Hong Kong (CUHK466011, 465112, 466613, CUHK2/CRF/11G, C4011-14R and AoE/M-05/12), NSFC/RGC (N_CUHK406/12), NSFC (31270226 and 31470294), and Shenzhen Peacock Project (KQTD201101) to L. Jiang.
Abbreviations
- AFE
allele frequency estimation
- DEX
dexamethasone
- ESCRT
endosomal sorting complex required for transport
- FREE1
FYVE domain protein required for endosomal sorting 1
- MVB
multivesicular body
- NGS
next generation sequencing
- sof
suppressor of free1
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgg.2015.03.012.
REFERENCES
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, Zhang S, Ecker JR, Guo H. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Bar M, Avni A. Endosomal trafficking and signaling in plant defense responses. Curr. Opin. Plant Biol. 2014;22:86–92. doi: 10.1016/j.pbi.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Bassham DC, Brandizzi F, Otegui MS, Sanderfoot AA. The secretory system of Arabidopsis. Arabidopsis Book. 2008;6:e0116. doi: 10.1199/tab.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhuang XH, Gao CJ, Wang XF, Jiang LW. The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 2014;165:1328–1343. doi: 10.1104/pp.114.238378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno-Pizarelli AM, Hervas-Aguilar A, Galindo A, Abenza JF, Penalva MA, Arst HN., Jr. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J. Cell Sci. 2011;124:4064–4076. doi: 10.1242/jcs.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L. Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell. 2014;26:2080–2097. doi: 10.1105/tpc.114.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F, Grunt M, Bezvoda R, Hala M, Kulich I, Rawat A, Zarsky V. Evolution of the land plant exocyst complexes. Front. Plant Sci. 2012;3:159. doi: 10.3389/fpls.2012.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang J, Wang JQ, Stierhof YD, Robinson DG, Jiang LW. Unconventional protein secretion. Trends Plant Sci. 2012;17:606–615. doi: 10.1016/j.tplants.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Ding Y, Wang J, Lai JHC, Chan VHL, Wang XF, Cai Y, Tan XY, Bao YQ, Xia J, Robinson DG, Jiang LW. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell. 2014a;25:412–426. doi: 10.1091/mbc.E13-10-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Robinson DG, Jiang L. Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 2014b;29:107–115. doi: 10.1016/j.ceb.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Galvao VC, Nordstrom KJ, Lanz C, Sulz P, Mathieu J, Pose D, Schmid M, Weigel D, Schneeberger K. Synteny-based mapping-by-sequencing enabled by targeted enrichment. Plant J. 2012;71:517–526. doi: 10.1111/j.1365-313X.2012.04993.x. [DOI] [PubMed] [Google Scholar]
- Gao C, Cai Y, Wang Y, Kang BH, Aniento F, Robinson DG, Jiang L. Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci. 2014a;19:508–515. doi: 10.1016/j.tplants.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 2014b;24:2556–2563. doi: 10.1016/j.cub.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Cui Y, Fu X, He Y, Zhao Q, Zeng Y, Shen J, Luo M, Jiang L. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA. 2015;112:1886–1891. doi: 10.1073/pnas.1421271112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lam SK, Tse YC, Robinson DG, Jiang L. Tracking down the elusive early endosome. Trends Plant Sci. 2007;12:497–505. doi: 10.1016/j.tplants.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Lam SK, Cai Y, Hillmer S, Robinson DG, Jiang L. SCAMPs highlight the developing cell plate during cytokinesis in tobacco BY-2 cells. Plant Physiol. 2008;147:1637–1645. doi: 10.1104/pp.108.119925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KF, Dacks 25 JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing, S The Sequence Alignment/Map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, Zhang F, Raikhel N, Jiang L, Chen X. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling QH, Huang WH, Baldwin A, Jarvis P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science. 2012;338:655–659. doi: 10.1126/science.1225053. [DOI] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151:859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U. The art and design of genetic screens: Arabidopsis thaliana. Nature. 2002;3:124–136. doi: 10.1038/nrg730. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Buono R, Otegui MS. Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 2011;14:666–673. doi: 10.1016/j.pbi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Samaj J, Muller J, Beck M, Bohm N, Menzel D. Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 2006;11:594–600. doi: 10.1016/j.tplants.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell. 2009;21:749–766. doi: 10.1105/tpc.108.064865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kitakura S, De Rycke R, De Groodt R, Friml J. Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 2009;19:391–397. doi: 10.1016/j.cub.2009.01.057. [DOI] [PubMed] [Google Scholar]
- Teh OK, Hofius D. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 2014;65:1297–1312. doi: 10.1093/jxb/ert441. [DOI] [PubMed] [Google Scholar]
- Wang J, Ding Y, Wang JQ, Hillmer S, Miao Y, Lo S, Wang X, Robinson DG, Jiang L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell. 2010;22:4009–4030. doi: 10.1105/tpc.110.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Madden TL. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Ji Y, Wen X, Zhang C, Zhao Q, Wang Y, Lu J, Guo H. Suppression of endogenous act-siRNA-mediated gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;348:120–123. doi: 10.1126/science.aaa2618. [DOI] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni JJ. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25:4596–4615. doi: 10.1105/tpc.113.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Jiang L. Autophagosome biogenesis in plants: roles of SH3P2. Autophagy. 2014;10:704–705. doi: 10.4161/auto.28060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.