ABSTRACT
We previously identified a second-messenger-regulated signaling system in the environmental bacterium Pseudomonas fluorescens which controls biofilm formation in response to levels of environmental inorganic phosphate. This system contains the transmembrane cyclic di-GMP (c-di-GMP) receptor LapD and the periplasmic protease LapG. LapD regulates LapG and controls the ability of this protease to process a large cell surface adhesin protein, LapA. While LapDG orthologs can be identified in diverse bacteria, predictions of LapG substrates are sparse. Notably, the opportunistic pathogen Pseudomonas aeruginosa harbors LapDG orthologs, but neither the substrate of LapG nor any associated secretion machinery has been identified to date. Here, we identified P. aeruginosa CdrA, a protein known to mediate cell-cell aggregation and biofilm maturation, as a substrate of LapG. We also demonstrated LapDG to be a minimal system sufficient to control CdrA localization in response to changes in the intracellular concentration of c-di-GMP. Our work establishes this biofilm signaling node as a regulator of a type Vb secretion system substrate in a clinically important pathogen.
IMPORTANCE Here, the biological relevance of a conserved yet orphan signaling system in the opportunistic pathogen Pseudomonas aeruginosa is revealed. In particular, we identified the adhesin CdrA, the cargo of a two-partner secretion system, as a substrate of a periplasmic protease whose activity is controlled by intracellular c-di-GMP levels and a corresponding transmembrane receptor via an inside-out signaling mechanism. The data indicate a posttranslational control mechanism of CdrA via c-di-GMP, in addition to its established transcriptional regulation via the same second messenger.
INTRODUCTION
Bacteria in nature exist as free-swimming motile organisms or as sessile communities adhered to solid surfaces that are enveloped in a self-produced matrix of adhesive proteins, polysaccharides, and nucleic acids (1). These biofilms protect bacterial communities from their surrounding environment, and as a result, infections caused by biofilm-forming pathogens are often tolerant to traditional antibiotic therapies and the immune system. Understanding the molecular mechanisms governing the regulation of bacterial biofilm formation is thus paramount to finding new avenues for treating such chronic infections.
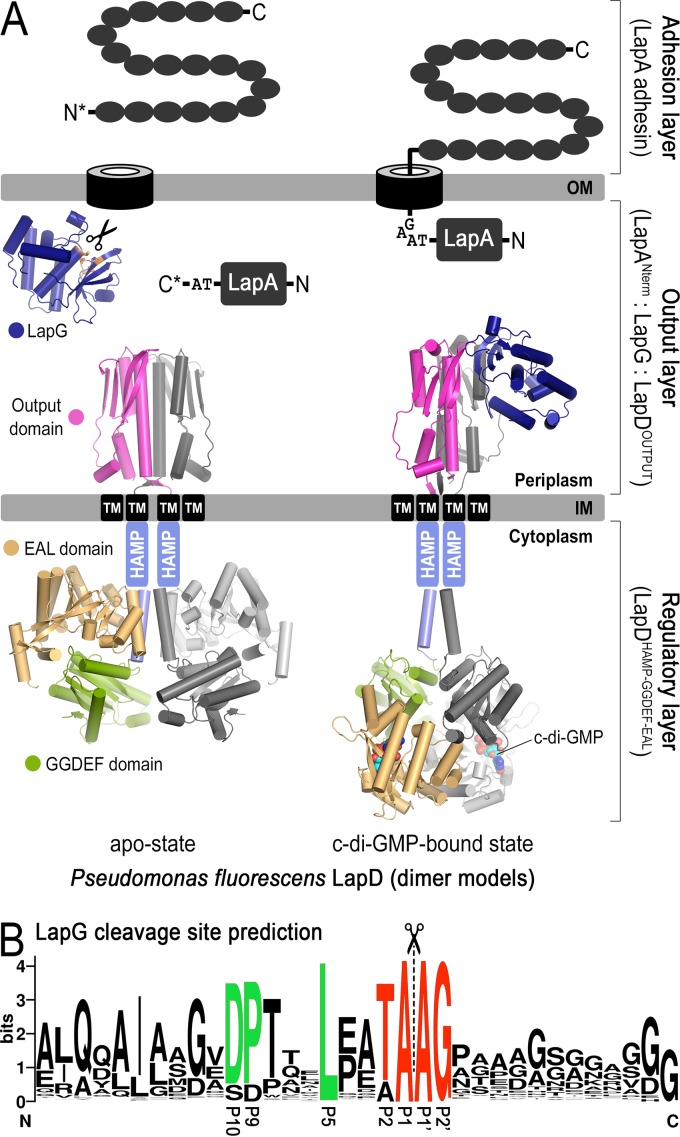
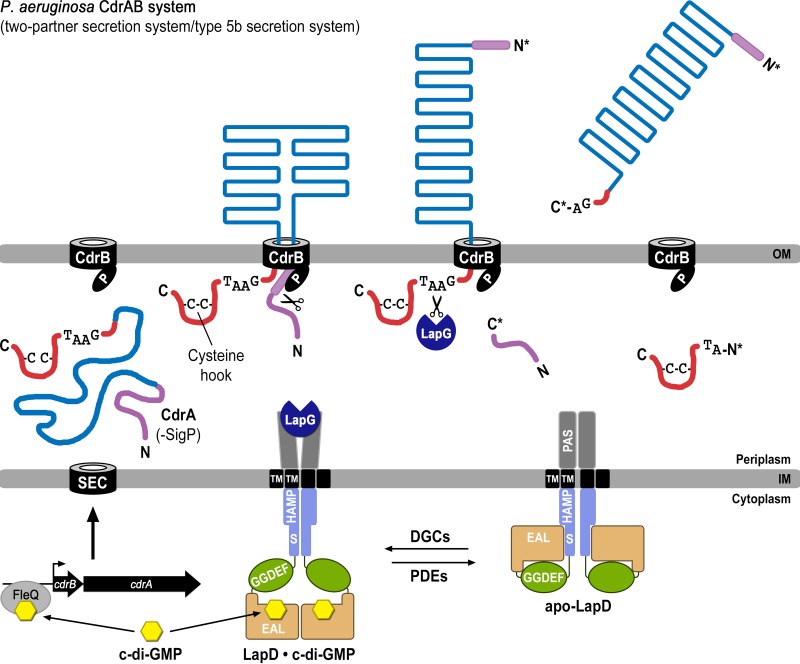
Biofilm formation is regulated via a bacterial second messenger, cyclic di-GMP (c-di-GMP), enzymes for its biosynthesis and degradation, and binding proteins that monitor levels of this cyclic dinucleotide. High levels of c-di-GMP are often associated with a switch to a sessile lifestyle through the binding of the second messenger to receptor proteins (2). Our previous studies identified a central c-di-GMP-specific receptor, LapD, and its associated signaling system in the environmental bacterium and model system for biofilm formation Pseudomonas fluorescens (3–6) (Fig. 1A). In particular, we found that the inner membrane-localized LapD receptor is autoinhibited at low cellular c-di-GMP levels but switches into a signaling-active state when c-di-GMP levels rise and the dinucleotide binds to the intracellular module of LapD. The associated conformational change extends to LapD's periplasmic output domain, which as a result recruits the periplasmic protease LapG. LapG's substrate in P. fluorescens is the large, cell surface-associated adhesin protein LapA. LapG cleaves the N-terminal domain of LapA, releasing this adhesin from the cell surface, which results in biofilm dispersal when c-di-GMP levels are low and LapG is not associated with LapD. When recruited to LapD under conditions of high c-di-GMP, LapG cannot process LapA, and LapA is in turn retained at the cell surface, stably anchoring cells to substrate during the early stages of biofilm formation.
FIG 1.
Mechanism of biofilm formation in P. fluorescens. (A) Overview of LapD- and LapG-mediated regulation of LapA localization to the outer membrane. Differential recruitment of LapG by LapD in response to cytoplasmic c-di-GMP levels controls LapA's surface attachment. When LapG is not sequestered to the inner membrane by LapD, LapG proteolytically cleaves the N terminus of LapA between two alanine residues of a TAAG motif, releasing the adhesin from the outer membrane, thus liberating the cell from the biofilm matrix. (B) Web Logo plot (24) of putative LapA-like substrates identified previously (4) reveals a series of conserved residues (green) upstream of the TAAG cleavage sequence (red).
LapA and LapDG orthologs could be predicted based on sequence conservation in a variety of biofilm-forming bacteria from environmental and pathogenic origins (4, 7). However, a particular knowledge gap was evident in regard to Pseudomonas aeruginosa, where we detected genes encoding LapD- and LapG-like proteins but failed to identify a corresponding LapA-like target protein or associated transporters in its genome. As one of the most prevalent opportunistic pathogens, P. aeruginosa accounts for ∼10% of hospital infections, notably in burn victims and surgical patients as well as those individuals with cystic fibrosis (8). These biofilm-based infections are notoriously difficult to treat, highlighting the importance of understanding the complex mechanisms controlling P. aeruginosa biofilm formation.
Here, we follow on previous work by revealing the target of the LapD/LapG system in P. aeruginosa biofilm regulation. We show that LapG targets the outer membrane-anchored adhesin protein CdrA for posttranslational processing and in doing so alters cell-cell interactions and biofilm formation. CdrA is the passenger protein of a two-partner secretion system (also known as type Vb secretion system), and its expression is regulated by c-di-GMP (9–11). Hence, our data reveal a second, c-di-GMP-regulated mechanism, in this case for the release of the adhesin from the cell surface, presumably when cellular second-messenger levels drop. This work validates the predicted conservation of the LapD/LapG receptor system but also uncovers a new class of adhesin proteins as targets for the LapG protease.
MATERIALS AND METHODS
Plasmid construction, protein expression, and protein purification. (i) CdrACterm and LapANterm.
A DNA fragment corresponding to residues 1844 to 2154 of CdrA (gene PA4625) was amplified from P. aeruginosa PAO1 genomic DNA (gDNA) with an N-terminal His6 tag by standard PCR and cloned into the pBAD/His A vector (Invitrogen) using NcoI and HindIII restriction sites. CdrACterm was expressed from this plasmid in TOP10 cells using ZYM-5052 autoinduction medium (12) supplemented with 100 μg/ml ampicillin at 37°C for 24 h. Cells were harvested by centrifugation and resuspended in IB wash buffer (25 mM Tris-HCl [pH 7.6], 100 mM NaCl, 5 mM β-mercaptoethanol, and 2 mM EDTA) supplemented with 0.5 mg/ml lysozyme and incubated for 15 min, at which time 1% (vol/vol) Triton X-100 was added and cells were lysed by sonication. Inclusion bodies were pelleted by centrifugation at 16,000 × g for 10 min, washed twice with IB buffer containing 1% Triton X-100, and washed two more times with IB buffer without detergent. Washed inclusion bodies were solubilized in denaturing buffer (100 mM Tris-HCl [pH 7.8], 8 M urea, and 5 mM β-mercaptoethanol) for 2 h at room temperature. Solubilized inclusion bodies were filtered through a 0.22-μm filter and loaded onto an Ni-nitrilotriacetic acid (NTA) His-Trap column (GE Healthcare). The column was washed with 50 column volumes of 25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 8 M urea, 5 mM imidazole, and 5 mM β-mercaptoethanol. CdrACterm was refolded on the Ni-NTA column with a linear gradient from 8 M to 0 M urea over 1.5 h, at which time soluble protein was eluted with 25 mM Tris-HCl (pH 7.6), 150 mM NaCl, and 300 mM imidazole. Imidazole was removed with a PD-10 desalting column (GE Healthcare), and CdrACterm was frozen in liquid nitrogen and stored at −80°C. LapANterm was expressed and purified as previously described (5).
(ii) LapG.
DNA fragments encoding P. aeruginosa PAO1 LapG (gene PA1434) and P. fluorescens Pf0-1 LapG (gene Pfl01_0130) lacking the signal peptide (residues 35 to 237 and 50 to 250, respectively) were amplified from gDNA and ligated into the pBrew vector (3) using NcoI and XhoI restriction sites, which generates in-frame superfolder green fluorescent protein (sfGFP) fusion proteins. These LapG-sfGFP fusion constructs were expressed in BL21ai cells (Invitrogen) with ZYM-5052 autoinduction medium supplemented with 50 μg/ml kanamycin at 37°C for 24 h, harvested by centrifugation, and lysed in buffer A (25 mM Tris-HCl [pH 7.6], 500 mM NaCl, and 20 mM imidazole). Insoluble debris was removed by centrifugation at 35,000 × g for 45 min. Ni-NTA Superflow resin (Qiagen) was incubated with the clarified supernatant and washed with buffer A, and the LapG-sfGFP was eluted with buffer A supplemented with 300 mM imidazole. The eluted protein was desalted into 25 mM Tris-HCl (pH 7.6)–250 mM NaCl with a PD-10 column, frozen in liquid nitrogen, and stored at −80°C.
(iii) Construction of pCleevR plasmids.
To create the pCleevR plasmid, residues 2078 to 2128 of CdrA were amplified from pBad-cdrACterm and fused N terminally with a His6-tagged small ubiquitin-like modifier (SUMO) and C terminally with sfGFP by standard overlap extension PCR protocols. The peptide inserts were linked to the fusion halves via short, flexible linkers (TGSTGS and GTSGTS, respectively) to ensure that individual domains folded autonomously. Encoded within these linkers were BamHI and KpnI restriction sites to permit convenient insertion of alternative substrate sequences. This triple-fusion construct was ligated into the pET28 vector (Novagen) using NcoI and XhoI restriction sites. The mutation A226R was then introduced into sfGFP to remove a potential LapG cleavage site. All mutations were made using the QuikChange kit (Agilent) or by overlap extension PCR. All pCleevR constructs were expressed and purified identically to LapG as described above.
(iv) Construction of LapD/LapG dual-expression plasmids.
A DNA fragment encoding P. aeruginosa PAO1 LapD (gene PA1433) was amplified from gDNA and ligated into the first open reading frame of the pRSFduet vector (Novagen) with a C-terminal His6 affinity tag using NcoI and HindIII restriction sites. Residues 1 to 34 (the periplasmic signal sequence) of LapG was amplified from gDNA, fused to the LapG-sfGFP fusion construct from the pBrew vector (lacking the His6 tag) by overlap extension PCR, and ligated into the second open reading frame of pRSFduet using NdeI and KpnI restriction sites. Mutations W125E, F222E, S229D, and K445A of LapD and C119A of LapG were introduced by overlap extension PCR prior to ligation. For these and all other expression constructs, endogenous restrictions sites required for ligation into their respective vectors were first mutated by overlap extension PCR so that the native amino acid sequence was maintained.
(v) Construction of the WspR expression plasmid.
A DNA fragment encoding P. aeruginosa PAO1 WspR (gene PA3702) was amplified from gDNA and ligated in the first open reading frame of the pACYCduet vector (Novagen) with a C-terminal His6 affinity tag using NcoI and HindIII restriction sites. To generate the catalytically inactive GGAAF variant of WspR, mutations E253A/E254A were introduced by overlap extension prior to ligation.
(vi) Constructs for pMQ72-based cdrAB overexpression.
pMJT-1 and pBAD-cdrAB were identical to those previously described (9) and were a generous gift of M. Parsek. Saccharomyces cerevisiae strain InvSci (Life Technologies) was used to construct plasmids for pMQ72-based overexpression studies as previously described (5, 13). Briefly, the pBAD plasmid was used as a template to amplify the cdrAB genes using primers 5′-cgaattcgagctcggtacccATAGGGAGATTTTCATGGTCCGTCCG-3′ and 5′-gcatgcctgcaggtcgactctagaggatccccTCAGAAGCGCGCCACCACGTTGAACAGG-3′, where lowercase nucleotides represent regions with sequence identity to SmaI-digested pMQ72. Similarly, the above-listed primers were used for site-directed mutagenesis to create the TRRG variant of cdrA, the former primer paired with 5′-ACCtcgacgGGTAGCCGGCAGCAGG-3′ and the latter paired with 5′-ACCcgtcgaGGTCCGGCGGAGGACGCCAGCGCCAA-3′, where lowercase nucleotides represent the site of mutagenesis.
LapG-substrate proteolysis assays. (i) CdrACterm and LapANterm cleavage by LapG.
LapG-sfGFP at 0.5 mg/ml was preincubated with 20 mM EGTA or CaCl2 in reaction buffer (25 mM Tris-HCl [pH 7.6], 250 mM NaCl) for 10 min, at which time it was diluted 10-fold into 1 mg/ml CdrACterm or LapANterm. Reaction mixtures were incubated for 30 min at room temperature, and then reactions were quenched by diluting into SDS-PAGE sample buffer. Reaction products were analyzed by SDS-PAGE and stained with Coomassie dye. N-terminal Edman sequencing of the 5-kDa CdrACterm proteolytic product was performed by the University of California at Davis Molecular Structure Facility. Samples were prepared according to facility-recommended protocols (http://msf.ucdavis.edu/edman-sequencing-analysis/).
(ii) LapG proteolysis of pCleevR substrates.
To assess the rate of cleavage of these purified pCleevR substrates, 5 μM LapG-sfGFP was preincubated for 10 min with either 20 mM EGTA or CaCl2 in reaction buffer. Reactions were initiated by diluting the pretreated LapG 10-fold into 20 μM pCleevR substrate in reaction buffer. Reactions were quenched at 2, 5, and 20 min by mixing an aliquot directly into SDS sample buffer, and products were analyzed by SDS-PAGE. Gels were imaged by fluorescence.
Analysis of LapG function in P. aeruginosa PAO1. (i) Strains and conditions.
Bacteria were grown overnight in lysogeny broth (LB) at 37°C with shaking at 250 rpm with the appropriate antibiotic(s). Carbenicillin was used at 300 μg/ml for P. aeruginosa and 150 μg/ml for Escherichia coli. Gentamicin was used at 30 μg/ml for P. aeruginosa and 10 μg/ml for E. coli. The pMQ72- and pMJT-1 based vectors (containing the pBAD promoter) were induced with arabinose at 0.2% (vol/vol) when indicated for the biofilm assays and at 1.0% (vol/vol) for the CdrA overexpression analysis.
(ii) Static biofilm formation assays.
Static biofilm formation assays were conducted as described previously (9) with minor modifications. Overnight cultures were subcultured and grown to mid-log growth phase in LB. A 96-well microtiter plate containing 95 μl of Vogel-Bonner minimal medium (VBMM) (14) per well was inoculated with 5 μl of a liquid culture. The plate was incubated for 20 h at 37°C under static conditions. Unattached bacterial cells and medium were removed by gently submerging the plate in deionized water. Each well was stained with 125 μl 0.1% crystal violet for 20 min and destained with 150 μl of a solution containing 30% methanol and 10% acetic acid for 20 min. Absorbance was measured at 550 nm.
(iii) CdrAB overexpression analysis.
Briefly, overnight cultures were subcultured until mid-log growth phase. Arabinose was added as described above and left for 3 h. Cells were harvested by centrifugation, and residual cells were removed from the supernatant by passing the entire supernatant volume through a 0.22-μm filter (Millipore). The supernatant was concentrated using a 100-kDa Amicon centrifugal filter unit (Millipore). For the whole-cell lysate, cell pellets were resuspended in 750 μl resuspension buffer (20 mM Tris-HCl [pH 8], 10 mM MgCl2) and sonicated on ice to lyse cells (30% power applied 4 times for a duration of 10 s, with a cooling period of 10 s after each treatment). Whole-cell and supernatant protein concentrations were determined using the Pierce bicinchoninic acid (BCA) protein assay (Thermo Scientific) according to the manufacturer's instructions, and 20 μg whole-cell lysate and 10 μg supernatant were used for SDS-PAGE, blotted, and incubated with anti-CdrA antibody (1:2,000) (9).
Reconstitution of the P. aeruginosa LapDG/CdrAB signaling node with WspR in E. coli. (i) Expression.
BL21ai cells freshly transformed with the dual-expressing pBAD-cdrAB and pRSFduet-lapDG plasmids were grown in ZYM-5052 autoinduction medium supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin at 37°C for 7 to 8 h, at which time the temperature was reduced to 18°C and cells were incubated for an additional 16 h. BL21ai cells harboring the pRSET-mKate vector (15) were expressed in an identical manner. To coexpress WspR in this system, BL21ai cells were transformed simultaneously with the pBAD-cdrAB, pRSFduet-lapDG, and pACYC-wspR plasmids by electroporation and expressed in an identical manner, except that the medium was supplemented with 50 μg/ml ampicillin, 25 μg/ml kanamycin, and 13 μg/ml chloramphenicol.
(ii) Assessment of autoaggregation by sedimentation.
One milliliter of cells from each culture was removed from shaking flasks and vortexed briefly, at which time 200 μl was removed and diluted 5-fold in saline buffer, and the optical density at 600 nm (OD600) was measured. The remaining cells were allowed to aggregate by gentle rocking at room temperature for 15 min and then centrifuged at 100 × g for 2.5 min. The OD600 of the top 200 μl was measured by 5-fold dilution. The propensity for the cells to sediment is reported as 1 − (OD600 after aggregation/OD600 before aggregation).
(iii) Microscopy of CdrA-mediated E. coli aggregates.
Live cells were allowed to autoaggregate exactly as described above for the sedimentation assay and then were imaged using a Zeiss Observer.Z1/Apotome inverted microscope with a CoolSNAP HQ2 camera. To assess whether cell aggregation was homotypic, cells expressing CdrAB and LapG-sfGFP were mixed with an equal volume of cells expressing only mKate and allowed to aggregate as described for the sedimentation assay prior to imaging.
(iv) Analysis of protein expression.
CdrA in the whole-cell and supernatant fractions was analyzed by Western blotting as described above. LapD-His6 was detected by Western blotting using an anti-His5 mouse antibody (Qiagen), while LapG-sfGFP was detected via in-gel fluorescence (3).
Bioinformatics analyses: identification of putative CdrA-like LapG substrates.
The Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) domain selection search tool was employed to generate a list of organisms that encode LapG- and CdrB-like gene products, and the protein sequences of genes adjacent to cdrB-like genes were analyzed for CdrA-like protein motifs in these organisms. CdrA-like candidates were defined as large proteins (>500 amino acids) possessing a “TAAG” or “GAAT” motif N terminal to at least one pair of C-terminal cysteine residues. This process was automated using the programming language R as described below.
First, two data sets were generated; one data set contained organisms whose genome coded for LapG-like proteins, and the other contained organisms encoding CdrB-like proteins. To accomplish this task, SMART was set to normal mode, and the architectural analysis option was utilized to perform a domain selection search with the input “Pfam:Peptidase_C93,” the motif found in the LapG protein. The bacterial node was selected from the results and the output action selected to download the protein sequences as a FASTA file. This process was repeated for architectural analysis with the domain input “Pfam:Bac_surface_Ag,” “Pfam:POTRA_2,” and “Pfam:Bac_surface_Ag AND Pfam:POTRA_2.” The results for these three searches were pooled into a single FASTA file and represent organisms that carry at least one cdrB-like gene.
Next, the organisms and UniProt identifiers (ID) were extracted from both FASTA files. Overlap between organisms carrying lapG- and cdrB-like genes was discerned with the intersect function in R, yielding a list of lapG/cdrB-like-carrying organisms to be analyzed for potential cdrA homologs. Each UniProt ID for cdrB-like genes was converted to the corresponding gene name. A value of 1 was added and subtracted from the cdrB-like gene name to investigate the putative upstream and downstream genes for cdrA-like features. For example, genes PA4625 and PA4623 were investigated for PA4624 in P. aeruginosa PAO1. Annotated genes (i.e., tpsB), which made up a small portion of the total gene names, were not recognized by the script and were discarded. If nearby genes were not returned, a value of 5 was added to the cdrB-like gene and the gene number investigated as before. For example, genes spanning loci K652_04573 to K652_04583 were investigated for K652_04578. This approach allowed increased coverage of cdrA-like candidates in the few genomes where loci are separated by a numerical distance of up to 5 instead of 1. The gene name, putative function, amino acid length, and final 300 C-terminal amino acids of all proteins possessing CdrA-like characteristics detected using this methodology were recorded and manually analyzed to verify proximity to the cdrB-like gene and the presence of CdrA-like sequence motifs.
RESULTS AND DISCUSSION
CdrA is proteolytically processed by LapG.
Previously, we identified putative substrates of LapG in bacterial species other than P. fluorescens by searching genomes containing LapD- and LapG-like operons for proteins possessing N-terminal sequences similar to that of P. fluorescens LapA that were associated with an ABC transporter system (4). Although this analysis revealed 20 putative LapG substrates from various bacterial species, no target was identified in P. aeruginosa. Alignment of the N-terminal sequences of these putative substrates revealed a set of modestly conserved residues immediately upstream of the dialanine site at which LapG cleaves LapA (5), which we hypothesized could be important for recognition by LapG (Fig. 1B). We expanded our search to proteins within the P. aeruginosa PAO1 proteome containing a similar short motif, ignoring any association with ABC transporters, the location of the motif within the protein sequence, or the location of the putative target gene within the genome.
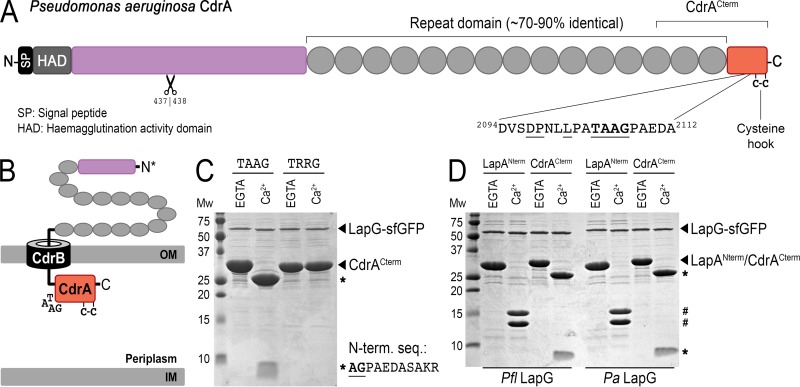
This search identified a protein in P. aeruginosa that possesses a putative LapG-like cleavage site 49 residues from its C terminus (Fig. 2A), in contrast to all other LapG cleavage sites (putative and established), which are located at their respective N termini. This protein, encoded by gene PA4625 and previously referred to as CdrA, is an adhesin-like protein responsible for cell-cell autoaggregation and biofilm maturation in P. aeruginosa (9). CdrA and its cognate outer membrane transporter CdrB (PA4624) constitute a two-partner secretion (TPS) system (also known as a type Vb secretion system) transcriptionally regulated by c-di-GMP (9, 11, 16) (Fig. 2B). cdrA encodes a protein of ∼220 kDa that is processed at the N terminus to produce a product with an apparent molecular mass of ∼150 kDa found associated with biofilm-resident cells (9). CdrA detected in the supernatant has an apparent molecular mass not significantly distinguishable by SDS-PAGE from the ∼150-kDa adduct observed in native biofilms (9); however, the mechanisms regulating CdrA processing and localization were unknown. We noticed that immediately downstream of the putative LapG cleavage site and immediately before the C terminus are the only two cysteine residues in CdrA (Fig. 2A). A previous report on the TPS adhesin protein HMW1a from Haemophilus influenzae showed that C-terminal anchoring of HMW1a to its cognate outer membrane transporter was dependent on an intact disulfide bond between its two C-terminal cysteines, and reduction (or mutation) of this disulfide resulted in release of the adhesin into the culture supernatant (17). However, the physiologic processes by which this class of adhesins is released from the outer membrane remained unclear. We therefore hypothesized that mature CdrA is anchored to the outer membrane with its N terminus in the extracellular environment and its C terminus exposed to the periplasm (Fig. 2B). We further proposed that regulation of CdrA localization would proceed through LapG proteolytically cleaving its small 5-kDa C-terminal domain, which would detach its putative “cysteine hook” and release the adhesin from the cell surface, ultimately resulting in the promotion of cell-cell disaggregation and biofilm disintegration.
FIG 2.
LapG proteolysis of P. aeruginosa CdrACterm. (A) Primary structure and domain organization of CdrA, depicting the location of the LapG cleavage site and the putative C-terminal “cysteine hook.” A second, N-terminal proteolytic site identified previously to be between residues 437 and 438 (9) is also depicted. (B) Cartoon model for the localization of CdrA at the outer membrane via its cognate TPS transporter CdrB, in which the C terminus of CdrA resides in the periplasm while the N terminus is excreted into the extracellular space. Such a topological orientation is the opposite of that of LapA-like proteins, in which the C terminus is extracellular (Fig. 1A). (C) Proteolysis of purified CdrACterm by purified LapG-sfGFP produces 23-kDa and 5-kDa fragments (indicated by asterisks) in the presence of calcium but not EGTA. The sequence of the first 10 residues of the 5-kDa proteolytic fragment determined by Edman degradation is shown. A variant of CdrACterm in which the double-alanine motif is mutated to arginine residues (TRRG) prevents proteolysis. (D) Proteolytic processing of CdrACterm and LapANterm by P. fluorescens (left) and P. aeruginosa (right) LapG-sfGFP demonstrates functional conservation across the two systems. LapANterm and CdrACterm proteolytic products are indicated by # and *, respectively.
To test this hypothesis, we expressed and purified a 28-kDa C-terminal construct of CdrA, CdrACterm, containing the predicted cleavage site and cysteine hook (Fig. 2A). Upon mixing this purified C-termina1 domain of CdrA with the calcium-dependent LapG from P. aeruginosa, CdrACterm was proteolytically processed into 23-kDa and 5-kDa fragments in the presence of calcium but not EGTA (Fig. 2C). N-terminal Edman sequencing of the 5-kDa proteolytic product confirmed that cleavage occurred between the two alanine residues of the TAAG motif of CdrA. Indeed, mutation of those two alanines to arginine (TAAG to TRRG) prevented LapG processing (Fig. 2C). Although CdrA and LapA share no detectable topological or evolutionary relatedness, CdrACterm and LapANterm (residues 1 to 235 of P. fluorescens LapA [5]) can be efficiently processed by either P. fluorescens or P. aeruginosa LapG orthologs in a calcium-dependent fashion (Fig. 2D), indicating overall conserved mechanisms and only a relatively short recognition motif being required for proteolysis.
CdrA residues upstream of the TAAG cleavage site are essential for LapG recognition.
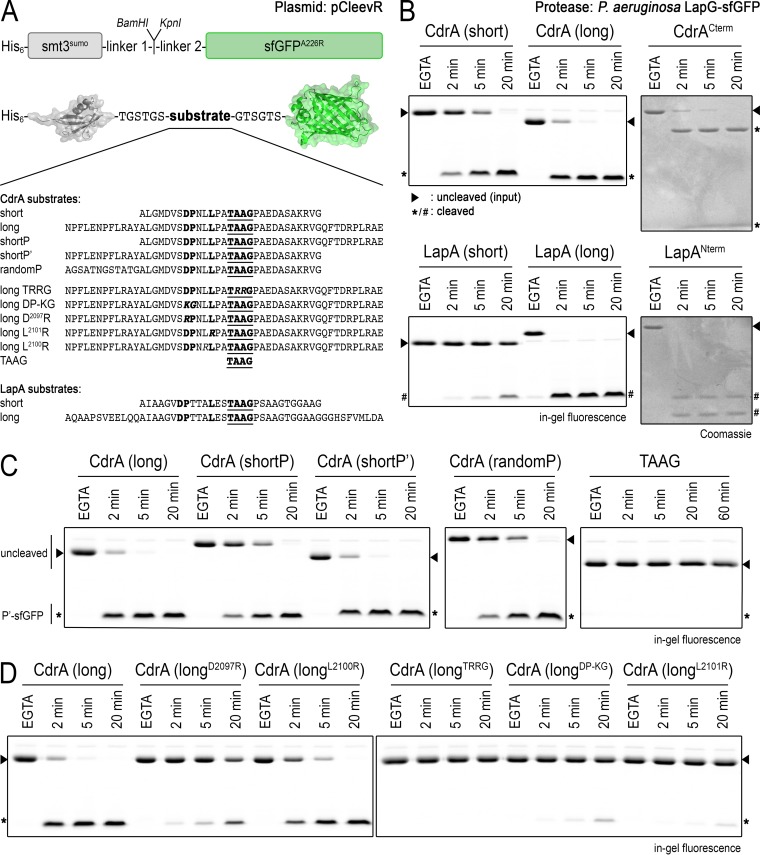
Although the TAAG-to-TRRG mutation in CdrA abolishes LapG cleavage, it is unclear exactly which residues upstream of the cleavage site, if any, are essential for LapG cleavage of CdrA. To validate the importance of the conserved residues used to identify CdrA as a substrate in P. aeruginosa, information which could be used to help identify additional putative substrates of LapG in other, related organisms, we developed the pCleevR expression vector, in which peptides of various length and sequence could be expressed between a SUMO and fluorescent superfolder GFP (sfGFP) tag (Fig. 3A). The approximate rate of proteolysis for each of the constructs by LapG was measured by SDS-PAGE in-gel fluorescence (3). Several conclusions can be drawn from this analysis. First, the length of the peptide sequence influences the rate of LapG cleavage (Fig. 3B). This enhanced rate of cleavage in the CdrAlong peptide is dependent on residues N terminal to the cleavage site (compare CdrAlong, CdrAshortP, and CdrAshortP′ in Fig. 3C), and this enhanced rate is sequence dependent (compare CdrAshortP′ and CdrArandomP in Fig. 3C), even though CdrA and LapA share no detectable sequence similarity in that region. While there is no obvious primary sequence motif in the extended peptide that could explain the increased level of proteolysis, we speculate whether secondary structures, not present in CdrArandomP but predicted to be in the faster-processed substrates, could be responsible for more efficient protease-substrate interactions. This notion is in agreement with prior suggestions that LapG recognizes helical structures upstream of its cleavage sites (18). In terms of key residues, Asp2097, Pro2098, and L2101 (but not L2100) of CdrA are critical for cleavage by LapG (Fig. 3D). Together, these results suggest that the motif DPX2–3LX2TAAG is crucial for LapG proteolysis, with a more cryptic motif just upstream of this sequence contributing to processing efficiency.
FIG 3.
Characterization of LapG specificity with the pCleevR assay. (A) Primary structure of pCleevR constructs in which various CdrA and LapA sequences containing their respective LapG cleavage sites are flanked N terminally by a His6-tagged SUMO protein and C terminally by sfGFP A226R. (B) Length of the pCleevR sequence influences rate of cleavage by LapG. CdrAshort/LapAshort (as defined in panel A) sequences are cleaved more slowly than CdrAlong/LapAlong sequences, which are processed by LapG at approximately the same rate as CdrACterm and LapANterm. Cleavage products are indicated by asterisks (for CdrA) and number symbols (for LapA). Intact substrates are indicated with triangles. (C) Assessment of the relative rates of cleavage of CdrAshortP′ and CdrAshortP substrates compared with the CdrAlong substrate indicates that residues N terminal to the cleavage site are responsible for this increased rate of proteolysis. The fact that CdrArandomP, which has the same length and number of residues flanking the cleavage site as CdrAshortP′ but is of unrelated sequence, is cleaved more slowly than CdrAshortP′ indicates that the enhanced rate is sequence dependent. The motif TAAG, in and of itself, is not sufficient for LapG targeting (right). Note that CdrAlong migrates with an aberrantly smaller size than CdrAshort in SDS-PAGE. (D) Single-amino-acid substitutions in CdrAlong identify residues D2097, P2098, and L2101 but not L2100 as crucial for LapG-mediated cleavage. As with the TRRG variant of CdrACterm (Fig. 2C), the analogous TRRG CdrAlong variant is not cleaved by LapG.
LapG mediates CdrA-dependent biofilm formation in P. aeruginosa PAO1.
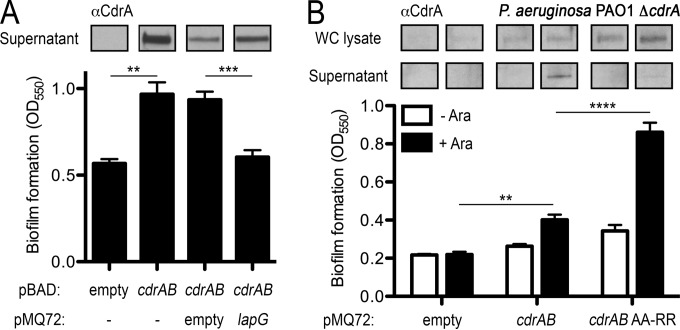
Having established CdrACterm as a bone fide substrate of P. aeruginosa LapG, we next sought to establish whether LapG could mediate CdrA-dependent biofilm formation in vivo. The CdrA and CdrB proteins are poorly detected in biofilms expressed under our assay and growth conditions (Fig. 4A). To overcome this limitation, we used a strategy similar to that reported by Borlee et al. in their previous work describing the function of CdrA in biofilm formation (9), and we conducted our studies with cdrA and cdrB cloned on an inducible multicopy plasmid in a P. aeruginosa PAO1 cdrA knockout strain. Cooverexpression of CdrA and CdrB in P. aeruginosa PAO1 resulted in an increase in biofilm mass compared to cells not expressing this two-partner adhesion system (Fig. 4A). However, expression of periplasmic LapG along with CdrA and CdrB returns biofilm formation to levels equivalent to that of the parent strain (Fig. 4A). This decrease in CdrAB-dependent biofilm formation when LapG is overexpressed is concomitant with an increase in an ∼150-kDa processed form of CdrA found in the culture supernatants (Fig. 4A). This form of CdrA is identical in size to supernatant CdrA as determined by previous measurements (9). Mutation of the TAAG cleavage site in full-length CdrA to TRRG increases biofilm formation with no detectable CdrA in the culture supernatants (Fig. 4B), consistent with prevention of LapG-catalyzed proteolysis of CdrA. Taken together, these results demonstrate that LapG cleaves CdrA and thus impairs CdrA-dependent biofilm formation in P. aeruginosa.
FIG 4.
Regulation of CdrAB-dependent biofilm growth by LapG in P. aeruginosa. (A) Assessment of biofilm formation in response to CdrAB and LapG expression in static cultures of the P. aeruginosa PAO1 ΔcdrA strain (9) as measured by crystal violet staining (see Materials and Methods). Top, Western blot of CdrA released into the supernatant of these cultures using an anti-CdrA antibody developed previously (9). (B) Biofilm formation in P. aeruginosa expressing CdrAB and LapG (+Ara), including a plasmid in which the TAAG cleavage site of CdrA has been mutated to TRRG. Above the plots are Western blots of CdrA from whole-cell and supernatant fractions of these cultures, demonstrating that the TRRG mutation prevents LapG-dependent release of CdrA into the supernatant fraction. Asterisks indicate P values of <0.01 (*), <0.001 (**), <0.0001 (***), and <0.00001 (****) between the indicated samples.
LapD regulates LapG proteolysis of CdrA.
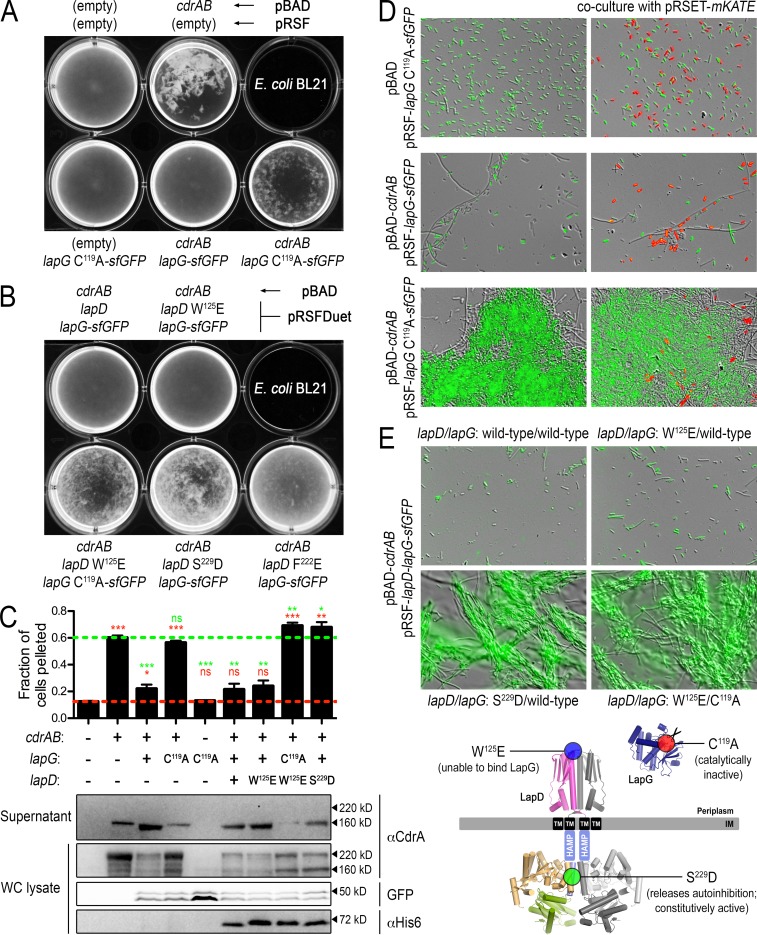
In P. fluorescens, LapG activity is controlled by the transmembrane receptor LapD, which in turn is regulated by levels of cytoplasmic c-di-GMP, thus linking c-di-GMP with biofilm formation (Fig. 1A). To establish whether the orthologous LapDG system of P. aeruginosa works in a conserved manner, we took advantage of the observation that overexpression of CdrAB promotes cell-cell autoaggregation and reconstituted the isolated signaling node (LapGD-CdrAB) in a heterologous strain background, E. coli BL21. Advantages of this approach include the controlled expression of the required factors in E. coli and the minimization of potential complexities associated with c-di-GMP regulation and biofilm formation present in the native host (e.g., potential cross talk to other c-di-GMP signaling pathways). To assess the relative amount of cell-cell aggregation in this E. coli expression system, three techniques were employed. First, the cell cultures themselves were imaged to visualize macroscopic phenotypes (Fig. 5A and B). Second, the relative amount of clumping was quantified by measuring the propensity of the cells to sediment (Fig. 5C). Lastly, cell aggregates were imaged using differential interference contrast and fluorescence microscopy (Fig. 5D and E).
FIG 5.
Regulation of CdrA-dependent cell autoaggregation by the LapDG receptor system in a heterologous host. (A) Direct visualization of E. coli BL21 cultures expressing CdrAB and periplasmic P. aeruginosa LapG-sfGFP. The pBAD and pRSF expression constructs for each culture are as indicated (written above for the top row and below for the bottom row). (B) Similar visualizations of E. coli cell autoaggregation as in panel A but with full-length LapD coexpression alongside CdrAB and LapG-sfGFP. (C) Top, propensity for the cultures shown in panels A and B with robust phenotypes to sediment (as described in Materials and Methods). Higher values indicate greater autoaggregation. Red and green dashed lines indicate values obtained for native, dispersed E. coli cells and those aggregating due to expression of only CdrAB. Red and green asterisks above each sample indicate statistical comparisons to the cells grown with empty plasmids and cells expressing only CdrAB, respectively. The numbers of asterisks indicate P values as described in the legend for Fig. 4. Those that are not statistically different are indicated with “ns.” Bottom, for each of the cultures in the top panel, protein levels of CdrA in supernatant and whole-cell lysates were analyzed by Western blotting. Also analyzed in the whole-cell lysates were LapG-sfGFP (by in-gel fluorescence) and LapD (by Western blotting with an anti-His5 antibody). Approximate molecular masses are shown to the right of each gel. (D) Cultures shown in the bottom row of panel A were imaged by differential interference contrast and GFP fluorescence at a magnification of ×40 to reveal microscopic features of the autoaggregated cells (left column). Cells expressing only the red fluorescent protein mKate did not aggregate with the CdrAB-expressing cells (right column). A small fraction of cells expressing CdrAB appear as long filaments, independent of the expression of active or inactive LapG. However, this appearance has no obvious impact on the overall autoaggregation phenotypes. Also, cells that appear unlabeled show a lower level of fluorescence that is detectable upon longer exposure times. (E) Similar overlays of differential interference contrast and GFP fluorescence images of cell aggregates from LapD-coexpressing strains displaying robust phenotypes shown in panel B. Bottom, mutations used to alter the functionality of LapD and LapG mapped onto their respective structural models.
E. coli BL21 cells expressing CdrA and CdrB formed large aggregates (Fig. 5A, C, and D), similar to P. aeruginosa (9) and consistent with the increased biofilm formation discussed above (Fig. 4). CdrA-dependent cell aggregation of P. aeruginosa relies on the secretion of, and CdrA's interaction with, the mannose-rich extracellular polysaccharide Psl (9), suggesting that E. coli expresses a matrix component that can substitute for Psl. When CdrAB was coexpressed in E. coli with wild-type LapG fused to sfGFP, the cells did not aggregate (Fig. 5A, C, and D). Aggregation was restored, however, when the catalytically inactive C119A variant of LapG-sfGFP was expressed. These sfGFP-labeled CdrA-dependent cell aggregates are homotypic, as E. coli cells that express only a red fluorescent protein (mKATE) do not cocluster with cells expressing CdrAB (Fig. 5D). Together, these results recapitulate analogous phenotypes observed in P. aeruginosa and further confirm the role of LapG in regulating CdrA-dependent cell autoaggregation.
To determine if LapD regulates LapG activity and thus CdrAB-dependent cell aggregation, we employed loss-of-function and gain-of-function alleles of LapD based on our insight into LapD regulation in P. fluorescens (3, 4), in addition to the wild-type receptor. In particular, the P. aeruginosa LapD variant W125E is predicted to prevent LapG binding to LapD, which would result in constitutive processing of CdrA by LapG. In contrast, the S229D (or F222E) variant should compromise the ability of LapD to adopt the autoinhibited state, creating a “constitutively active” LapD, thus mimicking the c-di-GMP-bound state that should efficiently sequester LapG and allow cell aggregates to form.
Cells expressing the wild type and W125E LapD variants with wild-type LapG-sfGFP and CdrAB did not form appreciable aggregates. Aggregation could be restored when LapD W125E was coexpressed with the catalytically inactive LapG-sfGFP C119A variant (Fig. 5B, C, and E). Cells expressing the gain-of-function S229D or F222E mutations in LapD formed significant cell aggregates when coexpressed with catalytically active LapG-sfGFP (Fig. 5B, C, and E), indicating that P. aeruginosa LapD regulates the activity of P. aeruginosa LapG in a manner analogous to the mechanisms described for the P. fluorescens orthologs. The facts that we observe LapG activity-dependent cell aggregation phenotypes and that the LapD mutants show the predicted relative effects on LapG function in the context of the heterologous E. coli host indicate functional reconstitution of this signaling node.
Western blot analysis of CdrA in these cultures indicates that cell aggregation correlates with CdrA being cell associated and having two distinct isoforms with apparent molecular masses of ∼220 kDa and ∼150 kDa, while in nonaggregated cells neither of these forms is as prominent in the cell-associated fractions. However, larger quantities of the ∼150-kDa CdrA species are found in the supernatants of these nonaggregated cultures (Fig. 5C). Although this is difficult to resolve by SDS-PAGE, we suspect that the cell-associated 150-kDa band has an intact C terminus while the supernatant species lacks the last 49 residues as a result of LapG proteolysis. This finding is consistent with subtle differences in electrophoretic mobility of the trace amounts of CdrA nonspecifically released into the supernatant in strains lacking active LapG compared to CdrA released in a LapG-dependent fashion (e.g., compare lanes 2, 3, and 4 of supernatant samples in Fig. 5C). Nevertheless, CdrA appears to be processed analogously in E. coli and in P. aeruginosa, in which catalytically active LapG is essential for release of CdrA from the cell surface. While the identity of the protease responsible for the N-terminal cleavage remains unknown, the result of LapG-dependent processing is cell disaggregation.
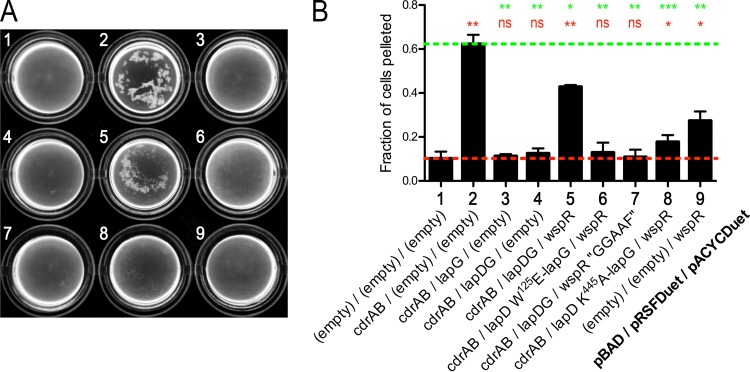
We next sought to establish whether c-di-GMP is able to regulate CdrA-dependent cell autoaggregation via the LapD/LapG signaling node. To do this, we coexpressed the diguanylate cyclase WspR (PA3702) with the LapDG-CdrAB expression system (19, 20) (Fig. 6). When wild-type WspR was expressed with wild-type LapD and LapG, significant cell autoaggregation was observed, indicating that c-di-GMP activates LapD, which in turn sequesters LapG. In contrast, when wild-type WspR is expressed with the W125E variant of LapD, the cells remain dispersed since W125E LapD cannot sequester LapG (3, 4). Cells also did not appreciably aggregate when wild-type WspR was expressed with a c-di-GMP-binding-deficient variant of LapD (K445A) (13). Catalytic activity of WspR is dependent on its GGEEF motif (21), and when the inactive GGAAF variant is used with wild-type LapD and LapG, the cells do not aggregate. These data strongly support the notion that P. aeruginosa LapD activity, and thus CdrA-dependent cell-cell autoaggregation, is controlled by c-di-GMP. Taken together, the results indicate that the LapDG-CdrAB signaling system adds a second, posttranscriptional layer of regulation of biofilm formation in P. aeruginosa, in addition to the established requirement of c-di-GMP for the transcription of the cdrAB operon (9, 20, 22).
FIG 6.
Regulation of CdrA-dependent cell autoaggregation by c-di-GMP. (A) Direct visualization of E. coli BL21 cultures expressing variants of CdrAB, LapG-sfGFP, LapD, and WspR. The pBAD, pRSFDuet, and pACYCDuet expression constructs used in each culture are as indicated in panel B. (B) Propensity for the cultures shown in panel A to sediment. Higher values indicate greater autoaggregation. Red and green dashed lines indicate values obtained for native, dispersed E. coli cells and those aggregating due to expression of only CdrAB. Red and green asterisks above each sample indicate statistical comparisons to the cells grown with empty plasmids and cells expressing only CdrAB, respectively. The numbers of asterisks indicate P values as described in the legend for Fig. 4. Those that are not statistically different are indicated with “ns.”
Finally, we asked how prevalent CdrA-like substrates are in the bacterial domain. Using our current understanding of LapG's specificity, we identified 7 unique species other than P. aeruginosa strains with a putative substrate that were also associated with a CdrB-like transporter and also possessed a LapG-like ortholog (see Table S1 in the supplemental material). Interestingly, four of these species are alphaproteobacteria that do not have detectable LapD-like orthologs and are likely subject to a different mode of posttranscriptional regulation. We suspect that a deeper understanding of LapG processing and regulation in these more divergent orthologs will reveal additional CdrA-like candidate substrates and signaling pathways controlling cell adhesion.
Concluding remarks.
Previous studies on c-di-GMP-regulated LapDG signaling focused on environmental bacteria (4, 5, 7, 13), and predictions of functional conservation in other bacteria awaited experimental validation. Here, we reveal the physiologic role of the LapDG signaling system in a pathogenic bacterium, P. aeruginosa, by identifying the protease substrate CdrA. By reconstituting a functional LapDG-CdrAB system in a heterologous expression host (i.e., E. coli), we have identified LapDG as the minimal regulatory system required to control localization of CdrA and thus CdrA-dependent cell aggregation. The collective data suggest the following model for two-tier, c-di-GMP-mediated control of CdrA. CdrA is initially controlled at the transcriptional level via the c-di-GMP-responsive transcription factor FleQ (Fig. 7) (11). High cellular c-di-GMP levels would also ensure an activated, LapG-binding-competent state of LapD (4, 5, 13), allowing CdrA to be anchored stably at the cell surface. However, when cellular c-di-GMP levels drop, LapD would revert to an autoinhibited state, concomitant with a release of LapG into the periplasm where it can cleave CdrA, terminating its function.
FIG 7.
Model for two-tier regulation of CdrA via c-di-GMP. Transcription of the cdrAB operon (bottom left) is initiated when c-di-GMP (yellow) binds the transcriptional regulator FleQ. Elevated c-di-GMP levels also result in second-messenger binding to the EAL domain of LapD, which in turn sequesters periplasmic LapG. Together these events allow CdrA to be secreted to and anchored within the outer membrane via its cognate outer membrane transporter CdrB. The N terminus of CdrA (purple) binds to the PORTA domain of CdrB and is processed by an unknown proteolytic activity, acting in the periplasm (as shown), within the transporter, or at the cell surface. When c-di-GMP levels are lowered by phosphodiesterases (PDEs), LapD adopts the autoinhibited state (bottom right), releasing LapG into the periplasm, where it cleaves the C-terminal tail of CdrA (red) at the TAAG cleavage site and releases this adhesin into the extracellular space, promoting cellular disaggregation.
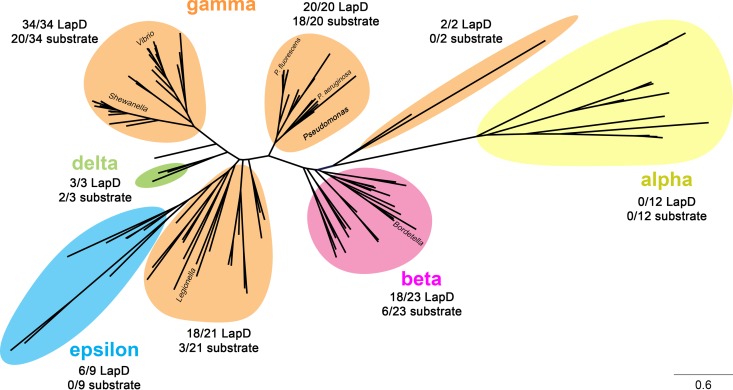
This work also expands the spectrum of substrates for LapG-like proteases. To date, all previously predicted substrates of LapG share some detectable amount of evolutionary or topological similarities to LapA. The discovery of CdrA as a substrate of the LapDG receptor-protease pair, which has no detectable homology or sequence similarity to LapA-like proteins apart from a few residues within their cleavage sites, suggest an unanticipated versatility of the LapG protease family. Though the functional purpose of CdrA's two C-terminal cysteines is not firmly established, these data provide a plausible physiologic mechanism by which related TPS adhesins with a C-terminal “cysteine hook,” such as HMW1a of H. influenzae (17), could be released from the outer membrane via regulated proteolytic processing. The versatility of LapG-like enzymes is supported by their broad distribution within the bacterial domain (Fig. 8). For example, in the genus Pseudomonas (to which P. aeruginosa and P. fluorescens belong) we identified 20 species with a LapG ortholog and a LapD ortholog. Of these 20, we could predict a substrate for 18 based on the cleavage site conservation reported above (Fig. 1B). On the other hand, our analysis revealed organisms that possess a LapG ortholog but no predictable substrate, and many of these (notably the alphaproteobacteria) even lack an obvious LapD ortholog. Toward a broader understanding of this diverse signaling system, which historically has been focused on the Pseudomonas genus, the functional importance of one such distant LapG ortholog, HvyA, has been recently established for the alphaproteobacterium Caulobacter crescentus by showing its crucial role in capsulation and phage resistance in this organism (23). However, its substrate specificity is currently unknown, and it remains to be shown whether HvyA is regulated following a mechanism analogous to the one we describe here for LapDG.
FIG 8.
Phylogenetic tree of LapG orthologs, displaying the diversity of the LapDG signaling system in bacteria. For this analysis, 5,220 bacterial genomes were downloaded from the NCBI database and initially searched for a putative LapG-like protease (identified by the conserved DX9WX12DCED[YF]X3KX20–40HX12–17LD motif). These genomes were subsequently searched for a LapD-like protein (motif F[DE]X[GS]X[YF]X20–40PXW[FLIVM]X16–18GWX47–51[RP]L) and a LapG-like substrate possessing a DPX2–3LX2[TA]AAG motif. LapG sequences were aligned by Clustal Omega (25) and analyzed phylogenetically by maximum likelihood with the JTT model for amino acid substitutions (26). Colored shadings highlight the class of bacteria to which individual species belong. Beside each shaded group, the corresponding number of species containing a LapD ortholog and a putative LapG substrate is indicated relative to the number of LapG orthologs within this group. Note that for many species, while LapD and LapG orthologs can be identified, no readily apparent LapA- or CdrA-like proteolysis motif was found. This observation suggests that other novel LapG targets may be present in these microbes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Parsek for providing the anti-CdrA antiserum, cdrAB expression plasmid, and P. aeruginosa PAO1 ΔcdrA strain.
ADDENDUM IN PROOF
While this article was at the proof stage, an independent, complementary study reported CdrA as a LapG target in P. aeruginosa biofilm formation (M. Rybtke et al., Microbiologyopen, 12 October 2015, http://dx.doi.org/10.1002/mbo3.301), supporting our conclusions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00369-15.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O'Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife 3:e03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinsa SM, O'Toole GA. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 7.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 8.Diekema D, Pfaller M, Jones R, Doern G, Winokur P, Gales A, Sader H, Kugler K, Beach M. 1999. Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis 29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 9.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filloux A. 2011. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol 2:155. doi: 10.3389/fmicb.2011.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 15.Shui B, Wang Q, Lee F, Byrnes LJ, Chudakov DM, Lukyanov SA, Sondermann H, Kotlikoff MI. 2011. Circular permutation of red fluorescent proteins. PLoS One 6:e20505. doi: 10.1371/journal.pone.0020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5060–5069. doi: 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buscher AZ, Grass S, Heuser J, Roth R, St Geme JW III. 2006. Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol Microbiol 61:470–483. doi: 10.1111/j.1365-2958.2006.05236.x. [DOI] [PubMed] [Google Scholar]
- 18.Boyd CD, Smith TJ, El-Kirat-Chatel S, Newell PD, Dufrene YF, O'Toole GA. 2014. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol 196:2775–2788. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardissone S, Fumeaux C, Berge M, Beaussart A, Theraulaz L, Radhakrishnan SK, Dufrene YF, Viollier PH. 2014. Cell cycle constraints on capsulation and bacteriophage susceptibility. eLife 3. doi: 10.7554/eLife.03587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.