ABSTRACT
High levels of the universal bacterial second messenger cyclic di-GMP (c-di-GMP) promote the establishment of surface-attached growth in many bacteria. Not only can c-di-GMP bind to nucleic acids and directly control gene expression, but it also binds to a diverse array of proteins of specialized functions and orchestrates their activity. Since its development in the early 1990s, the synthetic peptide array technique has become a powerful tool for high-throughput approaches and was successfully applied to investigate the binding specificity of protein-ligand interactions. In this study, we used peptide arrays to uncover the c-di-GMP binding site of a Pseudomonas aeruginosa protein (PA3740) that was isolated in a chemical proteomics approach. PA3740 was shown to bind c-di-GMP with a high affinity, and peptide arrays uncovered LKKALKKQTNLR to be a putative c-di-GMP binding motif. Most interestingly, different from the previously identified c-di-GMP binding motif of the PilZ domain (RXXXR) or the I site of diguanylate cyclases (RXXD), two leucine residues and a glutamine residue and not the charged amino acids provided the key residues of the binding sequence. Those three amino acids are highly conserved across PA3740 homologs, and their singular exchange to alanine reduced c-di-GMP binding within the full-length protein.
IMPORTANCE In many bacterial pathogens the universal bacterial second messenger c-di-GMP governs the switch from the planktonic, motile mode of growth to the sessile, biofilm mode of growth. Bacteria adapt their intracellular c-di-GMP levels to a variety of environmental challenges. Several classes of c-di-GMP binding proteins have been structurally characterized, and diverse c-di-GMP binding domains have been identified. Nevertheless, for several c-di-GMP receptors, the binding motif remains to be determined. Here we show that the use of a synthetic peptide array allowed the identification of a c-di-GMP binding motif of a putative c-di-GMP receptor protein in the opportunistic pathogen P. aeruginosa. The application of synthetic peptide arrays will facilitate the search for additional c-di-GMP receptor proteins and aid in the characterization of c-di-GMP binding motifs.
INTRODUCTION
Bis-(3′-5′)-cyclic di-GMP (c-di-GMP) is a universal bacterial secondary messenger and a key player in the decision between the motile planktonic mode of growth and the sessile biofilm-associated mode of growth (1–3). Intracellular c-di-GMP levels are tightly controlled in response to a broad range of environmental cues which impact the expression or the activity of two classes of enzymes, diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) (4–9). DGCs contain a conserved GGDEF domain and mediate the synthesis of c-di-GMP from two molecules of GTP, whereas PDEs are EAL or HD-GYP domain-containing proteins involved in c-di-GMP degradation. A common feature of most DGCs is allosteric feedback inhibition, as binding of c-di-GMP to an I-site motif within the GGDEF domain inhibits enzymatic activity (10, 11). The conserved GGDEF/EAL domains are associated with many diverse input and output domains that receive a great variety of extracellular signals to allow the cell to respond to a broad range of environmental cues with an adjustment of intracellular c-di-GMP levels (12, 13). In recent years, some extracellular signals, such as light, volatile compounds, and surface attachment, have been shown to regulate enzyme activity; however, knowledge of how elevated c-di-GMP levels are translated into a bacterial response on the single-cell level remains limited (14–20).
Insights into c-di-GMP effectors have come from the demonstration that proteins containing a PilZ domain can bind c-di-GMP and control phenotypes involved in motility, biofilm formation, and pathogenicity (21–24). Another class of c-di-GMP binding proteins contains degenerate GGDEF or EAL domains which lost their enzymatic activity but bind c-di-GMP and thus communicate intracellular c-di-GMP levels to output domains (14, 24–28). Furthermore, transcriptional regulators have been demonstrated to bind c-di-GMP and to directly impact gene expression (14, 29–32). These transcription factors do not seem to share a c-di-GMP binding motif. Additionally, conserved RNA domains, which reside in 5′ untranslated regions of different mRNAs, have been shown to be c-di-GMP targets and to affect gene expression via riboswitches (33, 34). Recently, a new c-di-GMP binding domain, the GIL domain, and a novel protein family, the YajQ family, were identified to be c-di-GMP effectors (35, 36).
Despite the recent advances in the identification of diverse c-di-GMP effectors, in many bacterial species only a few of them have been identified, and their low number does not seem to reflect the unprecedented range of effector components that are regulated by c-di-GMP. Thus, identification of further classes of c-di-GMP binding domains can be expected, and many questions on the nature of the targets of c-di-GMP action are still open. We have recently applied a chemical proteomics approach employing a functionalized c-di-GMP analog to uncover novel candidate c-di-GMP binding proteins in the opportunistic bacterial pathogen Pseudomonas aeruginosa (37).
In this study, we coupled chemically unmodified c-di-GMP to a Sepharose matrix and identified two not previously detected c-di-GMP binding proteins. One of those, PA3740, was confirmed to bind c-di-GMP by the use of surface plasmon resonance (SPR). To uncover the amino acid motif that binds its target c-di-GMP, we applied a peptide array approach (38) with a series of tiled overlapping peptides derived from the full amino acid sequence of PA3740. We identified a LKKALKKQTNLR peptide sequence to be a novel c-di-GMP binding motif. Interestingly, the leucine residues and the glutamate residue proved to be essential for c-di-GMP binding to the peptide as well as to the purified full-length PA3740 protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
P. aeruginosa PAO1 and Escherichia coli strains were cultured in lysogeny broth (LB) medium at 37°C and 180 rpm. If required, 100 μg/ml ampicillin was added. For all cloning steps, E. coli strain DH5α was used. A mutant harboring a transposon insertion within PA3740 was selected from the PAO1 mini-Tn5-lux transposon mutant library (of Robert E. W. Hancock [39]). Overexpression of PA3740 was realized by using pJN105::PA3740, by which the PA3740 gene was amplified with PA3740-specific forward primer fPr6 (5′-GATCGAATTCTAAGAAGGAGATATAATGACCATGTCCAATCAACAAC-3′) and PA3740-specific reverse primer rPr6 (5′-GATCTCTAGAGATCAGGGCCGCAGGCTGA-3′) and inserted between the EcoRI and XbaI restriction sites. The mutant and the PA3740-overexpressing strain were used for phenotypic assays.
For protein purification purposes, PA3740 was PCR amplified by the Pfu polymerase with the gene-specific primers PA3740fPr (5′-AAAACATATGATGACCATGTCCAATCAACAAC-3′) and PA3740rPr2 (5′-GATCAAGCTTGGGCCGCAGGCTGAT-3′). The PCR product was digested with the restriction enzymes NdeI and HindIII and cloned into pET21a+ (Novagen), which comprises a hexahistidine tag after the multiple-cloning site. The resulting plasmids were verified by sequencing. Alanine mutants of PA3740-His6 were generated by introducing respective point mutations into the pET21a+::PA3740 plasmid with the help of mutagenic primers and a QuikChange II site-directed mutagenesis kit (Agilent Technologies). The sequences of the mutagenic primers are available upon request.
Swimming and swarming motility.
Swimming and swarming assays were performed as previously described (56) with a slight modification: instead of 0.5% (wt/vol) Casamino Acids, only 0.1% (wt/vol) Casamino Acids was added to the swarm agar plates. To the motility plates, 0.5% arabinose and 50 μg/ml gentamicin were added. Bacteria were grown for 6 h at 37°C in LB medium supplemented with 50 μg/ml gentamicin at 180 rpm. Cells were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS), and adjusted in respect to cell number. Each motility plate was inoculated with 1 μl bacterial suspension. The plates were incubated at 33°C in a humid atmosphere for 17 h.
Growth curve.
The growth rates of wild-type planktonic cells with the pJN105 empty vector and wild-type planktonic cells overexpressing PA3740 from pJN105 were monitored using an automated growth analysis system (Bioscreen C MBR; iLF Bioserve, Langenau, Germany). Therefore, for each strain, triplicate precultures at an optical density at 600 nm (OD600) of 0.07 were inoculated in a 100-well honeycomb plate (iLF Bioserve) with LB and grown overnight at 37°C. From time zero, induction occurred with 0.5% arabinose. The plate was incubated at 37°C with continuous shaking in a computer-controlled Bioscreen incubator for 25 h. The turbidity of the bacterial suspensions (OD600) was automatically measured every 15 min.
Immobilization of c-di-GMP on Sepharose 6B.
c-di-GMP was synthesized as previously specified (40). Preparation of c-di-GMP affinity resin was performed as described for the immobilization of cyclic GMP (cGMP) (41) with some modifications: a total of 500 mg epoxy-activated Sepharose 6B (GE Healthcare) was treated 10 times with 25 ml cold deionized water. The washed resin was resuspended in 3 ml coupling buffer (100 mM Na2B4O7, 100 mM NaCl, pH 11) and incubated with 2.2 mg (3 μmol) c-di-GMP at 40°C in a thermomixer. The progression of immobilization was monitored by analytical high-pressure liquid chromatography (HPLC) using a 5% acetonitrile–20 mM triethylammonium formate buffer, pH 6.8, as the eluent. The HPLC system consisted of an L 6200 pump, an L 4000 variable-wavelength UV detector, and a D 2500 GPC integrator (all from Merck-Hitachi). The stationary phase was YMC ODS-A 120-10 (YMC, Kyoto, Japan) in a stainless steel column (250 by 4.6 mm). The coupling reaction proceeded for 144 h, and any unreacted epoxy groups were blocked with 100 μmol ethanolamine in coupling buffer for 24 h. After filtration and multiple washing steps (twice with 25 ml 20% ethanol, twice with 25 ml water, twice with 25 ml 30 mM NaH2PO4, pH 7), the affinity resin was stored in 30 mM NaH2PO4, 1% NaN3, pH 7, at 4°C. The ligand density was 0.45 μmol/100 mg (on a dry weight basis) resin, as determined by HPLC analysis during coupling. In a control experiment, c-di-GMP was incubated in coupling buffer without affinity resin at 40°C for 144 h. After this period, less than 15% degradation of c-di-GMP was detected by analytical HPLC.
To identify functional groups that react with epoxy-activated Sepharose, a model reaction was performed under the above-mentioned coupling conditions using the related cyclic nucleotide cGMP (Biolog, Bremen, Germany) and (S)-(−)-1,2-epoxy-butane (Aldrich). The reaction products obtained were analyzed by HPLC, nuclear magnetic resonance (NMR), and mass spectrometry (MS).
A control affinity resin was prepared by incubating epoxy-activated Sepharose 6B with 100 μmol ethanolamine in coupling buffer at 40°C for 48 h.
Affinity chromatography and MS analysis.
For isolation of c-di-GMP binding proteins, P. aeruginosa PAO1 was cultivated in 500 ml LB overnight at 37°C. After the bacteria were harvested by centrifugation (4°C, 6,000 × g, 10 min) and one washing step with PBS, the pellet was resuspended in 15 ml ice-cold PBS lysis buffer containing protease inhibitors (Roche) and 1 mM dithioerythritol. Cells were lysed by bead beating (three times for 20 s each time at power 4.5), and cell debris was removed by centrifugation at 4°C and 37,500 × g for 45 min. The supernatant was collected and subsequently submitted to affinity purification using 200 μl of the c-di-GMP affinity resin equilibrated with lysis buffer. Affinity chromatography was performed as previously described (37), except that six washing steps were performed with lysis buffer and proteins were eluted either by heat treatment or with 0.5 mM c-di-GMP in PBS. Eluted proteins were concentrated by trichloroacetic acid precipitation and subjected to SDS-PAGE analysis. Coomassie-stained protein bands were excised and analyzed by liquid chromatography (LC)-MS/MS essentially as described previously (37). The Scaffold program (version Scaffold_3_00_03; Proteome Software Inc.) was used to validate the MS/MS-based peptide and protein identifications. Proteins were accepted only when at least 1 unique peptide showed 95% confidence and the total protein confidence was at least 95%. Peptide probabilities were specified by the Peptide Prophet algorithm (42), and protein probabilities were assigned by the Protein Prophet algorithm (43).
PA3740 protein production and purification.
Despite several attempts, we were not able to produce soluble recombinant PA3740 protein in E. coli, Saccharomyces cerevisiae, or HEK293 cells and thus performed cell-free protein synthesis. In vitro translation of PA3740 was performed by the use of an EasyXpress protein synthesis kit (Qiagen). For protein production, 100 ng of the pET21a+::PA3740 construct and its mutated derivatives was used. Purification of the PA3740-His6 protein was carried out by affinity chromatography using Ni-nitrilotriacetic acid-magnetic agarose beads (Qiagen) following the manufacturer's instructions. The protein concentration was determined by the Bio-Rad protein assay (Bio-Rad), which is based on the Bradford method. The purified protein was captured at 4°C and immediately used.
c-di-GMP binding studies using SPR.
Surface plasmon resonance (SPR) interaction studies were performed using a Biacore 3000 instrument (Biacore GE Healthcare) as described by Düvel et al. (37). Briefly, 2′-O-(6-aminohexylcarbamoyl)-c-di-GMP (2′-AHC-c-di-GMP; 1 mM in 100 mM borate running buffer, pH 8.5; Biolog, Bremen, Germany) was immobilized via N-hydroxysuccinimide (NHS)–ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride amine coupling on a CM5 sensor chip. A reference cell was activated and deactivated without ligand immobilization.
All SPR analyses were performed in buffer containing 20 mM MOPS (morpholinepropanesulfonic acid), pH 8, 300 mM NaCl, 1 mg/ml CM-dextran, or 1 mg/ml bovine serum albumin plus 0.005% (vol/vol) surfactant P20 at 20°C. Association and dissociation were monitored for 4 min each. Nonspecific binding was subtracted by performing blank runs with the respective buffer on the reference cell.
Binding analyses were performed by injection of the PA3740 wild type or PA3740 alanine substitution mutants over the sensor surface at a flow rate of 30 μl/min. Competition experiments were performed in the presence of c-di-GMP, cyclic AMP (cAMP), or cGMP (1 μM or 10 μM, as indicated in the figure legends).
The half-maximal (50%) effective concentration (EC50) was determined in a solution competition assay (44). In brief, different concentrations of PA3740 (500 nM, 100 nM, and 50 nM) were preincubated with various amounts of free c-di-GMP (50 pM to 5 μM) for 30 min, and the mixture was injected over the 2′-AHC-c-di-GMP surface at a flow rate of 30 μl/min. The binding signals were collected at the end of the association phase and plotted against the logarithm of the free c-di-GMP concentration. An EC50 was calculated from the dose-response curve using GraphPad Prism software (version 5.01; GraphPad Software, San Diego, CA). After each binding experiment, the sensor surfaces were regenerated by two injections of 3 M guanidinium HCl and one injection of 0.05% SDS, and the drifting baseline was stabilized by one injection of 1 M NaCl.
Peptide array.
Peptide arrays were synthesized by the spot technique (38) on an amino-pegylated cellulose membrane (AIMS Scientific Products GmbH, Berlin, Germany) as described previously (45). Peptides are N-terminally acetylated and remain covalently attached to the membrane spot via their carboxyl terminus. For probing of the array, 10-ml volumes of the various solutions were applied to the membrane. The peptide arrays were conditioned in ethanol for 10 min and washed with TBS (20 mM Tris, 137 mM NaCl, pH 7.6) three times for 5 min each time. Inactivation of unspecific binding sites was achieved by incubation with blocking buffer (recipe, 20 ml casein-based blocking buffer [10×; catalog number B6429; Sigma], 80 ml TBS-Tween, pH 8, 5 g sucrose) for 2 h. The blocked arrays were scanned for background fluorescence. Eight hundred microliters of a 1 µM solution of the fluorescence-labeled 2′-O-(6-[fluoresceinyl]aminohexylcarbamoyl)-c-di-GMP (2′-Fluo-AHC-c-di-GMP) in blocking buffer was applied, and the membrane was incubated for 1 h 15 min in the dark. The arrays were then washed with TBS three times for 10 min each time. Fluorescence scanning was performed with an FLA9000 reader (Fujifilm) using the settings for Sybr green. For removal of bound 2′-Fluo-AHC-c-di-GMP, the peptide arrays were washed with water twice for 10 min each time, three times with buffer A (8 M urea, 1% SDS, 0.5% β-mercaptoethanol; the pH was adjusted to 7 with acetic acid) with 5 min of shaking followed by 5 min in a ultrasonic bath at 30°C, and finally three times with buffer B (10% [vol/vol] acetic acid, 50% [vol/vol] ethanol) for 10 min each time. For short-term storage, the arrays were kept in TBS at 4°C, and for long-term storage, they were washed twice in ethanol, air dried, sealed in a plastic bag, and stored at −20°C.
Dot blot c-di-GMP binding assay.
The dot blot assays were performed as described previously (37) but with the slight modification that the in vitro-translated protein was spotted onto the nitrocellulose membrane.
RESULTS AND DISCUSSION
Coupling of c-di-GMP to Sepharose beads.
Immobilized c-di-GMP has previously been used as the bait for cellular effector molecules for affinity pulldown assays in P. aeruginosa. More precisely, the functionalized c-di-GMP analog 2′-AHC-c-di-GMP, in which the 2′-OH group of one ribose ring was replaced by a 6-aminohexylcarbamoyl group, was coupled to NHS-activated Sepharose beads (37).
Here we used an alternative affinity resin for pulldown experiments, in which chemically unmodified c-di-GMP was coupled to epoxy-activated Sepharose under alkaline conditions, leading to a final ligand density of 0.45 μmol per 100 mg (on a weight dry basis) resin. The coupling conditions most likely resulted in one main product (more than 35%) and at least six by-products. For the main product, a monosubstitution at position N-1 of the guanine ring was demonstrated by NMR (data not shown). The by-products seemed to be partly multisubstituted at position N-7 (guanine ring) and position 2′OH (ribose ring), as depicted in Fig. 1. As c-di-GMP contains two guanosine moieties, the possibility that the compound is at least partially coupled to the Sepharose beads via both guanosine units cannot be excluded.
FIG 1.
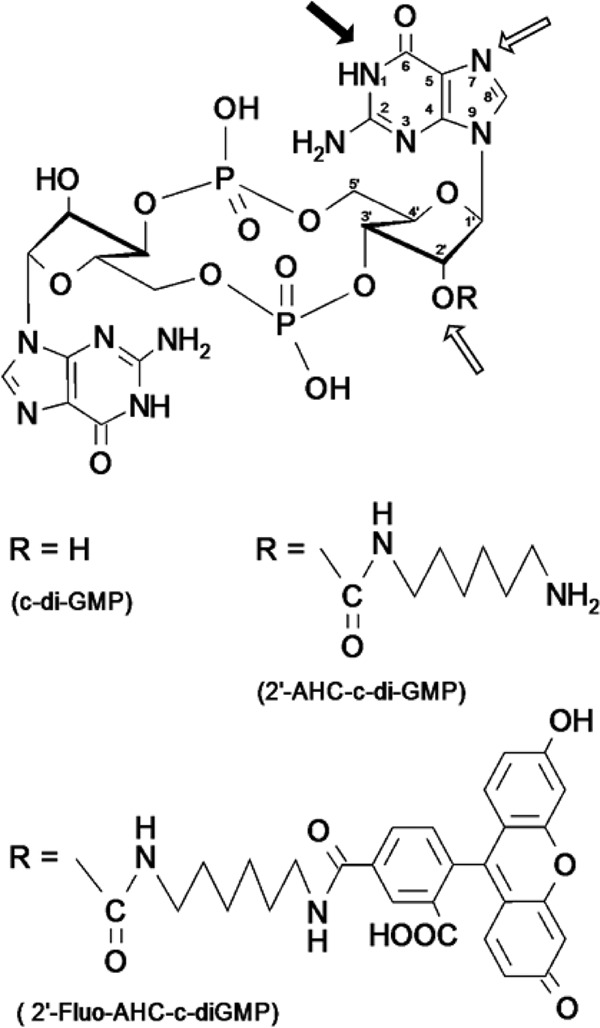
Chemical structure of c-di-GMP. Black arrow, the main position of c-di-GMP coupling to the Sepharose matrix; framed arrows, possible coupling positions of by-products. The possibility of multiple substitutions within the same guanosine moiety or with the second guanosine moiety cannot be excluded. In 2′-AHC-c-di-GMP, the 2′-OH group of one ribose ring is replaced by a 6-aminohexylcarbamoyl group. In 2′-Fluo-AHC-c-di-GMP, 5-carboxyfluorescein has been attached to the 2′-OH group of one ribose ring via an aminohexylcarbamoyl spacer.
Affinity chromatography in P. aeruginosa PAO1.
The c-di-GMP-coupled Sepharose served for the isolation of c-di-GMP binding proteins from P. aeruginosa PAO1. Epoxy-activated Sepharose transformed with ethanolamine was used as a negative control in order to identify proteins that bind to the Sepharose matrix nonspecifically. The freshly prepared cell lysates were incubated with the affinity resins, and following extensive washing steps, the enriched proteins were eluted by the addition of free c-di-GMP. Proteins that eluted in three independent pulldown experiments were analyzed following SDS-PAGE and automated peptide sequencing (LC-MS) as described previously (37). Seven proteins that were enriched by the c-di-GMP-coupled Sepharose in at least two out of three pulldown experiments but not by the respective control Sepharose were identified. Those included the methyltransferase PilK (PA0412), PA0174 (a protein probably involved in chemotaxis), the chemotaxis methyltransferase CheR1 (PA3348), the PilZ domain protein PA3353, PA2567 (which contains a GAF domain, a degenerate GGDEF domain, and an EAL domain) (4, 46), PA4396 (which contains a degenerate GGDEF domain with an intact I site) (4), and PA3740. With the exception of PA2567 and PA3740, all proteins have previously been identified in pulldown experiments with 2′-AHC-c-di-GMP coupled to NHS-activated Sepharose as the bait for the cellular effector molecules (37).
It was obvious that many known c-di-GMP binding proteins were missed by the pulldown approach used in this study. It seems that our alternative coupling approach had a lower overall binding efficiency than other approaches, which might have been due to the fact that in c-di-GMP, the N-7 and O-6 of the guanine nucleobases act as main hydrogen bond acceptors (10, 27, 47–50). However, a possible advantage of this alternative Sepharose over the one used previously is that in the majority of c-di-GMPs immobilized via position N-1, there are still two free 2′-OH groups for interaction with c-di-GMP binding proteins. Therefore, the alternative Sepharose might pull down a different and complementary set of c-di-GMP binding proteins in P. aeruginosa. Indeed, we identified two proteins (PA2567 and PA3740) that were not captured in our previous 2′-AHC-c-di-GMP-coupled Sepharose-based screen.
PA3740 showed a high affinity to c-di-GMP in surface plasmon resonance binding studies.
Whereas Rao et al. (46) showed that the EAL domain of PA2567 is catalytically active, PA3740 is a protein of unknown function which does not harbor any previously identified c-di-GMP binding domain. Furthermore, no putative nucleotide binding sites, such as those identified in GTPases and ATPases, ATP-binding sites of ABC transporters, or proteins binding the dinucleotide NAD or flavin adenine dinucleotide, were identified.
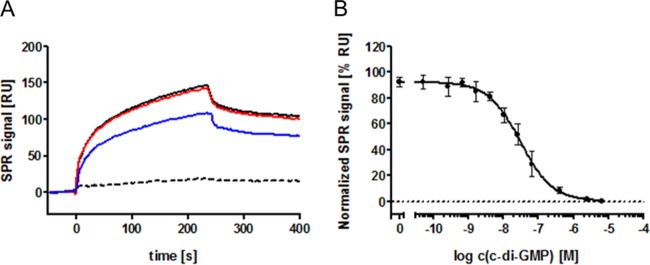
To quantify the c-di-GMP binding of PA3740, we applied SPR. 2′-AHC-c-di-GMP coupled via an amino linker to a CM 5 sensor chip, a strategy successfully applied in previous binding studies (37), was used. PA3740 with a C-terminal His tag was expressed in cell-free protein biosynthesis systems. An SDS gel showing protein production and purification is provided in Fig. S1 in the supplemental material. PA3740-His6 was tested for its ability to interact with the 2′-AHC-c-di-GMP sensor surface. The protein interacted specifically with immobilized 2′-AHC-c-di-GMP since this binding could be completely competed with free c-di-GMP (Fig. 2A). cAMP could not compete for binding to PA3740, whereas we observed a slight competition with cGMP (Fig. 2A). In a solution competition assay, a constant concentration of PA3740-His6 together with increasing amounts of free c-di-GMP was injected over the sensor chip. By plotting the residual SPR signal corresponding to each c-di-GMP concentration against the c-di-GMP concentration, an EC50 of 30 ± 12 nM (n = 3) (Fig. 2B) was determined. This concentration is within the range of intracellular c-di-GMP concentrations (50 nM to a few micromolar) (51). This indicates that PA3740 shows binding to c-di-GMP comparable to that of the high-affinity c-di-GMP receptors containing PilZ domains (1, 37, 51).
FIG 2.
Analysis of c-di-GMP binding to purified PA3740 protein by SPR. (A) Analysis of binding of purified PA3740-His6 demonstrates that PA3740 (800 nM; solid black line) binds specifically to a 2′-AHC-c-di-GMP sensor surface. This binding can be competed with c-di-GMP (1 μM; dashed black line) but not with cAMP (1 μM; red line). cGMP (1 μM; blue line) causes only a slight reduction in binding. (B) Solution competition assays of PA3740-His6 revealed an EC50 of 30 ± 12 nM (n = 3). Normalized data are shown. log c, log concentration.
Overexpression of PA3740 retards growth and affects swimming and swarming motility.
To investigate the cellular function of the c-di-GMP binding protein PA3740 in P. aeruginosa PAO1, we selected a PAO1 PA3740 transposon mutant (39) and monitored c-di-GMP-dependent motility phenotypes. Swimming, swarming, and twitching motility were not influenced in the PA3740 transposon mutant. Furthermore, the crystal violet assay determined that the PA3740 transposon mutant showed no altered biofilm formation.
We next tested whether overexpression of PA3740 has an effect on the bacterial phenotype. While biofilm formation and twitching motility did not differ between the wild type, the mutant, and the PA3740-overexpressing strain (data not shown), the overexpressing strain exhibited reduced swimming and swarming motility (Fig. 3A and B). The strain overexpressing the PA3740 protein reached the same final optical density as the wild type in stationary phase but exhibited retarded growth due to an extension of the lag phase (Fig. 3C). Thus, the possibility that the motility phenotype is linked to growth inhibition cannot be excluded.
FIG 3.
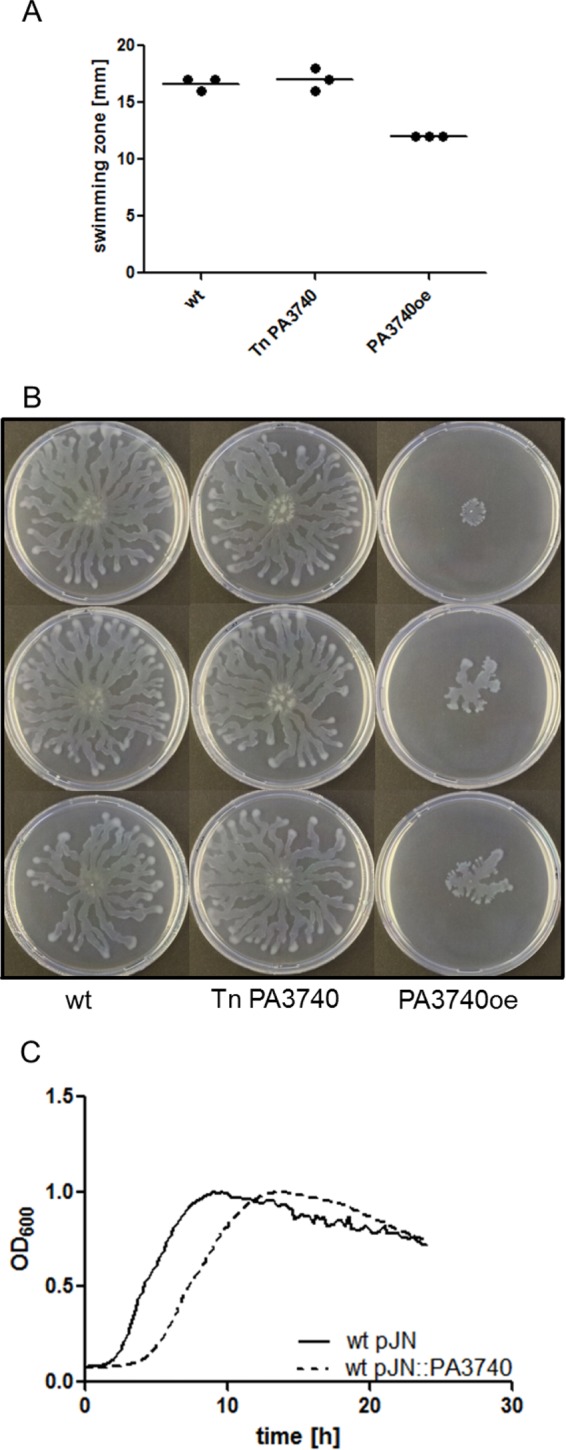
Phenotypic characterization. The swimming motility (A) and swarming motility (B) of wild-type PA3740 (wt), the PA3740 transposon mutant (Tn PA3740), and the PA3740-overexpressing strain (PA3740oe) show reduced swimming and a reduced swarming zone of the PA3740-overexpressing strain. The wild type and the PA3740 transposon mutant contain the empty pJN105 vector. (C) Growth curves of the wild type (wt) with an empty pJN105 vector (pJN) and the wild-type with PA3740 expressed from the vector show a growth delay of the PA3740-overexpressing strain, although both strains reached the same optical density.
Identification of a novel c-di-GMP binding domain.
In the last decade, many tools that serve for the characterization of samples of genome-wide origin have been developed and advanced. For example, spotting of oligonucleotide probes onto a planar surface provided the bases for the hybridization of labeled DNA or RNA target molecules and was exploited to characterize genomic regions for which the sequence is known (52). In this respect, oligonucleotide microarrays were used to define the transcriptional expression of particular genomic regions and also for the discovery of DNA and protein interactions (53, 54). Other methods rapidly complemented the approach of spotting of oligonucleotides, such as the synthesis of peptides on membrane supports. Peptide arrays are used for the analysis of immunological epitopes, protein-protein interactions, as well as enzyme substrate recognition and represent an attractive method for mapping and analysis of linear binding sites (55). Moreover, applications in other fields are steadily growing, and peptide arrays are increasingly used to identify peptides that selectively bind target molecules other than proteins.
In this study, we performed array-based oligopeptide scanning of the PilZ-domain protein PA3353 as a positive control and of PA3740 that exhibited neither a conserved I site nor a PilZ domain. The PilZ domain is a prototype for cellular c-di-GMP receptors, and binding of c-di-GMP involves conserved RXXXR and (D/N)XSXXG linear sequence motifs, whereas c-di-GMP binds to the primary I site in DGCs which contains a conserved RXXD motif (11, 21, 47). We designed peptide arrays which contained 15-mer peptides derived from the sequences of the PilZ domain protein PA3353 and of PA3740, for which the c-di-GMP binding site is unknown. The amino acid sequence of each of the peptides was shifted by 3 amino acids; thus, neighboring peptides overlapped by 12 amino acids. We screened the arrays for binding of fluorescently labeled 2′-AHC-c-di-GMP. As depicted in Fig. 4, the screen revealed two consecutive c-di-GMP binding peptides in both peptide arrays. As expected, in PA3353 the binding peptides corresponded to the previously identified RXXXR motif of the PilZ domain. Screening of the PA3740 peptide array revealed two peptides exhibiting a LKKALKKQTNLR consensus sequence.
FIG 4.
Array-based peptide scans for the identification of c-di-GMP binding motifs. The array contained 15-mer peptides derived from the amino acid sequence from positions 13 to 263 of the PilZ domain containing the PA3353 protein and the amino acid sequence from positions 1 to 228 of PA3740. Each peptide consisted of 15 amino acids overlapping with its neighboring peptides by 12 amino acids. (A) Peptides 46 and 47 containing the RXXXR (RNAYR) motif of the PilZ domain-containing protein PA3353 gave a positive signal with 2′-Fluo-AHC-c-di-GMP. (B) In PA3740, peptides 18 and 19 gave positive signals, the consensus sequence of which was LKKALKKQTNLR.
Importantly, the PA3353 peptide array also covered a region of the protein that exhibited a second RXXXR motif which does not belong to the PilZ domain and which also did not bind to fluorescence-labeled 2'-AHC-c-di-GMP. This result indicates that the mere presence of two arginine residues with a 3-amino-acid spacing is not sufficient for effective c-di-GMP binding.
Consensus sequence for the LKKALKKQTNLR peptide sequence.
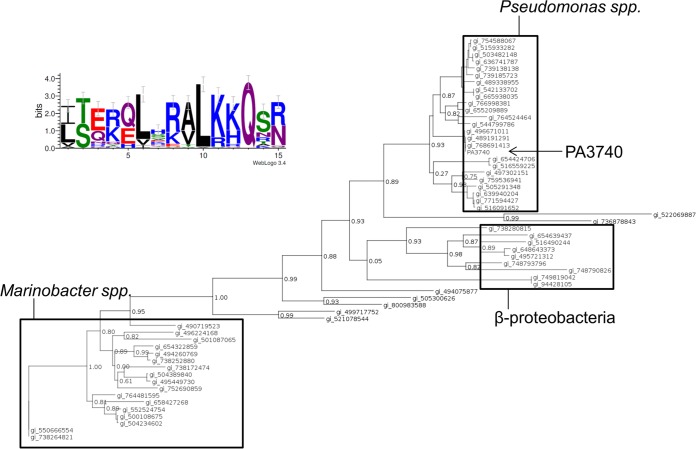
PA3740 is a protein with unknown function; however, its sequence is well conserved across different bacterial species. A protein BLAST (BLASTP) alignment of the full-length sequence of PA3740 against the sequences in the NCBI nonredundant protein collection delivered 57 hits of homologous sequences (outside the P. aeruginosa species with a cutoff value of <E−10) (Fig. 5; see also Table S1 in the supplemental material). Although the lengths of the phylogenetic tree branches indicate quite distant evolutionary relationships between the PA3740 homologs, their common motif shows conserved positions. As depicted in Fig. 5, two lysine residues (KK) of the LKKALKKQTNLR amino acid sequence (underlined) were conserved. Furthermore, both leucine (L) residues as well as the glutamine (Q) were highly conserved.
FIG 5.
Consensus sequence for the LKKALKKQTNLR motif. A BLASTP search aligning the PA3740 sequence against the sequences in the NCBI nonredundant protein collection was performed, with the sequences of the P. aeruginosa group (taxonomic identifier, 136841) being excluded from the search. This search delivered 57 hits (cutoff value, <E−10) of homologous sequences. Sequence alignment of the 57 PA3740 BLAST hits was performed using the ClustalW program, provided by the www.phylogeny.fr web service. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (version 3.1/3.0 aLRT), which was offered on the same server mentioned above. The WAG substitution model was selected, and an estimated proportion of invariant sites of 0.017 and 4 gamma-distributed rate categories were assumed to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data (gamma = 1.252). Reliability for internal branches was assessed using the approximate likelihood-ratio test (aLRT), and the output numbers are depicted on the tree branches. The three main groups of Pseudomonas spp. (without P. aeruginosa), Marinobacter spp., and betaproteobacteria are boxed and labeled accordingly. Table S1 in the supplemental material lists the indicated GI numbers. All 57 homologous sequences were used as an input for the creation of a consensus sequence. (Top left) The consensus sequence logo for the c-di-GMP binding motif was created by use of the WebLogo 3 web service. The overall height of the stack indicates the sequence conservation, while the height of the symbols within the stack indicates the relative frequency of each amino acid at that position.
In order to evaluate whether the c-di-GMP binding motif is present in proteins outside PA3740 and PA3740 homologs, we subsequently performed a BLASTP search against the sequences in the NCBI nonredundant protein collection by entering the LKKALKKQTNLR motif alignment extracted from the 57 PA3740 homologs. High-quality hits (E value, <E−7) belonged exclusively to PA3740 or its homologs, indicating that this motif is unique.
Binding of c-di-GMP to the full-length PA3740 protein confirms the LKKALKKQ motif.
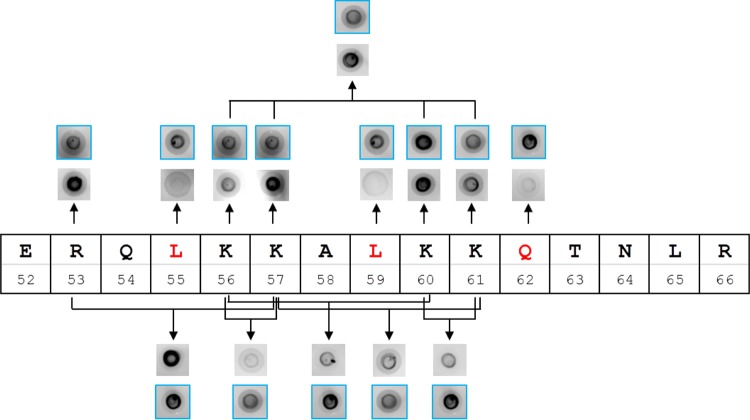
We next explored whether the LKKALKKQ amino acid sequence of PA3740 is important for c-di-GMP binding to the full-length protein and aimed at confirming the key residues of the binding motif. As depicted in Fig. 6, the PA3740 protein and variants thereof exhibiting variations of the putative c-di-GMP binding motif were translated in vitro, spotted onto a nitrocellulose membrane, and incubated with fluorescence-labeled 2′-AHC-c-di-GMP. Single amino acids at positions R53, L55, K56, K57, L59, K60, K61, and Q62 were exchanged to alanine. We also simultaneously exchanged to alanine two amino acids at positions R53 and K57, K56 and K57, K60 and K61, K56 and K60, and K57 and K61 and four amino acids at positions K56, K57, K60, and K61. Very clearly, the exchange of the leucine (L) residues at positions 55 and 59 and the exchange of glutamine (Q) at position 62 were detrimental. The simultaneous exchange of the two lysines (KK) at positions 56 and 57 as well as at positions 60 and 61 was also poorly tolerated.
FIG 6.
Dot blot assay for c-di-GMP binding. In vitro translation products with the indicated mutations were spotted onto the nitrocellulose membrane and incubated with fluorescence-labeled 2′-AHC-c-di-GMP. Highly conserved residues are depicted in red. For each mutant, the respective wild-type control blot is shown in a blue-framed picture.
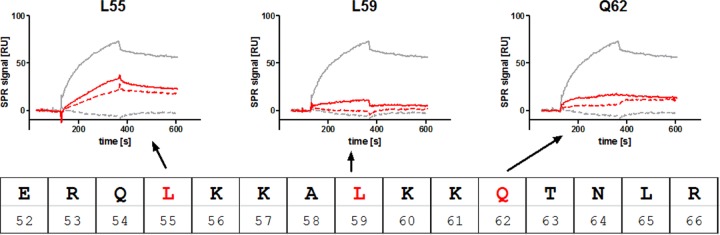
In a parallel experiment, we also applied SPR to test for c-di-GMP binding to the full-length wild-type protein and the mutants with alanine substitutions at L55, L59, and Q62 (Fig. 7). Therefore, a 100 nM concentration of the PA3740 wild type and a 100 nM concentration of each of the respective mutant proteins were injected over the 2′-AHC-c-di-GMP sensor surface. The wild-type protein yielded a binding signal of about 70 resonance units (RU), whereas the L55 mutant showed a signal of only about 30 RU. The L59 and Q62 mutants gave even smaller values (>17 RU). These results demonstrate the importance of L55 and, in particular, of L59 and Q62 for c-di-GMP binding in PA3740, supporting the idea of a novel c-di-GMP binding motif. In contrast to the wild-type protein, the binding signal of the mutant proteins could not be competed to the baseline with 10 μM c-di-GMP, again indicating severe disturbance of the binding motif.
FIG 7.
Surface plasmon resonance analysis of the PA3740 binding motif mutants. 2′-AHC-c-di-GMP was immobilized covalently to the sensor surface as described in Materials and Methods. The PA3740 wild type (gray) and mutant (red) proteins (100 nM each) were injected in the absence (solid lines) or presence (dashed lines) of 10 μM c-di-GMP. Association and dissociation phases were each monitored separately for 240 s.
Conclusion.
In this study, we identified PA3740 to be a novel c-di-GMP binding protein in P. aeruginosa and demonstrated the applicability of oligopeptide scanning for the identification of a novel c-di-GMP binding motif. This motif was demonstrated to be essential for the binding of c-di-GMP to the native full-length protein, and mutational analysis revealed a dominant role of the two leucine residues and the glutamine residue for c-di-GMP binding. The crystal structure of this binding domain in complex with c-di-GMP should uncover how the ligand is bound to the protein, including the involvement of the two leucine residues and the one glutamine residue and the extent to which the neighboring amino acids are important to the high-affinity binding. Whether c-di-GMP induces structural changes upon binding and thus affects downstream targets remains an open question, just as whether this c-di-GMP binding domain can also be expected to be present in other c-di-GMP effector proteins. The next challenge will be to unravel the cellular function of PA3740 in P. aeruginosa. Whether PA3740 inhibits motility directly or whether production of the protein causes an additional stress that leads to a slowdown of motility remains to be explored.
Supplementary Material
ACKNOWLEDGMENTS
The companies Biolog Life Science Institute and Biaffin contributed reagents (Biolog) and analysis tools (Biaffin GmbH & Co. KG). We are grateful to Robert E. W. Hancock (University of British Columbia) for donating PA3740 transposon mutant strains. We thank Susanne Daenicke (Helmholtz Center for Infection Research) for the synthesis of the peptide arrays and Johannes Spehr (Helmholtz Center for Infection Research) for testing different protein production and purification systems.
TWINCORE is a joint venture between the Hannover Medical School, Hannover, and the Helmholtz Center for Infection Research, Braunschweig, Germany.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00377-15.
REFERENCES
- 1.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 9.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem 281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 12.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 13.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol 15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 16.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 17.Tuckerman JR, Gonzalez G, Sousa EH, Wan X, Saito JA, Alam M, Gilles-Gonzalez MA. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 18.Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol 86:720–729. doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanka A, Düvel J, Dötsch A, Klinkert B, Abraham WR, Kaever V, Ritter C, Narberhaus F, Häussler S. 2015. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci Signal 8:ra36. doi: 10.1126/scisignal.2005943. [DOI] [PubMed] [Google Scholar]
- 21.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 22.Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 23.Ryan RP, Fouhy Y, Lucey JF, Jiang BL, He YQ, Feng JX, Tang JL, Dow JM. 2007. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol Microbiol 63:429–442. doi: 10.1111/j.1365-2958.2006.05531.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol 60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leduc JL, Roberts GP. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol 191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An SQ, Caly DL, McCarthy Y, Murdoch SL, Ward J, Febrer M, Dow JM, Ryan RP. 2014. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog 10:e1004429. doi: 10.1371/journal.ppat.1004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Düvel J, Bertinetti D, Möller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jänsch L, Herberg FW, Häussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Frank R. 1992. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217–9232. doi: 10.1016/S0040-4020(01)85612-X. [DOI] [Google Scholar]
- 39.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Römling U, Häussler S. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ Microbiol 9:2475–2485. doi: 10.1111/j.1462-2920.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- 41.Martins TJ, Mumby MC, Beavo JA. 1982. Purification and characterization of a cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues. J Biol Chem 257:1973–1979. [PubMed] [Google Scholar]
- 42.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 43.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 44.Moll D, Prinz A, Gesellchen F, Drewianka S, Zimmermann B, Herberg FW. 2006. Biomolecular interaction analysis in functional proteomics. J Neural Transm 113:1015–1032. doi: 10.1007/s00702-006-0515-5. [DOI] [PubMed] [Google Scholar]
- 45.Beutling U, Stading K, Stradal T, Frank R. 2008. Large-scale analysis of protein-protein interactions using cellulose-bound peptide arrays. Adv Biochem Eng Biotechnol 110:115–152. doi: 10.1007/10_2008_096. [DOI] [PubMed] [Google Scholar]
- 46.Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J 26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by PilZ domain proteins. J Mol Biol 398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. 2007. Structure of BeF3-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southern E. May 1988. Analysing polynucleotide sequences. US patent 20,050,032,048 A1.
- 53.Jin-Lee H, Goodrich TT, Corn RM. 2001. SPR imaging measurements of 1-D and 2-D DNA microarrays created from microfluidic channels on gold thin films. Anal Chem 73:5525–5531. doi: 10.1021/ac010762s. [DOI] [PubMed] [Google Scholar]
- 54.Roth A, Gill R, Certa U. 2003. Temporal and spatial gene expression patterns after experimental stroke in a rat model and characterization of PC4, a potential regulator of transcription. Mol Cell Neurosci 22:353–364. doi: 10.1016/S1044-7431(02)00039-8. [DOI] [PubMed] [Google Scholar]
- 55.Reineke U, Volkmer-Engert R, Schneider-Mergener J. 2001. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol 12:59–64. doi: 10.1016/S0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 56.Overhage J, Bains M, Brazas MD, Hancock REW. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.