ABSTRACT
The nucleotide-based second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is involved in regulating a plethora of processes in bacteria that are typically associated with lifestyle changes. Myxococcus xanthus undergoes major lifestyle changes in response to nutrient availability, with the formation of spreading colonies in the presence of nutrients and spore-filled fruiting bodies in the absence of nutrients. Here, we investigated the function of c-di-GMP in M. xanthus and show that this bacterium synthesizes c-di-GMP during growth. Manipulation of the c-di-GMP level by expression of either an active, heterologous diguanylate cyclase or an active, heterologous phosphodiesterase correlated with defects in type IV pilus (T4P)-dependent motility, whereas gliding motility was unaffected. An increased level of c-di-GMP correlated with reduced transcription of the pilA gene (which encodes the major pilin of T4P), reduced the assembly of T4P, and altered cell agglutination, whereas a decreased c-di-GMP level correlated with altered cell agglutination. The systematic inactivation of the 24 genes in M. xanthus encoding proteins containing GGDEF, EAL, or HD-GYP domains, which are associated with c-di-GMP synthesis, degradation, or binding, identified three genes encoding proteins important for T4P-dependent motility, whereas all mutants had normal gliding motility. Purified DmxA had diguanylate cyclase activity, whereas the hybrid histidine protein kinases TmoK and SgmT, each of which contains a GGDEF domain, did not have diguanylate cyclase activity. These results demonstrate that c-di-GMP is important for T4P-dependent motility in M. xanthus.
IMPORTANCE We provide the first direct evidence that M. xanthus synthesizes c-di-GMP and demonstrate that c-di-GMP is important for T4P-dependent motility, whereas we did not obtain evidence that c-di-GMP regulates gliding motility. The data presented uncovered a novel mechanism for regulation of T4P-dependent motility, in which increased levels of c-di-GMP inhibit transcription of the pilA gene (which encodes the major pilin of T4P), ultimately resulting in the reduced assembly of T4P. Moreover, we identified an enzymatically active diguanylate cyclase that is important for T4P-dependent motility.
INTRODUCTION
Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is a ubiquitous nucleotide-based second messenger involved in regulating a variety of processes that are typically associated with lifestyle changes in response to environmental cues in bacteria. These processes include motility, adhesion, exopolysaccharide (EPS) biosynthesis, biofilm formation, virulence, and cell cycle progression (for reviews, see references 1 to 5). Elevated c-di-GMP levels are generally associated with inhibition of motility, increased adhesion, and the increased EPS synthesis characteristic of biofilm formation. c-di-GMP is synthesized from two GTP molecules by diguanylate cyclases (DGCs) that contain catalytic GGDEF domains (6, 7) and degraded to pGpG or GMP by phosphodiesterases (PDEs) that contain either a catalytic EAL or HD-GYP domain (8–11). Changing c-di-GMP levels are sensed by effectors, which in turn interact with downstream targets. Not all GGDEF, EAL, and HD-GYP domain-containing proteins are active enzymes, and some of these proteins have been shown to bind c-di-GMP and function as c-di-GMP effectors (1, 2, 5, 12–16). Most bacterial genomes encode DGCs and PDEs, but the numbers vary dramatically (1).
Cells of the deltaproteobacterium Myxococcus xanthus undergo major lifestyle changes in response to nutrient availability (17). In the presence of nutrients, the rod-shaped cells grow, divide, and form colonies, where cells at the edge spread outwards in a highly coordinated manner. In response to nutrient limitation, cells initiate a developmental program that culminates in the formation of multicellular fruiting bodies inside which the rod-shaped motile cells differentiate into spherical spores. Spores germinate in the presence of nutrients and resume growth, division, and motility. The formation of spreading colonies as well as fruiting bodies depends on cellular motility. M. xanthus cells move by means of two motility systems. Gliding depends on motility complexes that are distributed along the cell length (18–22). Gliding motility also involves the deposition of slime, which has been proposed to enhance the adhesion of cells to the underlying substratum (23). The composition of the slime is not known, but it is thought to contain carbohydrates and outer membrane vesicles (23). The second motility system depends on type IV pili (T4P). T4P are highly dynamic structures that undergo cycles of extension, adhesion, and retraction, and during a retraction event a cell is pulled forward (for a review, see reference 24). In M. xanthus, T4P-dependent motility is cell-cell contact dependent because EPS stimulates T4P retractions (25) and mutants that have reduced or increased EPS accumulation are defective in T4P-dependent motility (13, 26–35). Regulators of EPS accumulation include the hybrid histidine protein kinase SgmT and its partner, DNA binding response regulator DigR (13, 31). DigR directly regulates the expression of genes for secreted proteins. Moreover, a lack of SgmT or DigR causes increased EPS accumulation by an unknown mechanism. SgmT contains a C-terminal GGDEF domain that is catalytically inactive but binds c-di-GMP in vitro (13). In vivo this binding has been suggested to be responsible for sequestering SgmT to one or two clusters that localize along the cell length (13).
Here, we systematically investigated the function of c-di-GMP as well as of proteins predicted to be involved in c-di-GMP metabolism in growth and motility in M. xanthus. We provide the first direct evidence that M. xanthus synthesizes c-di-GMP. Moreover, we demonstrate that manipulation of the c-di-GMP level in otherwise wild-type (WT) cells causes defects in T4P-dependent motility, whereas we did not obtain evidence that c-di-GMP is important for gliding motility. We also identified an enzymatically active DGC that is important for T4P-dependent motility.
MATERIALS AND METHODS
Strains, cell growth, and motility assays.
M. xanthus cells were grown in liquid 1% CTT medium or on 1% CTT–1.5% agar plates at 32°C (36). All M. xanthus strains are derivatives of the wild-type strain DK1622 (37). The M. xanthus strains and plasmids used in this work are listed in Tables 1 and 2, respectively. The in-frame deletions and insertion mutations were generated as described previously (38). Kanamycin and oxytetracycline were used at concentrations of 40 and 10 μg/ml, respectively. For motility assays, cells were grown in CTT medium to a density of 7 × 108 cells/ml, harvested, and resuspended in 1% CTT to a calculated density of 7 × 109 cells/ml. Five-microliter aliquots of cell suspensions were placed on 0.5% and 1.5% agar supplemented with 0.5% CTT and incubated at 32°C. After 24 h, colony edges were observed using a Leica MZ8 stereomicroscope or a Leica IMB/E inverted microscope and visualized using Leica DFC280 and DFC350FX charge-coupled-device cameras, respectively. T4P-dependent motility was quantified by determination of the increase in colony diameter in three technical replicates.
TABLE 1.
M. xanthus strains used in this work
| M. xanthus strain | Genotypea | Reference or source |
|---|---|---|
| DK1622 | Wild type | 37 |
| DK1217 | cglB2 | 78 |
| DK1300 | pilC | 78 |
| SA3502 | ΔsgmT | 13 |
| SW501 | difE::Kanr | 79 |
| DK10410 | ΔpilA | 80 |
| SA3535 | attB::pTP110 (PpilA-PA5295WT-Strep-tag II) | This study |
| SA3537 | attB::pTP112 (PpilA-PA5295E328A-Strep-tag II) | This study |
| SA3543 | attB::pTP114 (PpilA-dgcAWT-Strep-tag II) | This study |
| SA3559 | attB::pTP131 (PpilA-dgcAD164A-Strep-tag II) | This study |
| SA3524 | ΔMXAN2424 (5–256/275) | This study |
| SA3525 | ΔMXAN2530 (11–406/416) | This study |
| SA3544 | ΔMXAN4232 (6–407/412) | This study |
| SA3546 | ΔMXAN2061 (16–559/569) | This study |
| SA3554 | ΔtmoK (11–1101/1110) | This study |
| SA3533 | ΔMXAN5791 (11–338/348) | This study |
| SA3545 | ΔMXAN5199 (6–297/304) | This study |
| SA3548 | ΔMXAN4675 (11–352/372) | This study |
| SA3555 | ΔMXAN1525 (6–289/294) | This study |
| SA3556 | ΔMXAN2643 (11–355/265) | This study |
| SA3557 | ΔMXAN4029 (6–286/292) | This study |
| SA3558 | ΔMXAN2807 (6–655/661) | This study |
| SA3561 | dmxA::pTP133 | This study |
| SA3569 | ΔMXAN4257 (6–589/595) | This study |
| SA3599 | ΔactA (11–292/302) | This study |
| SA5524 | ΔMXAN2997 (9–641/646) | This study |
| SA5600 | ΔMXAN4463 (46–433/458) | This study |
| SA5605 | ΔMXAN3735 (10–310/319) | This study |
| SA5606 | ΔMXAN7362 (10–665/674) | This study |
| SA5607 | ΔMXAN5366 (10–312/321) | This study |
| SA5525 | ΔMXAN5340 (50–542/547) | This study |
| SA5526 | ΔMXAN5053 (6–612/617) | This study |
| SA5527 | ΔMXAN6098 (6–495/501) | This study |
| SA3568 | dmxA::pTP133 attB::pTP140 (Pnat-dmxA) | This study |
| SA5630 | ΔtmoK attB::pDJS57 (Pnat-tmoK) | This study |
For in-frame deletions, the numbers in parentheses indicate the codons deleted/the total number of codons in a given gene. For strains containing plasmids integrated at the Mx8 attB site, the gene expressed, including the promoter driving the expression, is indicated in parentheses.
TABLE 2.
Plasmids used in this work
| Plasmid | Description | Reference or source |
|---|---|---|
| pBJ114 | Kanr galK | 81 |
| pSWU30 | Tetr | 42 |
| pSW105 | PpilA Kanr | 44 |
| pBGS18 | Kanr | 82 |
| pET24b(+) | Kanr, expression vector | Novagen |
| pET28a(+) | Kanr, expression vector | Novagen |
| pTP110 | pSW105 PA5295WT-Strep-tag II Kanr | This study |
| pTP112 | pSW105 PA5295E328A-Strep-tag II Kanr | This study |
| pTP114 | pSW105 dgcAWT-Strep-tag II Kanr | This study |
| pTP131 | pSW105 dgcAD164A-Strep-tag II Kanr | This study |
| pTP120 | pBJ114, in-frame deletion construct MXAN1525 | This study |
| pTP125 | pBJ114, in-frame deletion construct MXAN2643 | This study |
| pNGS010 | pBJ114, in-frame deletion construct MXAN2997 | This study |
| pTP126 | pBJ114, in-frame deletion construct dmxA | This study |
| pDJS01 | pBJ114, in-frame deletion construct MXAN3735 | This study |
| pTP127 | pBJ114, in-frame deletion construct MXAN4029 | This study |
| pIH01 | pBJ114, in-frame deletion construct MXAN4463 | This study |
| pTP121 | pBJ114, in-frame deletion construct MXAN5199 | This study |
| pDJS02 | pBJ114, in-frame deletion construct MXAN5366 | This study |
| pIH03 | pBJ114, in-frame deletion construct MXAN5791 | This study |
| pDJS03 | pBJ114, in-frame deletion construct MXAN7362 | This study |
| pJH01 | pBJ114, in-frame deletion construct actA | This study |
| pTP122 | pBJ114, in-frame deletion construct MXAN4257 | This study |
| pTP123 | pBJ114, in-frame deletion construct tmoK | This study |
| pNGS009 | pBJ114, in-frame deletion construct MXAN5053 | This study |
| pNGS007 | pBJ114, in-frame deletion construct MXAN5340 | This study |
| pTP105 | pBJ114, in-frame deletion construct MXAN2424 | This study |
| pTP106 | pBJ114, in-frame deletion construct MXAN2530 | This study |
| pTP116 | pBJ114, in-frame deletion construct MXAN2061 | This study |
| pTP128 | pBJ114, in-frame deletion construct MXAN2807 | This study |
| pTP124 | pBJ114, in-frame deletion construct MXAN4232 | This study |
| pTP117 | pBJ114, in-frame deletion construct MXAN4675 | This study |
| pNGS008 | pBJ114, in-frame deletion construct MXAN6098 | This study |
| pTP133 | pBGS18, 500-bp fragment internal to dmxA | This study |
| pTP140 | pSWU30 Pnat-dmxA Tetr | This study |
| pDJS57 | pSWU30 Pnat-tmoK Tetr | This study |
| pDJS31 | pET24b(+) dgcAWT Kanr | This study |
| pTP33 | pET28a(+) sgmT Kanr | 13 |
| pTP137 | pET28a(+) dmxA223–722 Kanr | This study |
| pDJS53 | pET24b(+) tmoK654–1109 Kanr | This study |
Escherichia coli strains were grown in LB broth in the presence of relevant antibiotics (39). All plasmids were propagated in E. coli Mach1 [ΔrecA1398 endA1 tonA ϕ80ΔlacZΔM15 ΔlacX74 hsdR(rK− mK+)] unless otherwise stated.
Quantification of EPS accumulation using a trypan blue binding assay.
To quantify the binding of trypan blue, a liquid binding assay was adapted from the one described previously (40), except that 5 × 108 cells from exponentially growing cultures were harvested, washed, and resuspended in 900 μl 10 mM MOPS (morpholinepropanesulfonic acid), pH 7.5, 1 mM CaCl2 buffer. All experiments were done in biological triplicate. For plate-based assays, cells were grown in CTT medium to a density of 7 × 108 cells/ml, harvested, and resuspended in 1% CTT to a calculated density of 7 × 109 cells/ml. Twenty-microliter aliquots of the cell suspensions were placed on 0.5% agar supplemented with 0.5% CTT and 20 μg/ml trypan blue. The plates were incubated at 32°C for 24 h.
Cell agglutination assay.
Cell agglutination was measured as described previously (41) in agglutination buffer (10 mM MOPS, pH 7.0, 1 mM MgCl2, 1 mM CaCl2). Briefly, exponentially growing cells in 1% CTT were harvested; resuspended in agglutination buffer to a calculated density of 1 × 109 cells/ml, corresponding to an optical density (OD) at 550 nm (OD550) of ∼1; and kept at room temperature between OD readings. The OD550 was monitored every 20 min for 180 min. The relative absorbance was calculated by dividing the absorbance measured at each time point by the initial absorbance for each strain. Experiments were done in three biological replicates.
Immunoblot analysis.
Immunoblot assays were carried out as described previously (39) using a streptactin-horseradish peroxidase (HRP) conjugate (IBA Solutions for Life Sciences, Göttingen, Germany) that binds Strep-tag II. Alternatively, anti-PilA antibodies (42) or anti-PilC antibodies (43) were used together with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) as the secondary antibody. Blots were developed using the Luminata Crescendo Western HRP substrate (Millipore). T4P were sheared from cells that had been grown on 1% CTT–1.5% agar plates at 32°C, purified, and analyzed by immunoblotting with anti-PilA antibodies as described previously (42).
Transmission electron microscopy.
Cells growing exponentially in CTT were analyzed as described previously (44). Electron microscopy was performed on a Philips EM 301 electron microscope at calibrated magnifications.
qRT-PCR analysis.
RNA was isolated from cells grown on 1% CTT–1.5% agar plates using a hot phenol extraction method (31). RNA was treated with DNase I (Ambion) and purified with an RNeasy kit (Qiagen). RNA was confirmed to be free of DNA by PCR analysis. One microgram of RNA was used to synthesize cDNA with a high-capacity cDNA archive kit (Applied Biosystems) using random hexamer primers. Quantitative reverse transcription-PCR (qRT-PCR) was performed in a 25-μl reaction volume using SYBR green PCR master mix (Applied Biosystems) and 0.1 μM primers specific to the target gene in a 7300 real-time PCR system (Applied Biosystems). Experiments were done in two biological replicates. Relative gene expression levels were calculated using the comparative threshold cycle method.
c-di-GMP quantification.
Quantifications of c-di-GMP levels in vegetative M. xanthus cells were performed as described previously (45). Briefly, cells in exponential phase were grown overnight in CTT medium and then diluted. At the indicated time points after dilution, the cells were harvested by centrifugation at 4°C and 2,500 × g for 20 min. Cells were lysed in extraction buffer (high-pressure liquid chromatography [HPLC]-grade acetonitrile-methanol-water [2/2/1, vol/vol/vol]), and the supernatants were pooled and evaporated to dryness in a vacuum centrifuge. The pellets were dissolved in HPLC-grade water for analysis by liquid chromatography-coupled tandem mass spectrometry. As described in the appropriate figure legends, experiments were done in at least three biological replicates in which at least three independent cultures were grown in parallel. For all samples, protein concentrations were determined in parallel using a Bradford assay (Bio-Rad).
Protein purification.
For expression and purification of His6-tagged proteins, proteins were expressed in E. coli Rosetta 2(DE3) [F− ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2] at 18°C or 37°C. His6-tagged proteins were purified using Ni-nitrilotriacetic acid (NTA) affinity purification. Briefly, cells were resuspended in buffer A (50 mM Tris-HCl, 150 mM NaCl, 10 mM imidazole, 1 mM dithiothreitol, 10% glycerol, pH 8) and lysed using a Branson sonifier or a French pressure cell. After centrifugation (1 h, 48,000 × g, 4°C), the lysates were loaded on an Ni-NTA-agarose column (Qiagen) and washed with a 20× column volume using buffer B (50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, pH 8). Proteins were eluted with buffer C (50 mM Tris-HCl, 300 mM NaCl, 200 mM imidazole, pH 8).
In vitro DGC assay.
DGC activity was determined essentially as described previously (46, 47). Briefly, assays were performed with 10 μM (final concentration) purified proteins in a final volume of 40 μl. The reaction mixtures were preincubated for 5 min at 30°C in reaction buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM MgCl2). DGC reactions were initiated by adding 1 mM GTP and [α-32P]GTP (0.1 μCi/μl), and the reaction mixtures incubated at 30°C for the periods of time indicated below. The reactions were stopped by addition of 1 volume of 0.5 M EDTA. The reaction products were analyzed by polyethyleneimine-cellulose thin-layer chromatography (TLC) as described previously (11). Briefly, 2-μl aliquots were spotted on TLC plates (Millipore), dried, and developed in 2:3 (vol/vol) 4 M (NH4)2SO4–1.5 M KH2PO4 (pH 3.65). The plates were dried prior to exposure to a phosphorimaging screen (Molecular Dynamics). Data were collected and analyzed using a Storm 840 scanner (Amersham Biosciences) and Image Quant (version 5.2) software.
In vitro c-di-GMP binding assay.
c-di-GMP binding was determined using a differential radial capillary action of ligand assay (DRaCALA) with 32P-labeled c-di-GMP as described previously (48, 49). Briefly, 32P-labeled c-di-GMP was prepared in-house by incubating 10 μM His6-DgcA with 1 mM GTP and [α-32P]GTP (0.1 μCi μl−1) in reaction buffer (total volume, 200 μl) overnight at 30°C. The reaction mixture was then incubated with 5 units of calf intestine alkaline phosphatase (Fermentas) for 1 h at 22°C to hydrolyze the unreacted GTP. The reaction was stopped by incubation for 10 min at 95°C. The reaction mixture was centrifuged (10 min, 20,000 × g, 20°C), and the supernatant was used for the binding assay. 32P-labeled c-di-GMP was mixed with 20 μM protein, and the mixture was incubated for 10 min at room temperature in binding buffer (10 mM Tris, pH 8.0, 100 mM NaCl, 5 mM MgCl2). Ten microliters of this reaction mixture was transferred to a nitrocellulose filter, allowed to dry, and imaged as described above. For competition experiments, 0.4 mM unlabeled c-di-GMP (Biolog) was added.
RESULTS
M. xanthus accumulates c-di-GMP during vegetative growth.
To determine whether M. xanthus cells produce c-di-GMP, we quantified c-di-GMP levels in wild-type (WT) strain DK1622 cells during growth using a liquid chromatography-coupled tandem mass spectrometry method (45). For these experiments, three independent cultures were grown in parallel and then analyzed. c-di-GMP was detected at the same level throughout the exponential growth phase as well as in stationary phase (Fig. 1). While it was consistently observed that the relative level of c-di-GMP was similar in exponentially growing and stationary-phase cells, the absolute c-di-GMP levels varied up to 2-fold between experiments (cf. Fig. 1, 2B, and 5B). In the cultures for which the results are shown in Fig. 1, exponentially growing cells had a level of c-di-GMP of 4.4 ± 1.7 pmol/mg protein and stationary-phase cells had a level of 4.2 ± 0.7 pmol/mg protein. These data suggest that the level of c-di-GMP is not growth phase regulated in M. xanthus. This is in contrast to the findings for Escherichia coli, in which the level of c-di-GMP was shown to increase at entry into stationary phase from ∼1.2 pmol/mg protein to ∼2.1 pmol/mg protein and then decrease again during stationary phase (45).
FIG 1.
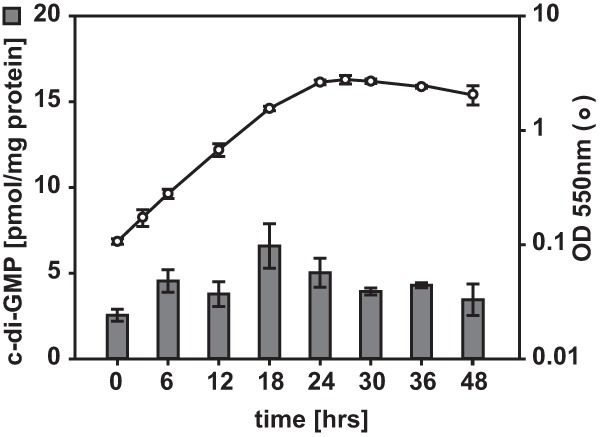
c-di-GMP accumulates in vegetatively growing M. xanthus cells. c-di-GMP levels were determined during vegetative growth of WT strain DK1622 in CTT medium in suspension culture. The levels of c-di-GMP are shown as the mean ± standard deviation (SD) calculated from three biological replicates in which three independent cultures were grown in parallel. Circles, growth measured by determination of the OD550.
FIG 2.
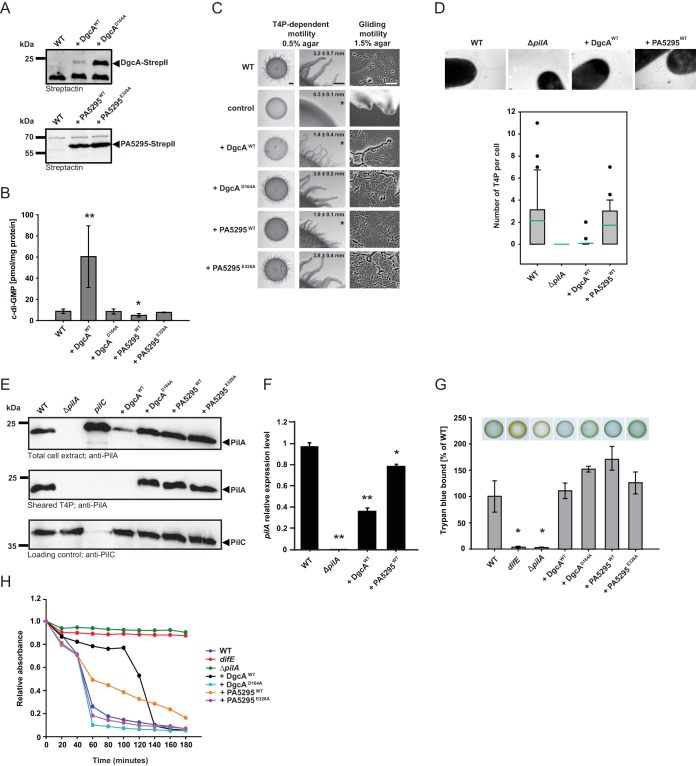
The c-di-GMP level is important for T4P-dependent motility. (A) Immunoblot detection of Strep II-tagged DgcA and PA5295 and their active-site variants. Total protein was isolated from exponentially growing cells expressing the indicated proteins. Total protein from the same number of cells (7 × 107) was loaded per lane, and blots were probed with streptactin. DgcAWT and PA5295WT have calculated molecular masses of 26.8 kDa and 63.6 kDa, respectively. (B) c-di-GMP level in exponentially growing cells expressing the indicated proteins. The levels of c-di-GMP are shown as the mean ± SD from six (WT as well as DgcAWT- and PA5295WT-expressing cells) or three (DgcAD164A- and PA5295E328A-expressing cells) biological replicates. *, P < 0.05 in a Student's t test; **, P < 0.001 in a Student's t test. (C) Motility assays. T4P-dependent motility and gliding motility were analyzed on 0.5% and 1.5% agar, respectively. DK1217 is deficient in gliding motility and DK1300 is deficient in T4P-dependent motility, and these strains were used as negative controls. T4P-dependent motility was quantified by the increase in colony diameter; numbers indicate the mean increase in colony diameter (in millimeters) ± SD from three biological replicates after 24 h. *, P < 0.05 in a Student's t test. Bars, 1 mm (T4P-dependent motility, left column), 500 μm (T4P-dependent motility, right column), and 50 μm (gliding motility). (D) (Top) Images obtained by transmission electron microscopy of exponentially growing cells expressing the indicated proteins. Cells were transferred to a grid, stained with 2% (wt/vol) uranyl acetate, and visualized by transmission electron microscopy. Bars, 100 nm. (Bottom) The box plots show the number of T4P per cell for at least 20 cells. Boxes indicate the 25th and 75th percentiles, the green line indicates the mean, whiskers indicate the 10th and 90th percentiles, and dots indicate outliers. (E) Immunoblot detection of PilA in total cell extract and in sheared T4P. (Top and bottom) Total protein was isolated from the indicated strains grown on 1% CTT agar plates; (middle) T4P were sheared off from the same number of cells used for the top and bottom panels and concentrated by MgCl2 precipitation. In all three blots, protein from the same number of cells was loaded per lane. The top and middle blots were probed with anti-PilA antibodies. The bottom blot was probed against PilC, which is important for T4P assembly and served as a loading control. PilA and PilC have calculated molecular masses of 23.4 kDa and 45.2 kDa, respectively. (F) qRT-PCR analysis of pilA expression. RNA was isolated from the indicated strains grown on 1% CTT–1.5% agar plates. The pilA transcript level relative to that of the WT is shown as the mean ± SD from two biological replicates, each with three technical replicates. *, P < 0.05 in a Student's t test; **, P < 0.001 in a Student's t test. (G) Quantification of EPS accumulation. Exponentially growing cells expressing the indicated proteins were assayed for EPS accumulation using a colorimetric assay. The percentage of trypan blue bound by a strain relative to the amount bound by the WT (100%) is indicated. The levels of trypan blue binding are shown as the mean ± SD from three biological replicates. *, P < 0.05 in a Student's t test. For the plate-based assay, 20-μl aliquots of cell suspensions at 7 × 109 cells/ml were spotted on 0.5% agar supplemented with 0.5% CTT and 20 μg/ml trypan blue and incubated at 32°C for 24 h. (H) Cell agglutination assay. Agglutination was monitored by measuring the decrease in absorbance at 550 nm for a suspension of cells in agglutination buffer. The relative absorbance was calculated by dividing the absorbance measured at each time point by the initial absorbance for each strain. The graph shows data from one representative experiment.
FIG 5.
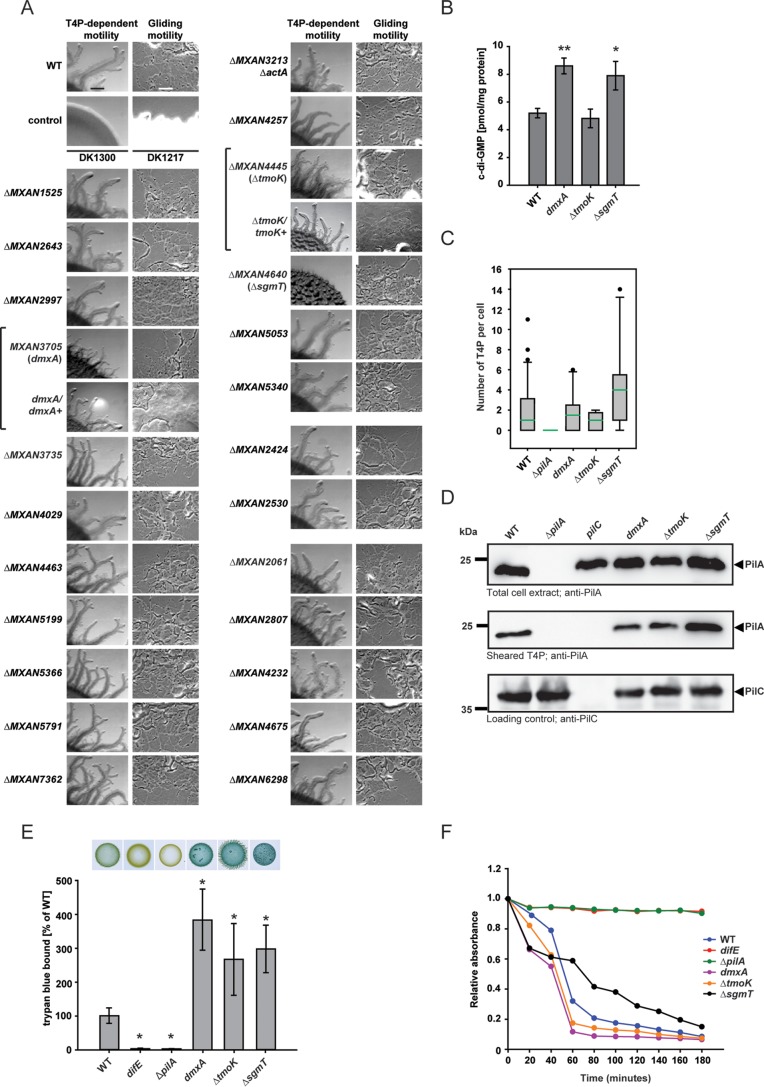
Systematic mutagenesis of M. xanthus genes encoding proteins containing a GGDEF, EAL, or HD-GYP domain. (A) Motility assays. Gliding and T4P-dependent motility was assessed as described in the legend to Fig. 2C. Bars, 500 μm (T4P-dependent motility) and 50 μm (gliding motility). Brackets indicate a mutant and the corresponding complemented strain. (B) c-di-GMP levels in exponentially growing cells of the indicated mutants. The levels of c-di-GMP are shown as the mean ± SD from three biological replicates. *, P < 0.05 in a Student's t test; **, P < 0.001 in a Student's t test. (C) T4P formation by exponentially growing cells of the indicated mutants. The experiment was performed as described in the legend to Fig. 2D. Note that the data for the WT and the ΔpilA mutant are the same as those in Fig. 2D. (D) Immunoblot detection of PilA in total cell extracts and in sheared T4P. The experiment was performed as described in the legend to Fig. 2E. (E) Quantification of EPS accumulation. Samples were analyzed as described in the legend to Fig. 2G. The percentage of trypan blue bound by a strain relative to the amount bound by the WT (100%) is indicated. Levels of trypan blue binding are shown as the mean ± SD from three biological replicates. *, P < 0.001 in a Student's t test. (F) Cell agglutination assay. The experiment was performed as described in the legend to Fig. 2H.
The c-di-GMP level is important for T4P-dependent motility.
To determine if the c-di-GMP level is important for growth or motility in M. xanthus, we manipulated the cellular level of c-di-GMP by overexpression of a heterologous DGC or a heterologous PDE in WT DK1622 cells as previously described for other bacteria (12, 50–52). As a DGC we used DgcA from a WT strain of Caulobacter crescentus (DgcAWT) (53) and as a PDE we used PA5295 from a WT strain of Pseudomonas aeruginosa (PA5295WT) (54). In parallel, we also expressed their active-site variants DgcAD164A, which contains a D-to-A substitution at position 164 (the active site in the WT protein is G162GDEF]), and PA5295E328A, which contains an E-to-A substitution at position 328 (the active site in the WT protein is E328AL), in the DK1622 WT strain. All four proteins were fused C terminally to Strep-tag II to enable their detection by immunoblotting.
In exponentially growing cells, DgcAD164A accumulated at a significantly higher level than DgcAWT, whereas PA5295WT and PA5295E328A accumulated at similar levels (Fig. 2A). The c-di-GMP level in exponential-phase DgcAWT-expressing cells was ∼7-fold higher than that in WT cells (60.4 ± 29.1 pmol/mg protein and 8.7 ± 2.1 pmol/mg protein, respectively; P < 0.001, Student's t test), and the c-di-GMP level in PA5295WT-expressing cells was ∼2-fold lower than that in WT cells (4.9 ± 1.6 pmol/mg protein and 8.7 ± 2.1 pmol/mg protein, respectively; P < 0.05, Student's t test) (Fig. 2B). Importantly, the c-di-GMP level in DgcAD164A- or PA5295E328A-expressing cells (8.5 ± 2.4 and 7.7 ± 0.3 pmol/mg protein, respectively) was not significantly different from that in WT cells (P > 0.2, Student's t test).
Cells expressing DgcA, DgcAD164A, PA5295, or PA5295E328A had the same growth rate as WT cells in suspension culture. On 1.5% agar, which is favorable to gliding, WT strain DK1622 displayed the single cells and slime trails characteristic of gliding motility at the edge of the colony, whereas the gliding-deficient control strain DK1217 did not (Fig. 2C). All four strains expressing DgcA or PA5295 variants displayed single cells and slime trails at the colony edges like the WT did and had the same increase in colony diameter on 1.5% agar as the WT, suggesting that the level of c-di-GMP is not important for gliding motility.
On 0.5% agar, which is favorable to T4P-dependent motility, the WT formed the long flares characteristic of T4P-dependent motility, whereas the control strain, DK1300, which is deficient in T4P-dependent motility, did not (Fig. 2C). The strains expressing DgcAWT or PA5295WT had reduced T4P-dependent motility, as quantified by the increase in the colony diameter and the length of the flares at the colony edges, whereas the two strains containing the active-site variants had WT T4P-dependent motility. We conclude that an increase as well as a decrease in the level of c-di-GMP in otherwise WT cells interferes with T4P-dependent motility.
T4P-dependent motility in M. xanthus depends on T4P and EPS. We used transmission electron microscopy to determine whether the reduced T4P-dependent motility in strains expressing DgcAWT and PA5295WT was due to the lack or reduced functionality of assembled T4P. WT cells assembled a mean of 2.1 T4P per cell in a unipolar pattern, whereas the ΔpilA control strain, which lacks the pilin subunit of T4P, did not assemble T4P (Fig. 2D). PA5295WT-expressing cells assembled T4P at WT levels of a mean of 1.7 T4P per cell in a unipolar pattern, whereas DgcAWT-expressing cells had strongly reduced amounts of assembled T4P, with less than 1 T4P per cell (mean, 0.1 T4P per cell) (Fig. 2D). To corroborate these observations, the total amount of cellular PilA as well as PilA assembled into T4P was quantified. In total cell extracts, the amount of PilA in WT cells and PA5295WT-, DgcAD164A-, or PA5295E328A-expressing cells was similar, whereas DgcAWT-expressing cells accumulated PilA at a significantly reduced level (Fig. 2E). As expected, PilA was not detected in the ΔpilA mutant. In the sheared T4P fraction, WT cells and PA5295WT-, DgcAD164A-, or PA5295E328A-expressing cells contained the same amount of PilA, whereas PilA was not detectable in the sheared T4P fraction from ΔpilA and DgcAWT-expressing cells (Fig. 2E). Thus, DgcAWT-expressing cells accumulate PilA and assemble T4P at a significantly reduced level.
To further understand the mechanism underlying the reduced accumulation of PilA in DgcAWT-expressing cells, we used qRT-PCR to determine the level of pilA transcription in WT cells and DgcAWT- and PA5295WT-expressing cells. As shown in Fig. 2F, the level of pilA mRNA in DgcAWT-expressing cells was approximately 2.5-fold lower than that in WT cells and slightly reduced in PA5295WT-expressing cells, suggesting that increased c-di-GMP levels in otherwise WT cells inhibit pilA transcription.
EPS accumulation in strains expressing variants of DgcA or PA5295 was determined using a colorimetric assay. For this assay, cells were grown in rich medium in suspension culture or on solid medium (0.5% agar). Under both conditions, no significant differences in the levels of EPS accumulation were observed for these four strains compared to the level of accumulation for the WT (Fig. 2G). In contrast, the level of EPS accumulation in the negative-control strain with a mutation in the difE gene, which encodes a component of the Dif chemosensory system that is important for EPS accumulation (26), was strongly reduced under both conditions. Because assembled T4P have been suggested to function upstream of the Dif chemosensory system to stimulate EPS accumulation (55), we also determined EPS accumulation in the ΔpilA mutant. As expected, this mutant also displayed strongly reduced EPS accumulation under both conditions (Fig. 2G).
M. xanthus cells agglutinate in the presence of the divalent cations Mg2+ and Ca2+ (41). Agglutination depends on T4P (41, 42) and EPS (56). In order to examine further the cell surface properties of the strains with altered levels of c-di-GMP, we determined their agglutination properties. As previously reported (41), WT cells started to agglutinate shortly after addition of Mg2+ and Ca2+, and the difE and ΔpilA mutants did not (Fig. 2H). The DgcWT-expressing strain as well as the PA5295WT-expressing strain showed delayed agglutination, with the DgcWT-expressing strain having a more severe defect than the PA5295WT-expressing strain, in agreement with the observation that the DgcWT-expressing strain has a reduced amount of assembled T4P. Importantly, the DgcAD164A- and PA5295E328A-expressing strains agglutinated similarly to the WT. In total, these data suggest that the defect in T4P-dependent motility in the DgcAWT-expressing strain is caused by reduced pilA expression, resulting in reduced PilA accumulation and reduced T4P assembly. On the other hand, our data suggest that the defect in T4P-dependent motility in the PA5295WT-expressing strain is likely not due to a difference in the level of assembled T4P or in the level of EPS accumulation. Because cell-cell cohesion is reduced among cells of the PA5295WT-expressing strain, we suggest that a reduced c-di-GMP level results in changes in cell surface properties that are not reflected either in the level of assembled T4P or in the level of EPS accumulation. Moreover, we suggest that these changes in cell surface properties negatively affect T4P-dependent motility.
M. xanthus proteins containing the GGDEF, EAL, or HD-GYP domain.
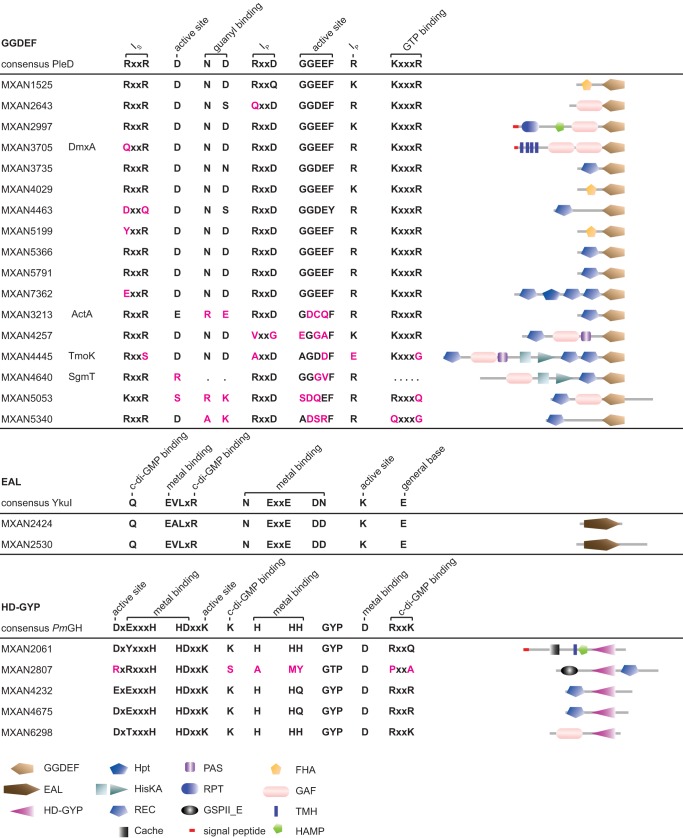
We previously identified 24 genes encoding proteins containing either a GGDEF, EAL, or HD-GYP domain (13). Seventeen proteins contain a GGDEF domain, two proteins contain an EAL domain, and five proteins contain an HD-GYP domain (Fig. 3). Composite proteins that contain a GGDEF domain as well as an EAL domain have been identified in many bacteria; however, among the 24 proteins identified in M. xanthus, none contain more than a single GGDEF, EAL, or HD-GYP domain. With the possible exception of the 2 proteins containing an EAL domain, all 24 proteins contain additional domains that are typically involved in signal sensing and signal transduction in bacteria (Fig. 3). Only 2 of the 24 proteins are predicted to be membrane proteins (MXAN3705 and MXAN2061), suggesting that most of these proteins are not involved in directly sensing extracellular signals.
FIG 3.
Domain organization of M. xanthus proteins containing a GGDEF, EAL, or HD-GYP domain. Locus tags and protein names are listed on the left. The domain composition is shown on the left. See the legend at the bottom for the annotation of the domains. The domains are not drawn to scale. Domain annotation was performed using the SMART web tool (70). For GGDEF domains, the GGDEF domain of PleD from C. crescentus was used as a reference for residues important for catalytic activity and allosteric inhibition, and their functions are indicated (Ip, primary I site; Is, secondary I site) (71, 72); for EAL domains, the EAL domain of YkuI from Bacillus subtilis was used as a reference for important residues, and their functions are indicated (73, 74); and for HD-GYP domains, the HD-GYP domain of PmGH from Persephonella marina EX-H was used as a reference for important residues, and their functions are indicated (75, 76). For M. xanthus proteins, the corresponding amino acid residues are indicated in black if they are conserved or replaced by a conservative substitution and in pink if they are nonconserved.
On the basis of the conservation of amino acid residues important for catalytic activity, 11 of the 17 GGDEF domain-containing proteins are predicted to have DGC activity (Fig. 3). Among the remaining six DGCs predicted to be catalytically inactive, four contain the residues for c-di-GMP binding and may function as c-di-GMP receptors. The two EAL domain-containing proteins and four of the HD-GYP domain-containing proteins contain all the residues important for catalytic activity, while the remaining protein (MXAN2807) lacks several of these conserved residues.
The 24 M. xanthus proteins containing a GGDEF, EAL, or HD-GYP domain are highly conserved in closely related fruiting myxobacteria (Myxococcus fulvus, Myxococcus stipitatus, Corallococcus coralloides, and Stigmatella aurantiaca). However, they are not highly conserved either in four isolates of the nonfruiting myxobacterium Anaeromyxobacter dehalogenans or in the more distantly related fruiting myxobacteria Sorangium cellulosum and Haliangium ochraceum (Fig. 4).
FIG 4.
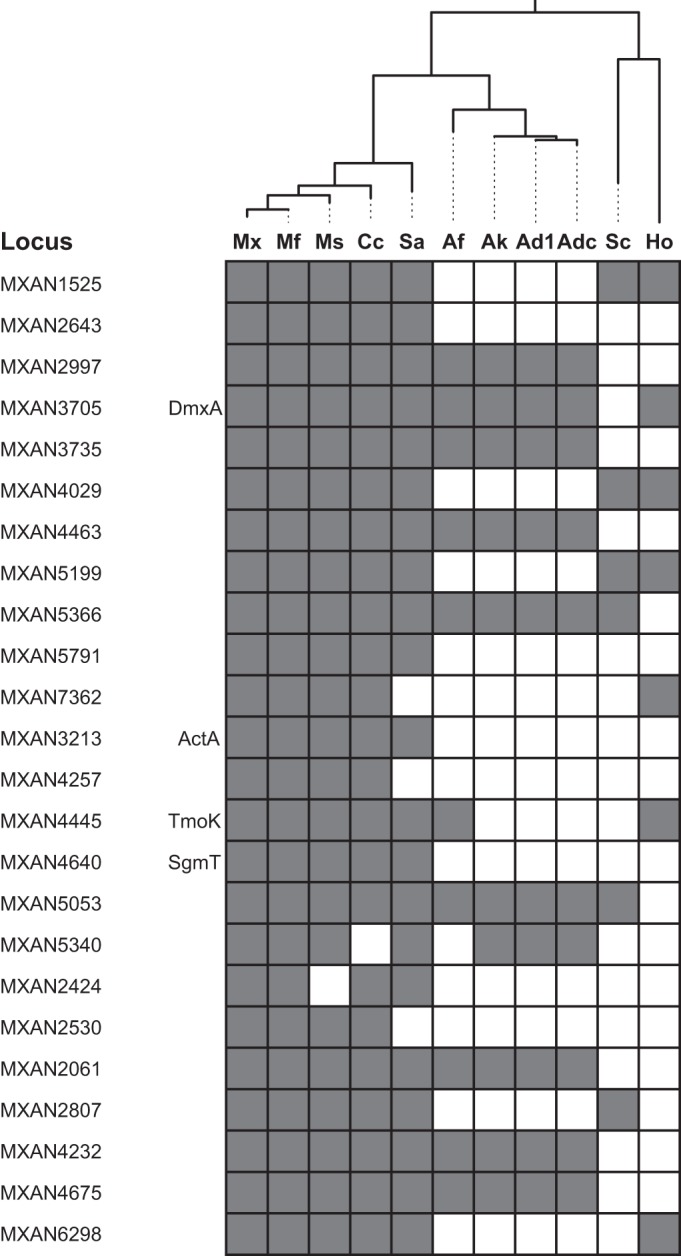
Conservation of M. xanthus proteins containing a GGDEF, EAL, or HD-GYP domain in other myxobacteria. The genomic distribution of M. xanthus proteins containing a GGDEF, EAL, or HD-GYP domain in other myxobacteria in sequenced myxobacterial genomes is shown. The phylogenetic tree was generated from 16S rRNA sequences. Abbreviations in the phylogenetic tree: Mx, M. xanthus DK1622; Mf, M. fulvus HW-1; Ms, M. stipitatus DSM 14675; Cc, Corallococcus coralloides DSM 2259; Sa, Stigmatella aurantiaca DW 3/4-1; Af, Anaeromyxobacter sp. strain Fw109-5; Ak, Anaeromyxobacter sp. strain K; Ad1, Anaeromyxobacter dehalogenans 2CP-1; Adc, A. dehalogenans 2CP-C; Sc, Sorangium cellulosum So ce 56; Ho, Haliangium ochraceum DSM 14365.
Identification of GGDEF and HD-GYP domain-containing proteins important for T4P-dependent motility.
To test whether any of the 24 proteins containing a GGDEF, EAL, or HD-GYP domain have a function in motility, we systematically generated in-frame deletion mutations in 23 of the 24 genes. For one of the genes (MXAN3705), we were not able to generate a deletion mutant, and therefore, an insertion mutation was generated.
Mutation of 21 of the 24 genes did not affect growth, gliding motility, or T4P-dependent motility (Fig. 5). In agreement with previous observations, mutation of actA did not have an effect on motility (57). We did not identify mutants with a defect in gliding motility but identified three mutants with defects in T4P-dependent motility. As previously reported, a lack of the cytoplasmic hybrid histidine protein kinase SgmT, which contains a C-terminal GGDEF domain that binds c-di-GMP but is catalytically inactive, causes strong defects in T4P-dependent motility (13). MXAN3705, which we have named dmxA (diguanylate cyclase from M. xanthus A), encodes a predicted integral membrane protein with a C-terminal GGDEF domain that is predicted to be catalytically active and to bind c-di-GMP (Fig. 3). Mutation of dmxA caused a reduction in T4P-dependent motility with the formation of flares shorter than those in the WT (Fig. 5A). MXAN4445, which we have renamed tmoK (T4P-motility kinase), encodes a predicted cytoplasmic hybrid histidine protein kinase with a C-terminal GGDEF domain which lacks residues important for catalytic activity and c-di-GMP binding. The ΔtmoK mutation caused a subtle defect in T4P-dependent motility with the formation of flares similar in length to those in the WT but less well separated than those in the WT (Fig. 5A). Importantly, the motility defects in the three mutants were complemented by ectopic expression of the relevant WT gene from its native promoter on a plasmid integrated at the Mx8 attB site (Fig. 5A) (13). We conclude that among the 24 genes analyzed, 3 code for proteins that are important for T4P-dependent motility (DmxA, SgmT, and TmoK).
DmxA has enzymatic activity and binds c-di-GMP in vitro.
To test for the enzymatic activity of SgmT, DmxA, and TmoK, we overexpressed His6-tagged full-length or truncated variants of these proteins (Fig. 6A) in Escherichia coli and purified them as soluble proteins. As positive and negative controls for enzyme activity, we purified full-length His6-tagged DgcAWT and SgmT, which we previously predicted does not have DGC activity (13), respectively.
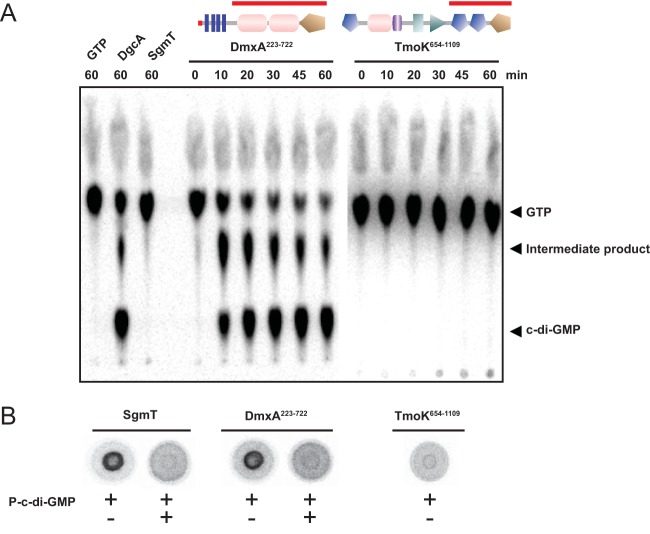
FIG 6.
n vitro assays for DGC activity and c-di-GMP binding. (A) In vitro DGC assay of DmxA and TmoK variants. The indicated DmxA and TmoK variants were incubated with [α-32P]GTP for the indicated periods of time, followed by separation of nucleotides by thin-layer chromatography. Full-length DgcAWT and SgmT were used as positive and negative controls, respectively. Domain architectures are shown as SMART images, as in Fig. 3, with the red bars indicating the part of each protein used in the assay. GTP and c-di-GMP are indicated. The intermediate product indicated was described to be a product formed during the DGC-dependent synthesis of c-di-GMP (77). (B) DRaCALA to detect specific c-di-GMP binding by purified proteins. Full-length SgmT or the DmxA and TmoK variants described in the legend to panel A were incubated with [α-32P]-labeled c-di-GMP with or without unlabeled c-di-GMP added.
DgcAWT as well as DmxA from positions 223 to 722 (DmxA223–722) produced c-di-GMP when incubated with [α-32P]GTP, as detected after separation of the nucleotides by thin-layer chromatography (Fig. 6A). TmoK from positions 654 to 1109 (TmoK654–1109) and SgmT did not detectably produce c-di-GMP (Fig. 6A). DGCs function as dimers, with two juxtaposed GGDEF domains forming the active site (58). In PleD of C. crescentus, dimer formation is mediated by the two receiver domains that are located N terminal to the GGDEF domain (59). For that reason, the truncated TmoK variant analyzed in vitro also contained the two receiver domains N terminal to the GGDEF domain. Although the possibility that TmoK654–1109 does not form a dimer under the conditions of the DGC assay cannot be ruled out, the observation that the GGDEF domain lacks several residues important for DGC activity (Fig. 4), taken together with the in vitro data, suggests that TmoK does not have DGC activity.
To test whether DmxA or TmoK binds c-di-GMP, we used a DRaCALA with α-32P-labeled c-di-GMP generated in an enzymatic reaction for detection of c-di-GMP binding by mixing purified DgcA from C. crescentus with α-32P-labeled GTP as described previously (47) and using SgmT as a positive control for c-di-GMP binding (13). In agreement with the predictions from sequence analyses, specific binding of α-32P-labeled c-di-GMP by DmxA was detected, whereas TmoK was not observed to bind α-32P-labeled c-di-GMP (Fig. 6B).
A lack of SgmT and DmxA causes an increase in the c-di-GMP level in vegetative M. xanthus cells.
To test if the effect of a lack of DmxA, TmoK, and SgmT on T4P-dependent motility involves changes in c-di-GMP levels, the c-di-GMP level was determined in the dmxA, ΔtmoK, and ΔsgmT mutants. Unexpectedly, exponentially growing cells of the dmxA and ΔsgmT mutants had slightly (approximately 1.5-fold) but significantly higher c-di-GMP levels than the WT (Fig. 5B), whereas the ΔtmoK mutant had a c-di-GMP level similar to that of the WT (Fig. 5B).
To understand the mechanism underlying the reduced T4P-dependent motility in the dmxA mutant and the subtle defect in T4P-dependent motility in the ΔtmoK mutant, T4P formation, EPS accumulation, and agglutination were quantified in these two mutants. The dmxA mutant assembled T4P similarly to the WT (Fig. 5C and D) and accumulated ∼4-fold more EPS than the WT in suspension cultures as well as on solid medium (Fig. 5E). The ΔtmoK mutant also assembled T4P similarly to the WT (Fig. 5C and D) and accumulated ∼3-fold more EPS than the WT in suspension cultures as well as on solid medium (Fig. 5E). As previously reported, the ΔsgmT mutant assembled slightly more T4P than the WT and also accumulated ∼3-fold more EPS than the WT in suspension cultures as well as on solid medium (Fig. 5C and E) (13). In support of these data, we observed that all three mutants assembled the same total level of PilA as the WT; however, the ΔsgmT mutant had slightly more PilA in the sheared T4P fraction (Fig. 5D). Moreover, all three mutants exhibited changed agglutination, with the ΔsgmT mutant displaying delayed agglutination compared with the time of agglutination for the WT and the dmxA and the ΔtmoK mutants displaying faster agglutination than the WT (Fig. 5F).
DISCUSSION
Here, we show that the second messenger c-di-GMP is important for T4P-dependent motility in M. xanthus. This conclusion is based on three observations. First, c-di-GMP accumulates in growing M. xanthus cells. Second, expression in otherwise WT cells of a heterologous DGC (DgcAWT) or a heterologous PDE (PA5295WT) but not the corresponding variants with substitutions in the active sites allowed the manipulation of the c-di-GMP level in growing cells. In these cells, an increase as well as a decrease in the c-di-GMP level caused defects in T4P-dependent motility. Because the enzymatically inactive variants of DgcA and PA5295 did not interfere with T4P-dependent motility, these effects are caused by changes in the c-di-GMP level. Third, in an approach in which all 24 genes potentially encoding active DGCs or PDEs in M. xanthus were systematically inactivated, it was observed that a lack of the active DGC DmxA or the c-di-GMP binding histidine protein kinase SgmT caused a defect in T4P-dependent motility.
During growth, reduced T4P-dependent motility was observed as a consequence of both increased and decreased levels of c-di-GMP in otherwise WT cells. T4P-dependent motility in M. xanthus depends on T4P and EPS. An ∼7-fold increase in the c-di-GMP level in otherwise WT cells caused by expression of the heterologous DgcAWT resulted in a significant reduction in the level of transcription of the pilA gene, which codes for the major pilin; in PilA accumulation; and in the amount of assembled T4P. It has previously been shown that there is a correlation between cell-cell cohesion and T4P-dependent motility in M. xanthus and that this cohesion requires T4P and EPS (41, 56, 60). Consistent with the significantly reduced formation of T4P in DgcAWT-expressing cells, they displayed delayed agglutination. Elevated c-di-GMP levels are often associated with increased EPS synthesis (for reviews, see references 1 to 5). However, the DgcAWT-expressing cells did not exhibit differences in EPS accumulation. In M. xanthus, assembled T4P have been suggested to function upstream of the Dif chemosensory system to stimulate EPS accumulation (55) (Fig. 2G), whereas a lack of EPS does not affect T4P assembly (26). Taken together, these observations suggest that the primary defect caused by a highly increased c-di-GMP level in otherwise WT cells during growth is reduced pilA transcription, leading to reduced PilA accumulation and, as a result, reduced T4P formation. As discussed below, in M. xanthus mutants that lack either DmxA or SgmT, increased c-di-GMP levels correlate with increased EPS accumulation. Therefore, we speculate that the increased c-di-GMP level in otherwise WT cells may also stimulate EPS accumulation, but this effect is possibly confounded by the lack of assembled T4P in DgcAWT-expressing cells. Previously, pilA expression has been shown to depend on the transcriptional regulator PilR, which is an NtrC-like transcription regulator (42). How c-di-GMP regulates PilA accumulation remains to be shown; however, it is interesting to note that NtrC-like transcriptional regulators have been identified to be c-di-GMP effectors (see below).
During growth, an ∼2-fold reduction in the c-di-GMP level by expression of the heterologous PA5295WT in otherwise WT cells did not cause significant differences in PilA accumulation, T4P assembly, or EPS accumulation. Nevertheless, these cells had reduced T4P-dependent motility. Interestingly, these cells displayed delayed agglutination, suggesting that a reduced c-di-GMP level results in changes in cell surface properties that are not reflected either in the level of assembled T4P or in the level of EPS accumulation. We speculate that these changes in cell surface properties cause the defect in T4P-dependent motility.
Manipulation of the c-di-GMP level in otherwise WT cells documents that the c-di-GMP level is important for T4P-dependent motility. By systematically inactivating the 24 genes encoding GGDEF, EAL, or HD-GYP domain-containing proteins, we identified three such proteins that are important for T4P-dependent motility. TmoK is a hybrid histidine protein kinase with a catalytically inactive C-terminal GGDEF domain that, on the basis of sequence analysis and experimental data obtained using DRaCALA, does not bind c-di-GMP. A lack of TmoK caused a subtle defect in T4P-dependent motility but did not affect the level of c-di-GMP or T4P, while EPS accumulation was increased and cells agglutinated faster than WT cells. Because the c-di-GMP level was unchanged in the ΔtmoK mutant, the effects of a lack of TmoK on EPS, T4P-dependent motility, and agglutination are likely independent of c-di-GMP. SgmT is a hybrid histidine protein kinase with a catalytically inactive C-terminal GGDEF domain that binds c-di-GMP (13), and DmxA contains a catalytically active GGDEF domain that also binds c-di-GMP. Unexpectedly, a lack of either SgmT or DmxA caused slight but significant ∼1.5-fold increases in the c-di-GMP levels. A lack of SgmT or DmxA also caused 3- to 4-fold increases in the level of EPS accumulation, had no or only a small effect on T4P assembly, and also altered cell-cell cohesion, as measured in the agglutination assay. EPS biosynthesis in M. xanthus depends on proteins encoded by the eps locus (35); however, little is known about the function of the corresponding proteins. Several regulators of EPS synthesis have been identified, including the Dif chemosensory system, T4P, the SgmT/DigR two-component system, NtrC-like transcription regulators, the MasK tyrosine protein kinase, the FrzS response regulator, and DnaK homologs (13, 26–35). Also for these regulators, little is known about how they function in regulating EPS accumulation. Response regulators and NtrC-like transcriptional regulators have been shown to be c-di-GMP effectors (61–63). Thus, it remains a possibility that c-di-GMP directly regulates the activity of one or more of the regulators previously identified to be important for correct EPS accumulation.
A reduced level of c-di-GMP in otherwise WT cells caused by an unknown mechanism correlates with reduced T4P-dependent motility. The increased c-di-GMP levels in otherwise WT cells caused by expression of DgcAWT and in cells lacking SgmT or DmxA also correlate with reduced T4P-dependent motility. Clearly, however, for these three strains the underlying mechanisms leading to this defect in T4P-dependent motility are different; i.e., in the case of the DgcAWT-expressing cells, our data suggest that this defect is caused by a lack of T4P, and in the case of the dmxA and ΔsgmT cells, our data suggest that this defect is caused by increased EPS accumulation. In total, while altered levels of c-di-GMP correlate with an effect on T4P-dependent motility, there is no simple relationship between a change in the c-di-GMP level and the effect on T4P-dependent motility, suggesting that the relationship between changing levels of c-di-GMP and T4P-dependent motility is complex. Also, while it is clear that DmxA and SgmT are important for T4P-dependent motility, it remains to be clarified how they impact c-di-GMP levels and T4P-dependent motility.
It is an open question how the c-di-GMP produced by different DGCs can elicit different responses. In one model for c-di-GMP-dependent regulation, distinct signaling systems with dedicated functions regulate c-di-GMP levels in highly localized and insulated pools rather than contribute to a shared cellular pool of c-di-GMP (2). Based on the observation that the mechanism(s) underlying the defects in T4P-dependent motility in the dmxA and ΔsgmT mutants is different from that in DgcAWT-expressing cells, we suggest that DmxA and SgmT are embedded in signaling systems that contribute to local c-di-GMP pools. Along the same lines, because SgmT does not have DGC activity and DmxA does, the higher c-di-GMP levels in the mutants lacking one or the other of these two proteins are not simply caused by the lack of either protein but likely involve an indirect effect(s) on other DGCs or PDEs. We previously showed that SgmT is sequestered in one or more clusters localized along the cell length in a manner that depends on c-di-GMP binding by the GGDEF domain and suggested that a catalytically active DGC(s) is present in these clusters and would function to sequester SgmT (13). Thus, in the case of the ΔsgmT mutant, it is possible that a lack of SgmT would cause an increase in the activity of this hypothetical DGC(s).
In several other bacteria, low c-di-GMP levels are associated with reduced EPS accumulation and high levels are associated with increased EPS accumulation (for a review, see reference 1), as reported here for the dmxA and ΔsgmT mutants. Also, c-di-GMP-dependent inhibition of motility is commonly observed. A well-understood example involves the PilZ domain protein YcgR in E. coli and Salmonella that upon c-di-GMP binding interacts with the flagellum basal body to interfere with flagellum rotation (64, 65). c-di-GMP has also been reported to regulate gliding motility in Bdellovibrio bacteriovorus (66). Finally, c-di-GMP has been implicated in the regulation of T4P-dependent motility in P. aeruginosa and Xanthomonas axonopodis pv. citri by binding to the catalytically inactive EAL domain of the FimX protein that stimulates T4P assembly (16, 67–69). In Xanthomonas axonopodis, FimX interacts with a PilZ domain protein that in turn interacts with the PilB ATPase that is required for T4P assembly (68, 69). The inhibition of pilA transcription by high levels of c-di-GMP observed here thus represents a novel mechanism for how c-di-GMP may regulate T4P-dependent motility. The M. xanthus genome encodes at least 15 PilZ domain proteins but does not encode a FimX homolog.
The data presented here provide evidence that c-di-GMP is important for T4P-dependent motility in M. xanthus. While the identification of 3 proteins—among 24 proteins predicted to be involved in c-di-GMP metabolism—that are important for T4P-dependent motility provides a basis for beginning to understand how c-di-GMP regulates T4P-dependent motility in M. xanthus, it also raises the question of the function of the remaining 21 proteins. Most of these 21 proteins are conserved in related fruiting myxobacteria (Fig. 4), suggesting strong selective pressure to maintain these genes, yet no phenotypic differences between the WT and any of the 21 mutants were evident, suggesting that these 21 proteins are not active under the conditions tested, function in the regulation of fruiting body formation, or have partially redundant functions. Future research will focus on understanding how c-di-GMP accumulation is regulated and how c-di-GMP regulates T4P-dependent motility and possibly also development in M. xanthus.
ACKNOWLEDGMENTS
We gratefully acknowledge Urs Jenal and the late Alex Böhm for providing plasmids, Annette Garbe for perfect technical assistance in performing the c-di-GMP quantifications, Nuria Gomez-Santos and Isabel Hinderberger for the construction of strains and plasmids, and Kristin Wuichet for help with bioinformatics.
This work was supported by the German Research Council within the framework of the Collaborative Research Center 987 Microbial Diversity in Environmental Signal Response and the Max Planck Society.
REFERENCES
- 1.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 4.Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci 21:929–948. doi: 10.1002/pro.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem 280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He Y-W, Zhang L-H, Heeb S, Cámara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 12.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, Hoppert M, Jenal U, Søgaard-Andersen L. 2012. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol Microbiol 84:147–165. doi: 10.1111/j.1365-2958.2012.08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y, Chuah MLC, Dong X, Xie K, Luo Z, Tang K, Liang Z-X. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J Biol Chem 286:2910–2917. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konovalova A, Petters T, Søgaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol Rev 34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 18.Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T. 2011. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet 7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. 2011. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A 108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nan BY, Mauriello EMF, Sun IH, Wong A, Zusman DR. 2010. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol 76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. 2011. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A 108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. 2012. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci U S A 109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelicic V. 2008. Type IV pili: pluribus unum? Mol Microbiol 68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A 100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol 182:5793–5798. doi: 10.1128/JB.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J Bacteriol 185:6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berleman JE, Vicente JJ, Davis AE, Jiang SY, Seo Y-E, Zusman DR. 2011. FrzS regulates social motility in Myxococcus xanthus by controlling exopolysaccharide production. PLoS One 6:e23920. doi: 10.1371/journal.pone.0023920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomasson B, Link J, Stassinopoulos AG, Burke N, Plamann L, Hartzell PL. 2002. MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus. Mol Microbiol 46:1399–1413. doi: 10.1046/j.1365-2958.2002.03258.x. [DOI] [PubMed] [Google Scholar]
- 30.Lancero H, Caberoy NB, Castaneda S, Li Y, Lu A, Dutton D, Duan X-Y, Kaplan HB, Shi W, Garza AG. 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology 150:4085–4093. doi: 10.1099/mic.0.27381-0. [DOI] [PubMed] [Google Scholar]
- 31.Overgaard M, Wegener-Feldbrügge S, Søgaard-Andersen L. 2006. The orphan response regulator DigR is required for synthesis of extracellular matrix fibrils in Myxococcus xanthus. J Bacteriol 188:4384–4394. doi: 10.1128/JB.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weimer RM, Creighton C, Stassinopoulos A, Youderian P, Hartzell PL. 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol 180:5357–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dana JR, Shimkets LJ. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J Bacteriol 175:3636–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancero HL, Castaneda S, Caberoy NB, Ma X, Garza AG, Shi W. 2005. Analysing protein-protein interactions of the Myxococcus xanthus Dif signalling pathway using the yeast two-hybrid system. Microbiology 151:1535–1541. doi: 10.1099/mic.0.27743-0. [DOI] [PubMed] [Google Scholar]
- 35.Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol Microbiol 55:206–220. [DOI] [PubMed] [Google Scholar]
- 36.Hodgkin J, Kaiser D. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A 74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A 76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi X, Wegener-Feldbrugge S, Huntley S, Hamann N, Hedderich R, Søgaard-Andersen L. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J Bacteriol 190:613–624. doi: 10.1128/JB.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Black WP, Yang Z. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol 186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimkets LJ. 1986. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol 166:837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu SS, Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol 179:7748–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Höne A, Maier B, Hoppert M, Søgaard-Andersen L. 2009. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol 74:691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J Bacteriol 190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
- 47.Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun 79:3273–3283. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A 108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Romling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levi A, Folcher M, Jenal U, Shuman HA. 2011. Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. mBio 2:e00316-10. doi: 10.1128/mBio.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem 281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 54.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Black WP, Xu Q, Yang Z. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol 61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- 56.Arnold JW, Shimkets LJ. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol 170:5771–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gronewold TM, Kaiser D. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol Microbiol 40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 58.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem 282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 60.Shimkets LJ. 1986. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol 166:842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Böhm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 66.Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol 60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guzzo CR, Dunger G, Salinas RK, Farah CS. 2013. Structure of the PilZ–FimXEAL–c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol 425:2174–2197. doi: 10.1016/j.jmb.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 69.Guzzo CR, Salinas RK, Andrade MO, Farah CS. 2009. PilZ protein structure and interactions with PilB and the FimX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol 393:848–866. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 70.Letunic I, Doerks T, Bork P. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. 2007. Structure of BeF3-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 72.Vorobiev S, Neely H, Yu B, Seetharaman J, Xiao R, Acton T, Montelione G, Hunt J. 2012. Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product. J Struct Funct Genomics 13:177–183. doi: 10.1007/s10969-012-9136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Baslé A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem 284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao F, Yang Y, Qi Y, Liang Z-X. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bellini D, Caly DL, McCarthy Y, Bumann M, An S-Q, Dow JM, Ryan RP, Walsh MA. 2014. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol 91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. 2011. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio 2:e00163-11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427. doi: 10.1099/mic.0.053892-0. [DOI] [PubMed] [Google Scholar]
- 78.Hodgkin J, Kaiser D. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet 171:177–191. doi: 10.1007/BF00270004. [DOI] [Google Scholar]
- 79.Yang Z, Geng Y, Xu D, Kaplan HB, Shi W. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol Microbiol 30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- 80.Wu SS, Kaiser D. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J Bacteriol 178:5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A 97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spratt BG, Hedge PJ, Ste Heesen Edelman A, Broome-Smith JK. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]