Abstract
The formation of the organized bacterial community called biofilm is a crucial event in bacterial physiology. Given that biofilms are often refractory to antibiotics and disinfectants to which planktonic bacteria are susceptible, their formation is also an industrially and medically relevant issue. Pseudomonas aeruginosa, a well-known human pathogen causing acute and chronic infections, is considered a model organism to study biofilms. A large number of environmental cues control biofilm dynamics in bacterial cells. In particular, the dispersal of individual cells from the biofilm requires metabolic and morphological reprogramming in which the second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) plays a central role. The diatomic gas nitric oxide (NO), a well-known signaling molecule in both prokaryotes and eukaryotes, is able to induce the dispersal of P. aeruginosa and other bacterial biofilms by lowering c-di-GMP levels. In this review, we summarize the current knowledge on the molecular mechanisms connecting NO sensing to the activation of c-di-GMP-specific phosphodiesterases in P. aeruginosa, ultimately leading to c-di-GMP decrease and biofilm dispersal.
PSEUDOMONAS AERUGINOSA: A MODEL SYSTEM FOR BIOFILM RESEARCH
The ubiquitous Gram-negative bacterium Pseudomonas aeruginosa is an opportunistic pathogen responsible for both acute and chronic infections. P. aeruginosa chronic lung infections are the major cause of death in cystic fibrosis (CF) patients, a genetic disease affecting 1/2,500 newborns in Europe (1). P. aeruginosa is frequently resistant to conventional antibiotic therapy and to host antimicrobial effector mechanisms and forms biofilms in many infection sites, including CF lung chronic infections (2). Therefore, this bacterium has become a model system for biofilm research.
The life span of a P. aeruginosa biofilm can be divided into several stages, starting with the reversible interaction of a planktonic cell with a surface (3). The cells subsequently attach to the surface and mature into three-dimensional mushroomlike structures, also known as microcolonies (4). This process is accompanied by the production of an extracellular hydrated matrix, also known as extracellular polymeric substance (EPS), composed of exopolysaccharides, extracellular DNA (eDNA), proteins, amyloid fibers, and bacteriophages, in which the bacteria are embedded (5, 6). P. aeruginosa produces three secreted polysaccharides, Pel, Psl, and alginate, each of which provides different physiological properties to the matrix (7). eDNA is also considered an important component of the bacterial matrix, particularly during early stages of biofilm formation (3). Overall, the extracellular matrix facilitates the formation of three-dimensional structures that give the bacteria increased access to nutrients and advantages of multicellular living and protects the bacteria from outside predators and toxic chemicals, including antibiotics.
Many adaptive traits may arise during P. aeruginosa infections, which may favor biofilm formation: these novel traits include nonmotility, small-colony variant formation, deficiency in the population density-sensing system known as quorum sensing (QS), changes in the chemical structure of the membrane, and increased mutation rates (8). Bacteria within a biofilm are significantly more tolerant of antibiotic treatment than their planktonic counterparts (9, 10).
During dispersal, motile cells from microcolonies leave the biofilm to colonize new locations (4, 11). Dispersed cells are often considered identical to planktonic ones; however, recent reports support earlier findings that dispersed cells represent a specific intermediate state between biofilm and planktonic lifestyles in P. aeruginosa (4, 12). This proposal is mainly based on the observation that the physiology and virulence of dispersed cells are highly different from those of biofilm and planktonic ones (12, 13). The dispersal phenotype can be achieved and maintained by either the addition of a dispersing agent or the overexpression of a phosphodiesterase (PDE) (see below) (12).
BIOFILM DISPERSAL
While passive detachment of bacterial cells is triggered by external physical forces and can occur during all stages of biofilm development, active dispersal is initiated by the bacteria themselves and can be specifically triggered by certain environmental cues in various bacterial species. These include the availability of nutrients (i.e., carbon and iron) (11, 14–18) and oxygen (19), changes in temperature (20, 21), and low levels of exogenous and endogenous nitric oxide (NO) (22–25). In addition, native dispersal has also been described which involves the self-synthesized signaling molecule cis-2-decenoic acid (26). Active dispersal is specific and tightly regulated and involves modulation of the intracellular concentration of the second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), which also plays a role in biofilm formation (5, 10).
ROLE OF c-DI-GMP IN BIOFILM FORMATION AND DISPERSAL
In recent years, it became clear that the second messenger c-di-GMP is a key player in the regulation of biofilm formation and dispersal (27). While high levels of c-di-GMP promote the sessile lifestyle and the formation of biofilms, low levels of c-di-GMP lead to biofilm dispersal and favor the planktonic lifestyle. In response to environmental cues, the intracellular level of this second messenger is modulated by enzymes that build (diguanylate cyclases [DGCs]) or break (phosphodiesterases [PDEs]) c-di-GMP. These enzymes possess characteristic domains with conserved amino acid motifs: GGDEF domains in DGCs and EAL or HD-GYP domains in PDEs (27). GGDEF and EAL/HD-GYP domains are often part of multidomain proteins that harbor additional signal input or transduction domains (i.e., PAS [Per Arnt Sim], GAF [found in cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA], and REC [signal receiver] domains) (27). This organization implies that the upstream signal input domains may regulate the activity of the catalytic domains, and indeed, several reports support this hypothesis (28, 29). The GGDEF and EAL domains are not only linked to signal input domains but are often found fused in a single polypeptide chain. These proteins are either bifunctional or one of the domains is catalytically inactive and adopts a new regulatory function (30, 31). Interestingly, one single organism usually harbors more than one each of putative DGCs and PDEs, indicating a certain amount of redundancy; it is also evident that specific DGCs and PDEs may control distinct phenotypes. DGC and PDE activities are able not only to control the global intracellular level of c-di-GMP but also to regulate localized pools of the second messenger, for example, during cell division (32, 33); the presence of target receptors (like PilZ domains) thus leads to the specific cellular responses (34).
Given that NO is able to induce dispersal of biofilms formed by several bacterial species, including P. aeruginosa, Escherichia coli, Vibrio cholerae, Staphylococcus epidermidis, Bacillus licheniformis, Serratia marcescens, Legionella pneumophila, Nitrosomonas europaea, and Neisseria gonorrhoeae (23, 35–38), a wealth of studies has been conducted in the last 5 years addressing the effectiveness of NO-based antibiofilm strategies. Several classes of NO-releasing compounds active in biofilm modulation have been found, whose properties were very recently reviewed by Barraud et al. (39).
Within this minireview, we focus mainly on the effect of NO on P. aeruginosa biofilm dispersal, the origin and perception of NO, and the subsequent effect of NO on c-di-GMP levels, trying to summarize what is known on the molecular mechanisms underlying this complex biological process.
PROPERTIES AND SOURCES OF NO
Nitric oxide is a colorless diatomic gas with the chemical formula NO. Due to an unpaired electron, NO is a free radical with a half-life of only a few seconds, causing lipid peroxidation, nitration, and S-nitrosylation of proteins (40). NO readily interacts with transition metals found in metalloproteins (41). This topic has been intensely studied, for instance, for the interaction of NO with the heme-iron of hemoglobin (42). In addition, NO is a signaling molecule in vertebrates, controlling a plethora of biological processes, such as muscle relaxation leading to vasodilation and increased blood flow. In the latter example, the receptor for NO is also a heme-containing protein, i.e., soluble guanylate cyclase, which upon NO binding is activated to produce cyclic GMP (cGMP) (43).
In bacterial systems, including P. aeruginosa, NO can be produced directly from the microorganism itself, as an intermediate or by-product of specific metabolic pathways, such as denitrification, a form of anaerobic respiration. NO may also come from outside the bacterial cell, produced by eukaryotic host cells that attack pathogens with NO (44, 45). To characterize the response of biofilms formed by several bacteria to exogenous NO, different NO-releasing compounds have been employed, mostly under aerobic conditions. These compounds release variable amounts of NO, ranging from low nanomolar to micromolar concentrations, with different half-lives, mainly depending on their kinetics of NO release. In P. aeruginosa, biofilm dispersal has been shown to occur in the presence of exogenous compounds spontaneously releasing NO, such as sodium nitroprusside (SNP), 6-(2-hydroxy-1-methyl-2-nitrosohydrazino)-N-methyl-1-hexanamine (MAHMA NONOate) or aminoxyl free radicals (nitroxides) (reference 39 and references therein). SNP as an NO donor has been shown to deliver an effective NO concentration about 1,000-fold lower than the concentration of the donor over several minutes (up to 30 min) (22), whereas the half-life of MAHMA NONOate is much shorter (1 to 2 min) (39). More recently, prodrugs releasing NO after an activation step in the bacterium, such as diethylamine (DEA) NONOate-cephalosporin prodrug (DEACP), were also shown to promote dispersal (46, 47).
ENDOGENOUS SOURCES OF NO IN P. AERUGINOSA AND THEIR EFFECTS ON BIOFILM
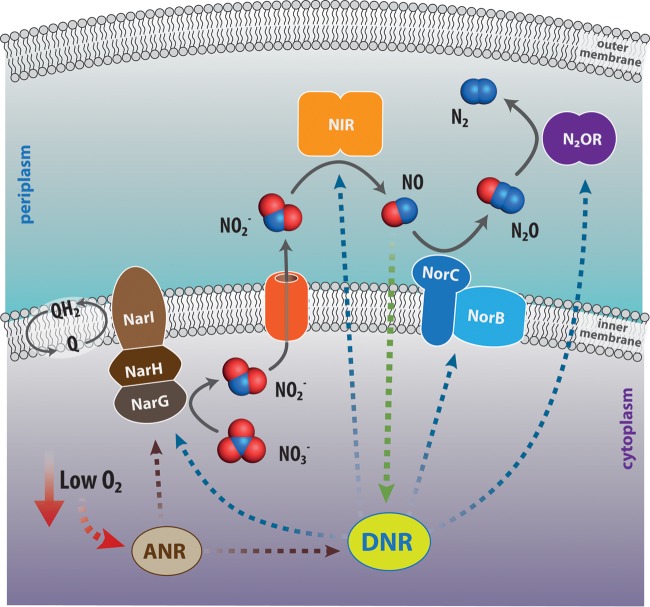
P. aeruginosa is a facultative anaerobe, able to respire under anaerobic conditions in the presence of the alternative electron acceptors nitrate and nitrite, employing denitrification. In this metabolic pathway, nitrate is reduced to NO and then to N2 in four reaction steps, each catalyzed by a specific reductase (Fig. 1) whose expression is tightly controlled, mainly by the arginine nitrate regulation (ANR) and dissimilative nitrate respiration regulator (DNR) transcription factors (48–53). Two putative nitrate reductases are present in P. aeruginosa, i.e., the inner membrane-bound nitrate reductase NarGHI (Fig. 1), encoded within the narK1K2GHJI operon, and the periplasmic nitrate reductase NapAB, encoded within the napEFDABC operon. NarGHI has been shown to be the predominantly expressed nitrate reductase under anaerobic conditions in the presence of nitrate (54). Nitrite reductase, the bacterial enzyme that reduces nitrite to NO during denitrification, may contain either copper or heme. The corresponding nirK and nirS genes are found in both Gram-negative and Gram-positive bacteria, with nirS being the most frequent one. Denitrification has been implicated in the virulence of several bacterial species, including members of the Brucella, Pseudomonas, and Neisseria genera (55–57).
FIG 1.
Scheme of the denitrification pathway in Pseudomonas aeruginosa. In denitrification, a form of anaerobic respiration, nitrate (NO3−) is reduced to molecular nitrogen (N2) in four reductive steps, each catalyzed by a specific reductase, namely, nitrate reductase (NAR), nitrite reductase (NIR), NO reductase (NOR), and nitrous oxide reductase (N2OR). The expression of these enzymes is mainly controlled by two transcriptional regulators that sense low-oxygen conditions (ANR, an iron-sulfur protein) and NO (DNR, an heme-containing protein), respectively. Q, coenzyme Q or ubiquinone (2,3-dimethoxy-5-methyl-6-multiprenyl-1,4-benzoquinone); QH2, reduced form of ubiquinone, i.e., ubiquinol.
In P. aeruginosa, the enzyme responsible for nitrite reduction to NO (Fig. 1) is cytochrome cd1 nitrite reductase (hereinafter NIR), a homodimer containing one c-heme and one d1-heme group in each subunit (58), belonging to the NirS family of nitrite reductases (59). The d1-heme is a partially saturated macrocycle, unique to this enzyme and synthesized by a specialized pathway present only in denitrifiers (strongly induced in P. aeruginosa upon nitrite treatment): this specialized cofactor is required for the efficient release of NO (60).
The last two enzymes in the denitrification chain are the membrane-bound NO reductase (NorCB) (61) and the periplasmic copper-containing N2O reductase (62), reducing nitrous oxide (N2O) to molecular nitrogen (Fig. 1).
Several studies suggest the involvement of the NO produced by denitrification in biofilm dynamics and virulence in P. aeruginosa; these studies have been carried out under different environmental conditions, including oxygen availability. Under aerobic conditions, nitrate sensing and metabolism control swarming motility, a type of motility which requires the production of flagella, type IV pili, and the biosurfactant rhamnolipid (encoded by the rhlAB operon) and affect biofilm architecture, showing a central role for NIR-derived NO in these events (57, 63). In addition, biofilm dispersal under aerobic conditions requires NIR activity and NO production (22, 38, 57): a P. aeruginosa strain lacking NIR does not disperse in the presence of SNP, whereas a NO reductase mutant exhibits a hyperdispersal phenotype (38). Under these experimental conditions, NO affects the expression or activity of proteins related to motility (such as those encoded by pilA and rhlAB) (22, 63).
The overall picture is somewhat different under strict anaerobic conditions, when NO accumulation may ultimately favor biofilm formation (64). As shown by Yoon and coworkers and confirmed in a more recent study (64, 65), P. aeruginosa PAO1 cells grown anaerobically are more elongated than that grown aerobically and easily form highly cohesive clumps, yielding a robust biofilm. Cell elongation is dependent on the presence of NIR and is repressed in the presence of an NO antagonist. Under low-oxygen tension, the nonelongated NIR-deficient mutant failed to form biofilm, while the wild-type PAO1 was highly elongated and formed robust biofilm.
Very recently, NIR was also shown to control flagellum production and swimming motility under anaerobic conditions, independently from its ability to produce NO, by serving as a scaffold to form a ternary complex with the chaperone DnaK and the flagellar protein FliC in the periplasm (66). In the same study, the nirS mutant showed a partial restoration of swimming ability under aerobic conditions, whereas swarming mobility was previously shown to be affected under aerobiosis (63). An intriguing possibility suggested by the latter study, which will require further investigation, is that NIR promotes motility in different ways, depending on oxygen availability, either by producing NO to increase rhamnolipid synthesis or activate other signaling pathways, or by directly controlling flagellum formation by protein-protein interactions. As underlined by Borrero-de Acuna et al. (66), to fully understand these phenomena, the assembly of flagella under aerobic and anaerobic conditions will have to be analyzed in further detail. Since it is known that NO transcriptionally activates nirS expression, the production of a catalytically inactive version of NIR may also be a route for motility control by exogenously added NO donors.
Another interesting link between denitrification and biofilm modulation in P. aeruginosa involves the cell-to-cell communication signaling systems collectively known as quorum sensing (QS) systems that coordinate gene expression in response to population density. P. aeruginosa possesses at least three different QS systems: two N-acyl-l-homoserine lactone (AHL) signals, the LasR-LasI (las) and RhlR-RhlI (rhl) systems (67), and a third one, 2-heptyl-3-hydroxy-4-quinolone, referred to as the Pseudomonas quinolone signal (PQS) (68). LasI directs the synthesis of the AHL signal N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), and RhlI directs the synthesis of another AHL signal, N-butyryl-l-homoserine lactone (C4-HSL). The transcription regulatory proteins LasR and RhlR are specifically activated by 3-oxo-C12-HSL and C4-HSL, respectively.
Regulation of denitrification by QS may thus be important in both aerobically and anaerobically grown biofilms. The denitrification pathway is regulated by both the AHL and PQS systems via different mechanisms, including transcriptional regulation and iron chelation (38, 69–72). C4-HSL represses denitrification via its cognate regulator RhlR (69); on the other hand, the addition of PQS to the growth medium specifically promotes NO accumulation (70), thus suggesting a possible mechanism of endogenous regulation of biofilm dispersal (38). Anr, the oxygen-responsive transcription factor controlling the expression of denitrification genes, also controls QS in biofilms under low-oxygen conditions (1%) (73).
It is noteworthy that bacterial species devoid of nirK and nirS are also able to reduce nitrite to NO anaerobically; this capability likely involves products of fnr and hmp genes, as shown for Salmonella (74). Dispersal of Salmonella biofilms by NO donors was shown to depend on the presence of the recA-hydN genomic region, suggesting its involvement in NO sensing (75).
An alternative group of bacterial enzymes capable of producing NO are the bacterial nitric oxide synthases (bNOS), only found in Gram-positive bacteria. These enzymes catalyze the oxidation of the amino acid l-arginine in the presence of oxygen (O2) as an essential substrate (76). Mammalian NOS are composed of both oxygenase and reductase domains, whereas bNOS from Bacillus and Staphylococcus contain only an oxygenase domain. In Gram-positive bacteria, bNOS-produced NO modulates several aspects of bacterial physiology, including protection from oxidative stress and antimicrobials (77, 78). Interestingly, although NOS-derived NO appears to inhibit biofilm dispersal of Bacillus subtilis 3610 (79), heterologous expression of B. subtilis bNOS in Pseudomonas putida increases motility and decreases biofilm formation (80), suggesting that the same signal (NO) may exert different effects depending on the cellular background.
DISPERSAL OF P. AERUGINOSA BIOFILMS INDUCED BY EXOGENOUS NO SOURCES
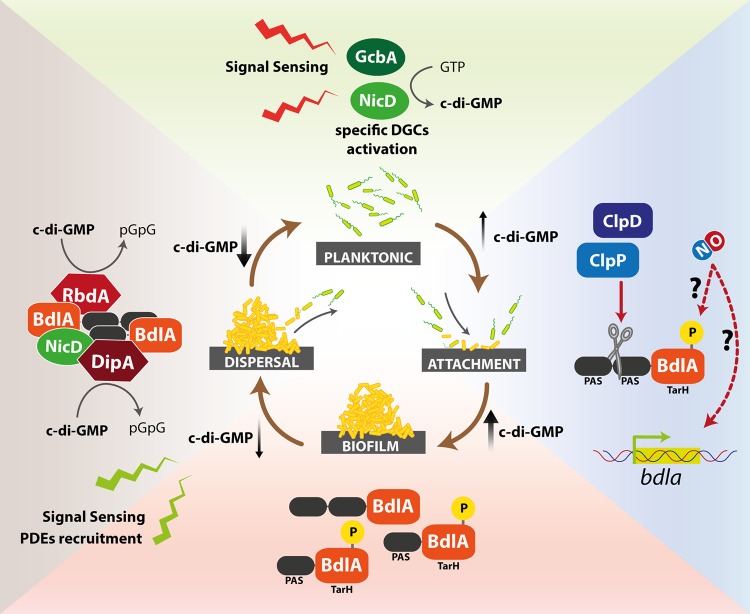
The molecular events leading to biofilm dispersal in P. aeruginosa are still far from being completely understood. One of the first proteins identified to play a role is the chemotaxis transducer protein BdlA (biofilm dispersion locus A), now considered a key player in this process. BdlA was identified during a search for a chemotaxis transducer protein involved in sensing different environmental cues that might trigger biofilm dispersal (81). It is a multidomain protein of ∼47 kDa consisting of two adjacent Per Arnt Sim (PAS)-domains, putatively involved in binding a heme cofactor, followed by a C-terminal TarH domain (82), related to ligand binding domains of methyl-accepting chemotaxis receptors. Although BdlA lacks domains directly responsible for modulating c-di-GMP levels, strains devoid of bdlA are impaired in dispersal of P. aeruginosa biofilms in response to several environmental cues, including NO (22, 81). Functional BdlA requires an unusual nonprocessive ClpP/ClpD-dependent proteolytic cleavage, which is stimulated by elevated c-di-GMP levels and BdlA phosphorylation (83) (Fig. 2).
FIG 2.
Nitric oxide activation of the BdlA signaling cascade. The BdlA protein is a key player in the modulation of biofilm formation and dispersal. It may be activated by nutrient and/or other signals (NO); the complex events occurring during and after BdlA activation are summarized in this figure (clockwise). Signal sensing initially leads to a transient increase in c-di-GMP, possibly triggered by specific DGCs, such as NicD or GcbA. Activation of BdlA by site-specific proteolysis by ClpD/ClpP at the level of the PAS domains or by phosphorylation leads to a further increase in c-di-GMP levels and biofilm formation. Other signaling events may trigger the recruitment of c-di-GMP-specific PDEs (RbdA and DipA), ultimately leading to a decrease in c-di-GMP levels and biofilm dispersal.
In P. aeruginosa, the response to nutrient (glutamate)-induced dispersal was recently proposed to be governed by a signal transduction mechanism involving a multiprotein complex composed of BdlA together with the DGC NicD and the PDE DipA, ultimately leading to the modulation of the cellular c-di-GMP pool (Fig. 2) (82, 83). Upon signal (i.e., glutamate) sensing, the DGC NicD, a seven-transmembrane receptor, becomes dephosphorylated, leading to enhanced DGC activity and resulting in a transient increase in c-di-GMP levels (84). NicD contributes to BdlA activation in two ways, by elevating c-di-GMP levels and via phosphorylation. Once activated, BdlA stimulates the PDE DipA and recruits a second PDE (RbdA), which ultimately leads to decreased c-di-GMP levels and biofilm dispersal (82, 83). RbdA itself contains sensory PAS and PAC domains, in addition to GGDEF and EAL domains. The PAS domain of RbdA appears to be involved in sensing low-oxygen stress. The EAL domain was shown to be catalytically active as a PDE, while the GGDEF domain controls PDE activation through GTP binding (19).
Very recently, an additional player in the BdlA signal transduction system has been identified, i.e., the catalytically active DGC GcbA (85). GcbA was shown to contribute to the regulation of BdlA cleavage shortly after initial cellular attachment to surfaces. It was shown that the levels of both proteins are inversely regulated, depending on the mode of growth (motile versus sessile), through the cellular c-di-GMP level. Therefore, both GcbA and BdlA translate different cues received by sensor proteins into the molecular cascade of events ultimately leading to dispersal (Fig. 2).
Although the involvement of BdlA and its cognate DGCs and PDEs in controlling c-di-GMP levels and biofilm dispersal is becoming increasingly clear, much less is known on the nature of other P. aeruginosa proteins which may directly sense NO and activate downstream response pathways. The features of possible candidates are summarized in the next two paragraphs.
NO SENSOR PROTEINS
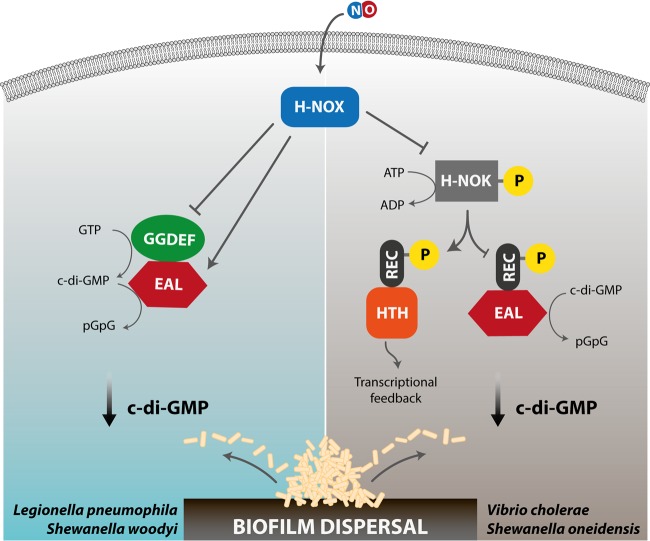
Almost a decade ago, the exposure of bacteria to NO was linked to changes in intracellular levels of c-di-GMP (38). In the meantime, heme-based sensor domains (H-NOX [heme-nitric oxide/oxygen]), involved in detecting NO, have been identified and linked to c-di-GMP signaling in a number of species. The H-NOX domain is a member of a large family of hemoproteins, including mammalian soluble guanylate cyclase, which sense diatomic gases (86, 87). In many bacteria, H-NOX domains act as NO sensors that regulate biofilm formation by modulating the intracellular level of c-di-GMP (88); these domains are often found adjacent to GGDEF and EAL domains, yielding a simplified one-component signaling which may either increase or decrease the c-di-GMP levels. In Legionella pneumophila, NO binding to the Hnox1 protein (composed of a fused H-NOX–GGDEF-EAL polypeptide) directly inhibits DGC activity and, thus, biofilm formation (Fig. 3) (35). The two-component system present in Shewanella woodyi comprises the SwH-NOX (H-NOX of S. woodyi) and SwGGDEF-EAL (HaCE [H-NOX-associated c-di-GMP-processing enzyme]) proteins: NO binds to SwH-NOX and regulates the partner protein by simultaneously downregulating its cyclase activity and upregulating the phosphodiesterase activity (89, 90). In general, NO binding events in bacterial H-NOX domains seem to cause conformational changes involving the N-terminal helices, which mediate interaction with partner enzymes, such as HaCEs (see above), or control the activity of a dedicated histidine kinase (H-NOX-associated histidine kinase [HnoK]), as seen in Shewanella oneidensis (91), ultimately resulting in changes of c-di-GMP within the cell (88) (Fig. 3).
FIG 3.
Nitric oxide-induced signaling events mediated by the heme-based sensor domain H-NOX. The binding of NO to the heme moiety of H-NOX may result in biofilm dispersal following different signaling cascades in different bacterial species harboring this protein domain. Left, in Legionella pneumophila or Shewanella woodyi, the NO derivative of the H-NOX protein interacts with the GGDEF-EAL (HaCE) protein, thus lowering c-di-GMP levels by stimulation of the PDE activity of the HaCE EAL domain. Right, in Vibrio cholerae or Shewanella oneidensis, interaction of the NO-bound H-NOX domain with a coupled histidine kinase (H-NOK) controls the phosphorylation activity of the kinase. Specific phosphorylation events lead to a decrease in c-di-GMP levels, either by stimulating the hydrolysis of c-di-GMP to pGpG by a cognate PDE (REC-EAL) via the fused REC domain or by controlling the transcriptional response through a dedicated transcription regulator (REC-HTH).
Although this signaling system is quite well understood, several bacteria, including P. aeruginosa, lack H-NOX proteins but are still able to respond to NO (38). Interestingly, in P. aeruginosa, the response to NO has been linked to increased PDE activity (22, 24). Thus far, only one PDE (i.e., NbdA) specifically involved in the NO-specific reduction of c-di-GMP has been identified (24). While two additional PDEs (i.e., DipA and RbdA) have been identified to be involved in the dispersal response to various environmental cues (including NO) and are shown to be part of the BdlA signal transduction system (see previous paragraph), it is still not completely understood how NbdA is interwoven in this signal transduction network. This is especially intriguing since the overexpression of the E. coli PDE YhjH in P. aeruginosa leads to biofilm dispersal, as does the addition of a dispersal-inducing agent (12). Similar behavior has been observed for other bacteria, including Shewanella oneidensis, where overexpression of yhjH also led to rapid cellular detachment from biofilms (92).
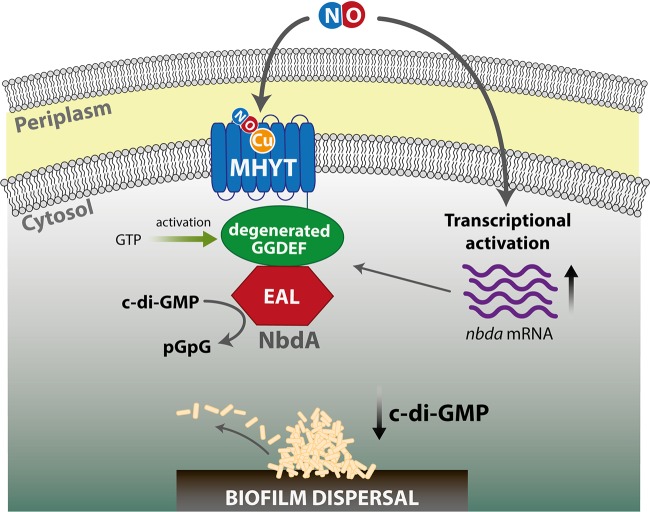
NbdA possesses a so-called MHYT-domain (93), a transmembrane domain composed of seven predicted membrane spanning helices, proposed to possess a putative sensory function for diatomic gases like oxygen, carbon monoxide, or NO through protein-bound copper ions (93) (Fig. 4). In addition to the MHYT domain, NbdA harbors cytoplasmic GGDEF and EAL domains. Biochemical studies revealed that NbdA has PDE activity, stimulated by GTP bound to the degenerated GGDEF domain. Lack of NbdA resulted in biofilms impaired in the dispersal response to NO but not glutamate and could be linked to changes in c-di-GMP levels (24). In NO-dispersed biofilms, nbdA mRNA levels are increased, suggesting that an (additional) event involving transcriptional regulation is taking place (Fig. 4) (24). However, whether one of the NO-responsive transcriptional regulators is involved in this process and whether the MHYT-domain itself has a NO-sensory function has to be determined. In this context, it is also worth mentioning that P. aeruginosa possesses a second c-di-GMP-modulating enzyme with a domain organization similar to that of NbdA. MucR was shown to be involved in regulating alginate synthesis through its DGC activity (94). However, weak PDE activity could also be demonstrated in vitro (24). The lack of MucR resulted in biofilms that did not disperse either in response to NO or to glutamate.
FIG 4.
Scheme of NbdA activation. Possible mechanism of NO-induced biofilm dispersal via the MHYT domain-containing NbdA protein. This protein is composed of three domains, a membrane-spanning MHYT domain, a degenerated GGDEF domain, and a PDE-EAL domain. Binding of NO is supposed to occur via copper ions (Cu) located in the MHYT domain. The signal is then transmitted via the GGDEF domain to the EAL domain, triggering its PDE activity and, thus, c-di-GMP hydrolysis to pGpG. GTP may increase the PDE activity of the EAL domain by binding to the degenerated GGDEF. In parallel, NO may also activate the transcription of NbdA by a yet-unknown mechanism.
In addition to the c-di-GMP-modulating enzymes, a preliminary report suggests that the periplasmic protease LapG may be involved in the dispersal response to NO, as P. aeruginosa biofilms devoid of LapG did not show a dispersal response upon NO addition (39). LapG is a membrane-bound protease that, in a c-di-GMP-dependent fashion via an inside-out signaling mechanism through the membrane receptor LapD, cleaves the surface adhesion LapA in Pseudomonas putida to mediate dispersal (95, 96). Since a LapA homolog is absent in P. aeruginosa, it is yet to be determined how the Lap system is integrated in the dispersal response in this pathogenic Pseudomonas species.
NO RESPONSIVE TRANSCRIPTIONAL REGULATORS
As summarized above, the addition of NO often also leads to biofilm dispersal by modulating the transcriptional profile of the bacterium. NO is an established signaling molecule in bacteria, interacting with many bacterial regulatory components, such as OxyR, SoxR, NsrR, NorR, FhpR, DNR, and other regulators of the fumarate nitrate reductase transcriptional regulator (FNR) family (97, 98). These regulators display iron-containing cofactors (iron–sulfur clusters, mononuclear iron, heme) or reactive cysteine thiols that react with NO, thereby changing their affinity to the target DNA. Many molecular studies have elucidated the role of these regulators in NO metabolism and NO detoxification. On the other hand, although it has been shown that NO sensing during biofilm dispersal requires transcriptional activation of selected members of signaling or metabolic pathways, ultimately leading to c-di-GMP decrease, there is little evidence on which regulators are involved in this process.
In P. aeruginosa, during biofilm dispersal, low doses of NO were shown to modulate the expression of a subset of genes, including nirS and other components of the denitrification chain and genes involved in attachment (12, 22). The expression of the denitrification genes is controlled by the activity of the DNR transcription factor (52), which is also highly expressed under biofilm dispersal conditions (12) and may play a role in this process. DNR is a heme-based gas sensor of the cAMP receptor protein (CRP)-FNR superfamily, which positively responds to NO (48, 49, 51). The affinity of the DNR-NO complex for its cognate DNA was recently determined to be in the nanomolar range (50).
A recent study has correlated the appearance after biofilm dispersal of a superinfective (SI) version of the Pseudomonas aeruginosa filamentous phage Pf4 with oxidative and nitrosative stress response mediated by OxyR, a LysR-type transcriptional regulator (99). Although studies reporting S-nitrosylation of regulatory proteins are not available for P. aeruginosa, it is known that OxyR is a master regulator of S-nitrosylation under anaerobic conditions in E. coli (100). In P. aeruginosa, the OxyR regulon includes genes involved in iron homeostasis and the production of cytochromes (cyoA and snr1) (101), all found to be significantly altered in dispersed cells (12), suggesting that an OxyR-dependent mechanism could also be envisaged.
CONCLUSIONS AND OPEN QUESTIONS
Dispersed cells are physiologically different from planktonic cells, as previously suggested (4, 13) and remarked in a recent study showing that freshly dispersed cells (induced by either NO or overexpression of a PDE) are highly virulent and sensitive to iron stress (12). Gene expression patterns are different between planktonic and dispersed cells, but the SNP- (NO donor) and PDE-induced dispersal patterns overlap. As summarized in this minireview, although the dispersed cells show a defined physiological profile, distinct from those of the planktonic and biofilm cells, all available evidences suggest that the bacterium may employ different routes to reach this dispersed state after sensing external or internal stimuli, including NO. A summary of the possible effects of NO as a signaling molecule leading to biofilm dispersal in P. aeruginosa is presented in Fig. 5.
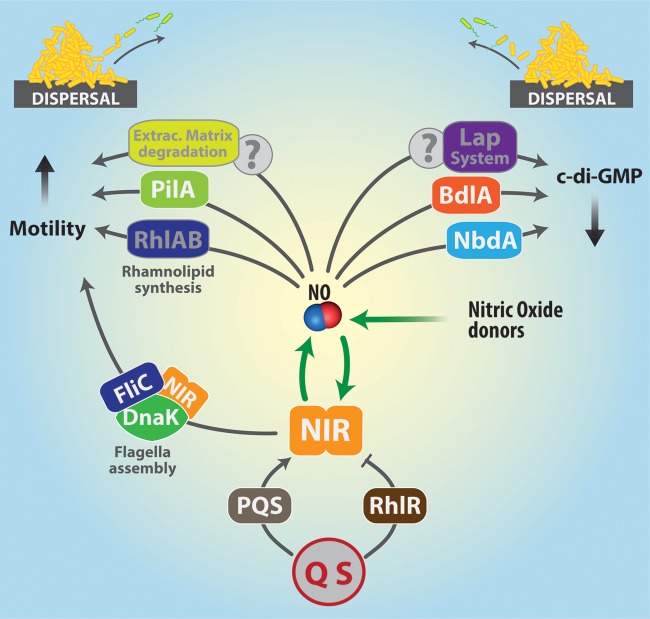
FIG 5.
Summary of the possible effects of NO leading to biofilm dispersal in P. aeruginosa. In P. aeruginosa, NO can be produced by the endogenous enzyme nitrite reductase (NIR) or come from external sources (NO donors). Since NO is known to stimulate NIR synthesis, the two sources (endogenous and exogenous) may also cooperate, as shown in the figure. Biofilm dispersal is triggered by events that increase motility by stimulating synthesis of pili (PilA) or rhamnolipids (RhlAB). Flagellum assembly can also be stimulated by the formation of a ternary complex of NIR (in the active and inactive form), FliC (the major flagellar subunit), and DnaK (a chaperone). NO-induced biofilm dispersal also involves the decrease of c-di-GMP intracellular levels. Two major signaling pathways are known to lower c-di-GMP concentrations, namely, those involving the chemotaxis receptor BdlA and the membrane protein NbdA. Another possible effector of c-di-GMP decrease is the Lap system, whose mechanism of action is yet to be fully elucidated. Many details of these processes will require further investigation, to clarify how NO may affect remodeling of the extracellular matrix to facilitate the escape of individual motile cells and to identify which NO-sensitive transcriptional regulators are involved in regulating gene expression during dispersal.
NO can be either endogenously produced as a metabolic intermediate of denitrification or come from exogenous NO donors (such as SNP). The NO molecule can increase motility directly, by acting on flagella, pili, and/or rhamnolipid production, or indirectly stimulate NIR expression to produce more NO or to facilitate flagellum assembly (Fig. 5). NIR expression or activity is also controlled by QS (RhlR and PQS), which thereby modulate NO levels in response to population clues. The response to NO in terms of biofilm formation/dispersal is, however, different under aerobic and anaerobic conditions: anaerobiosis favors the accumulation of NO, ultimately leading to biofilm formation as a stress defense mechanism. Thus, investigations of the effects of endogenously generated NO and/or those obtained with NO donors (such as SNP), should take oxygen availability into account more carefully, and such studies should be done under anaerobic or microaerobic conditions in both nonbiofilm (planktonic and dispersed) and biofilm cells.
A common theme in the dispersal of P. aeruginosa biofilms is the observed decrease of c-di-GMP intracellular levels: the major signaling pathways are those involving BdlA (an intracellular effector) and NbdA (a membrane-bound effector) (Fig. 5). However, many other molecular mechanisms of NO sensing and the downstream signaling events are still to be fully clarified. This includes not only the events leading from perception of various environmental signals to decreased cellular c-di-GMP levels but also the events following thereafter. How is the c-di-GMP concentration transduced into physiological responses, such as secretion of extracellular enzymes that degrade the matrix to release individual motile cells?
The other open questions to be answered concern the type and role of transcriptional regulators involved and the mechanism of NO sensing by individual receptors (metals, prosthetic groups, and active thiols). To shed more light on the role of single transcriptional regulators in biofilm dispersal, it would be interesting to compare the behavior of genetic mutants of some of the proteins mentioned above (DNR and OxyR) with respect to dispersal. Moreover, it would be highly desirable to enlarge our knowledge regarding dispersal by investigating, in P. aeruginosa (as well as other denitrifying bacteria), the influence of quorum sensing. Last but not least, the effects of the growth medium should also be considered, using, instead of a rich undefined medium, other media, possibly more closely resembling the growth conditions encountered by the pathogen during colonization of host tissues, which were recently shown to greatly influence gene expression patterns (102).
ACKNOWLEDGMENTS
We gratefully acknowledge the contribution of Giorgio Giardina (Sapienza University of Rome, Rome, Italy) in producing the figures.
Research in the laboratory of N.F.-D. was supported by the Ruhr University Bochum Protein Research Department. F.C. gratefully acknowledges the support of Sapienza University of Rome, Rome, Italy.
Biographies

Francesca Cutruzzolà is full professor of molecular biology at the Sapienza University of Rome (Italy). She has spent several periods abroad, both in Europe (ETH, U. Leiden, MRC United Kingdom) and in the United States (Illinois, California). Since 2014, she has been a member of the Scientific Board of Directors of Institute Pasteur-Fondazione Cenci Bolognetti (Italy) and the ERC (European Research Council) grant panel (LS6). She has authored over 100 papers and one patent; she has been invited to over 30 international meetings. In 2012, her research group was chosen as one of the 3 best groups in the European Life Science Awards. The main goal of her research is to understand how proteins function and control the behavior of prokaryotic and eukaryotic cells. Her ongoing research is focused on small molecules (nucleotides) involved in the formation of bacterial biofilm in chronic infections, on nitric oxide and redox metabolism in bacterial pathogens, and more recently, on metabolic reprogramming in cancer cells.

Nicole Frankenberg-Dinkel is a full professor of microbiology at the Technical University Kaiserslautern, Germany. After obtaining a diploma in biology from the University of Regensburg and a Ph.D. in biochemistry from the University of Freiburg, she joined the laboratory of J. Clark Lagarias at UC Davis, California, to work on open-chain tetrapyrrole biosynthesis. She returned to Germany in 2003 to start an independent Emmy-Noether research group. In 2006, she was appointed as professor of microbial physiology at the Ruhr University Bochum, and just recently, she joined the faculty of biology of the Technical University Kaiserslautern. She has authored over 50 papers and three patents and is an editorial board member of the Journal of Biological Chemistry. Her research focuses on enzymes involved in open-chain tetrapyrrole biosynthesis and on how tetrapyrroles are used within proteins to sense environmental cues. More recently, parts of her research have been focused on nitric oxide-mediated biofilm dispersal in Pseudomonas aeruginosa.
REFERENCES
- 1.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Parkins MD, Glezerson BA, Sibley CD, Sibley KA, Duong J, Purighalla S, Mody CH, Workentine ML, Storey DG, Surette MG, Rabin HR. 2014. Twenty-five-year outbreak of Pseudomonas aeruginosa infecting individuals with cystic fibrosis: identification of the prairie epidemic strain. J Clin Microbiol 52:1127–1135. doi: 10.1128/JCM.03218-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tolker-Nielsen T. 2014. Pseudomonas aeruginosa biofilm infections: from molecular biofilm biology to new treatment possibilities. APMIS Suppl 2014(138):1–51. doi: 10.1111/apm.12335. [DOI] [PubMed] [Google Scholar]
- 4.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 6.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penesyan A, Gillings M, Paulsen IT. 2015. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20:5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 11.Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chua SL, Liu Y, Yam JK, Chen Y, Vejborg RM, Tan BG, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. 2014. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog 10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol 7:894–906. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol 70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. 2009. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS One 4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musk DJ, Banko DA, Hergenrother PJ. 2005. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem Biol 12:789–796. doi: 10.1016/j.chembiol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.An S, Wu J, Zhang LH. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol 76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan JB, Fine DH. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol 68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. d-Amino acids trigger biofilm disassembly. Science 328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. 2009. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol 2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlag S, Nerz C, Birkenstock TA, Altenberend F, Gotz F. 2007. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol 189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies DG, Marques CN. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Gilles-Gonzalez MA. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420–3426. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 29.Savakis P, De Causmaecker S, Angerer V, Ruppert U, Anders K, Essen LO, Wilde A. 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol 85:239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 30.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 31.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 32.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 35.Carlson HK, Vance RE, Marletta MA. 2010. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol Microbiol 77:930–942. doi: 10.1111/j.1365-2958.2010.07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt I, Steenbakkers PJ, op den Camp HJ, Schmidt K, Jetten MS. 2004. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J Bacteriol 186:2781–2788. doi: 10.1128/JB.186.9.2781-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potter AJ, Kidd SP, Edwards JL, Falsetta ML, Apicella MA, Jennings MP, McEwan AG. 2009. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J Infect Dis 199:227–235. doi: 10.1086/595737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barraud N, Kelso MJ, Rice SA, Kjelleberg S. 2015. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des 21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 40.Stamler JS, Singel DJ, Loscalzo J. 1992. Biochemistry of nitric oxide and its redox-activated forms. Science 258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 41.Heinrich TA, da Silva RS, Miranda KM, Switzer CH, Wink DA, Fukuto JM. 2013. Biological nitric oxide signalling: chemistry and terminology. Br J Pharmacol 169:1417–1429. doi: 10.1111/bph.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doyle MP, Hoekstra JW. 1981. Oxidation of nitrogen oxides by bound dioxygen in hemoproteins. J Inorg Biochem 14:351–358. doi: 10.1016/S0162-0134(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 43.Friebe A, Koesling D. 2003. Regulation of nitric oxide-sensitive guanylyl cyclase. Circ Res 93:96–105. doi: 10.1161/01.RES.0000082524.34487.31. [DOI] [PubMed] [Google Scholar]
- 44.Zumft WG. 1993. The biological role of nitric oxide in bacteria. Arch Microbiol 160:253–264. doi: 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]
- 45.Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest 99:2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barraud N, Kardak BG, Yepuri NR, Howlin RP, Webb JS, Faust SN, Kjelleberg S, Rice SA, Kelso MJ. 2012. Cephalosporin-3′-diazeniumdiolates: targeted NO-donor prodrugs for dispersing bacterial biofilms. Angew Chem Int Ed Engl 51:9057–9060. doi: 10.1002/anie.201202414. [DOI] [PubMed] [Google Scholar]
- 47.Yepuri NR, Barraud N, Mohammadi NS, Kardak BG, Kjelleberg S, Rice SA, Kelso MJ. 2013. Synthesis of cephalosporin-3′-diazeniumdiolates: biofilm dispersing NO-donor prodrugs activated by beta-lactamase. Chem Commun (Camb) 49:4791–4793. doi: 10.1039/c3cc40869h. [DOI] [PubMed] [Google Scholar]
- 48.Castiglione N, Rinaldo S, Giardina G, Cutruzzolà F. 2009. The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 155:2838–2844. doi: 10.1099/mic.0.028027-0. [DOI] [PubMed] [Google Scholar]
- 49.Giardina G, Castiglione N, Caruso M, Cutruzzolà F, Rinaldo S. 2011. The Pseudomonas aeruginosa DNR transcription factor: light and shade of nitric oxide-sensing mechanisms. Biochem Soc Trans 39:294–298. doi: 10.1042/BST0390294. [DOI] [PubMed] [Google Scholar]
- 50.Lobato L, Bouzhir-Sima L, Yamashita T, Wilson MT, Vos MH, Liebl U. 2014. Dynamics of the heme-binding bacterial gas-sensing dissimilative nitrate respiration regulator (DNR) and activation barriers for ligand binding and escape. J Biol Chem 289:26514–26524. doi: 10.1074/jbc.M114.571398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinaldo S, Castiglione N, Giardina G, Caruso M, Arcovito A, Longa SD, D'Angelo P, Cutruzzolà F. 2012. Unusual heme binding properties of the dissimilative nitrate respiration regulator, a bacterial nitric oxide sensor. Antioxid Redox Signal 17:1178–1189. doi: 10.1089/ars.2011.4226. [DOI] [PubMed] [Google Scholar]
- 52.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 53.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, Schobert M, Jahn D. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189:4310–4314. doi: 10.1128/JB.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baek SH, Rajashekara G, Splitter GA, Shapleigh JP. 2004. Denitrification genes regulate Brucella virulence in mice. J Bacteriol 186:6025–6031. doi: 10.1128/JB.186.18.6025-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barth KR, Isabella VM, Clark VL. 2009. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology 155:4093–4103. doi: 10.1099/mic.0.032961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. 2007. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun 75:3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cutruzzolà F, Arese M, Grasso S, Bellelli A, Brunori M. 1997. Mutagenesis of nitrite reductase from Pseudomonas aeruginosa: tyrosine-10 in the c heme domain is not involved in catalysis. FEBS Lett 412:365–369. doi: 10.1016/S0014-5793(97)00583-8. [DOI] [PubMed] [Google Scholar]
- 59.Castiglione N, Rinaldo S, Giardina G, Stelitano V, Cutruzzolà F. 2012. Nitrite and nitrite reductases: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal 17:684–716. doi: 10.1089/ars.2011.4196. [DOI] [PubMed] [Google Scholar]
- 60.Rinaldo S, Sam KA, Castiglione N, Stelitano V, Arcovito A, Brunori M, Allen JW, Ferguson SJ, Cutruzzolà F. 2011. Observation of fast release of NO from ferrous d(1) haem allows formulation of a unified reaction mechanism for cytochrome cd(1) nitrite reductases. Biochem J 435:217–225. doi: 10.1042/BJ20101615. [DOI] [PubMed] [Google Scholar]
- 61.Hino T, Nagano S, Sugimoto H, Tosha T, Shiro Y. 2012. Molecular structure and function of bacterial nitric oxide reductase. Biochim Biophys Acta 1817:680–687. doi: 10.1016/j.bbabio.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 62.Pauleta SR, Dell'Acqua S, Moura I. 2013. Nitrous oxide reductase. Coord Chem Rev 257:332–349. doi: 10.1016/j.ccr.2012.05.026. [DOI] [Google Scholar]
- 63.de la Fuente-Nunez C, Reffuveille F, Fairfull-Smith KE, Hancock RE. 2013. Effect of nitroxides on swarming motility and biofilm formation, multicellular behaviors in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4877–4881. doi: 10.1128/AAC.01381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon MY, Lee KM, Park Y, Yoon SS. 2011. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One 6:e16105. doi: 10.1371/journal.pone.0016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamada M, Toyofuku M, Miyano T, Nomura N. 2014. cbb3-type cytochrome c oxidases, aerobic respiratory enzymes, impact the anaerobic life of Pseudomonas aeruginosa PAO1. J Bacteriol 196:3881–3889. doi: 10.1128/JB.01978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borrero-de Acuna JM, Molinari G, Rohde M, Dammeyer T, Wissing J, Jansch L, Arias S, Jahn M, Schobert M, Timmis KN, Jahn D. 2015. A periplasmic complex of the nitrite reductase NirS, the chaperone DnaK and the flagellum protein FliC is essential for flagellum assembly and motility in Pseudomonas aeruginosa. J Bacteriol 197:3066–3075. doi: 10.1128/JB.00415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pesci EC, Pearson JP, Seed PC, Iglewski BH. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179:3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toyofuku M, Nomura N, Fujii T, Takaya N, Maseda H, Sawada I, Nakajima T, Uchiyama H. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J Bacteriol 189:4969–4972. doi: 10.1128/JB.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. 2008. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol 190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 73.Hammond JH, Dolben EF, Smith TJ, Bhuju S, Hogan DA. 2015. Links between Anr and quorum sensing in Pseudomonas aeruginosa biofilms. J Bacteriol 197:2810–2820. doi: 10.1128/JB.00182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilberthorpe NJ, Poole RK. 2008. Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J Biol Chem 4:11146–11154. doi: 10.1074/jbc.M708019200. [DOI] [PubMed] [Google Scholar]
- 75.Marvasi M, Chen C, Carrazana M, Durie IA, Teplitski M. 2014. Systematic analysis of the ability of nitric oxide donors to dislodge biofilms formed by Salmonella enterica and Escherichia coli O157:H7. AMB Express 4:42. doi: 10.1186/s13568-014-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crane BR, Sudhamsu J, Patel BA. 2010. Bacterial nitric oxide synthases. Annu Rev Biochem 79:445–470. doi: 10.1146/annurev-biochem-062608-103436. [DOI] [PubMed] [Google Scholar]
- 77.Gusarov I, Nudler E. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci U S A 102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schreiber FBM, Enning D, Lamprecht-Grandio M, Zafra O, González-Pastor JE. dBD 2011. The role of nitric-oxide-synthase-derived nitric oxide in multicellular traits of Bacillus subtilis 3610: biofilm formation, swarming, and dispersal. BMC Microbiol 11:111. doi: 10.1186/1471-2180-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu P, Huang Q, Chen W. 2012. Heterologous expression of bacterial nitric oxide synthase gene: a potential biological method to control biofilm development in the environment. Can J Microbiol 58:336–344. doi: 10.1139/w11-141. [DOI] [PubMed] [Google Scholar]
- 81.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. 2004. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci U S A 101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, Marletta MA. 2006. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase beta1 H-NOX domain. J Biol Chem 281:21892–21902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- 88.Plate L, Marletta MA. 2013. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci 38:566–575. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick JA, Boon EM. 2012. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry 51:2087–2099. doi: 10.1021/bi201753f. [DOI] [PubMed] [Google Scholar]
- 90.Lahiri T, Luan B, Raleigh DP, Boon EM. 2014. A structural basis for the regulation of an H-NOX-associated cyclic-di-GMP synthase/phosphodiesterase enzyme by nitric oxide-bound H-NOX. Biochemistry 53:2126–2135. doi: 10.1021/bi401597m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell 46:449–460. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galperin MY, Gaidenko TA, Mulkidjanian AY, Nakano M, Price CW. 2001. MHYT, a new integral membrane sensor domain. FEMS Microbiol Lett 205:17–23. doi: 10.1111/j.1574-6968.2001.tb10919.x. [DOI] [PubMed] [Google Scholar]
- 94.Hay ID, Remminghorst U, Rehm BH. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 97.Spiro S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev 31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 98.Arai H, Hayashi M, Kuroi A, Ishii M, Igarashi Y. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J Bacteriol 187:3960–3968. doi: 10.1128/JB.187.12.3960-3968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hui JG, Mai-Prochnow A, Kjelleberg S, McDougald D, Rice SA. 2014. Environmental cues and genes involved in establishment of the superinfective Pf4 phage of Pseudomonas aeruginosa. Front Microbiol 5:654. doi: 10.3389/fmicb.2014.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seth D, Hausladen A, Wang YJ, Stamler JS. 2012. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science 336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei Q, Minh PN, Dotsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Haussler S, Cornelis P. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]