Abstract
The cyclic dinucleotides cyclic 3′,5′-diguanylate (c-di-GMP) and cyclic 3′,5′-diadenylate (c-di-AMP) have emerged as key components of bacterial signal transduction networks. These closely related second messengers follow the classical general principles of nucleotide signaling by integrating diverse signals into regulatory pathways that control cellular responses to changing environments. They impact distinct cellular processes, with c-di-GMP having an established role in promoting bacterial adhesion and inhibiting motility and c-di-AMP being involved in cell wall metabolism, potassium homeostasis, and DNA repair. The involvement of c-dinucleotides in the physiology of the filamentous, nonmotile streptomycetes remained obscure until recent discoveries showed that c-di-GMP controls the activity of the developmental master regulator BldD and that c-di-AMP determines the level of the resuscitation-promoting factor A(RpfA) cell wall-remodelling enzyme. Here, I summarize our current knowledge of c-dinucleotide signaling in Streptomyces species and highlight the important roles of c-di-GMP and c-di-AMP in the biology of these antibiotic-producing, multicellular bacteria.
INTRODUCTION
Streptomyces, the largest genus of actinobacteria, consists of species of Gram-positive bacteria with high G+C content in their DNA. Streptomyces species inhabit diverse natural environments such as terrestrial and aquatic ecosystems but are mostly found in soil, where they recycle carbon and other nutrients trapped in insoluble organic debris from plants and fungi (1). These are multicellular bacteria with branched vegetative filaments consisting of long chains of joined single, multinucleoid cells. They are nonflagellated, sessile organisms during their mycelial growth phase but can spread to new habitats during reproductive growth via the dispersal of spores. Streptomycetes are the most abundant source of antibiotics and other bioactive molecules and are therefore among the most important taxa of clinical and industrial microorganisms. Morphological differentiation and antibiotic production are temporally and genetically coordinated in Streptomyces, and much progress has been made in uncovering the regulatory mechanisms that govern developmental processes in these bacteria (2, 3).
The first suggestion that the nucleotide second messenger cyclic 3′,5′-diguanylate (c-di-GMP) might be involved in the control of differentiation in Streptomyces came when several c-di-GMP metabolizing enzymes were found to be under developmental control (4). c-di-GMP was discovered in the late 1980s as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus (5) and is now the best-known representative of the c-dinucleotide family. Three distinct protein domains are responsible for the synthesis and degradation of this second messenger: the GGDEF, EAL, and HD-GYP domains. These domain names are based on amino acid motifs that crucially contribute to the active sites of these enzymes. Homodimeric GGDEF domains with diguanylate cyclase (DGC) activity bind two GTP substrate molecules to their active site and catalyze the formation of c-di-GMP (6, 7). On the other hand, bacteria have evolved two domains with phosphodiesterase (PDE) activity for c-di-GMP degradation. EAL domains attack one ester bond of the circular dinucleotide, yielding the linear dinucleotide 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) (8, 9), and the less frequent HD-GYP domains hydrolyze c-di-GMP into two GMP molecules (10). A unique and remarkable feature of c-di-GMP is its conformational flexibility, which enables the molecule to bind to different types of effectors but limits bioinformatic prediction of c-di-GMP binding sites. In complex with effector proteins, c-di-GMP has been shown to exist as a linear monomer (11), as an intercalated dimer (7), and as a tetramer (12). To date, 6 classes of c-di-GMP effectors have been described: degenerate GGDEF/EAL domains (13–16), PilZ domains (17), GIL domains (18), riboswitches (19), histidine kinases (20), and transcription factors. Of these, transcriptional regulators represent the most diverse class of c-di-GMP effectors, comprising members of the TetR (21), cyclic AMP (cAMP) receptor protein (CRP)/FNR (22–24), NtrC (25, 26), and FixJ/LuxR/CsgD (27) protein families. Through its ability to control the activity of such a broad range of effectors, c-di-GMP regulates diverse physiological processes, but its pivotal role in Gram-negative bacteria is to control transitions from motile-planktonic single cells to sedentary multicellular communities (28).
While c-di-GMP is highly distributed among the members of the bacterial kingdom, the more recently discovered c-dinucleotide c-di-AMP is predominantly found in Gram-positive genera, including Streptomyces. DAC (diadenylate cyclase) domains, bearing no amino acid or structural similarity to the GGDEF domain, synthesize c-di-AMP from two ATP molecules (29), and the DHH-DHHA1 domain containing the Asp-His-His motif or the HD domain with a catalytic His-Asp motif hydrolyzes the cyclic molecule to pApA (30, 31). Multiple studies have provided evidence that a well-balanced level of c-di-AMP is vital for bacterial physiology and that it is the first essential second messenger in bacteria, as shown for Staphylococcus aureus, Bacillus subtilis, Streptococcus pneumoniae, and Listeria monocytogenes (32–36). c-di-AMP signaling is mediated by a large variety of effectors, such as transcriptional regulators (37), potassium transporters, cation-proton antiporters, and histidine kinases (38), PII-like proteins (39), pyruvate carboxylases (40), and riboswitches (41), and is involved in the control of sporulation, DNA repair, cell wall and potassium homeostasis, virulence, and metabolic functions (42, 43).
In this review I summarize and discuss recent discoveries that have established important roles for c-dinucleotides in the control of key cellular processes in Streptomyces, placing c-di-GMP and c-di-AMP signaling in the context of multicellular differentiation.
A BACTERIAL LIFE CYCLE UNDER c-DI-GMP CONTROL
The Streptomyces life cycle is defined by a progression through several distinct developmental stages, each characterized by a particular morphology of the bacterial colony (Fig. 1). The vegetative phase of growth initiates with swelling of a free spore and the emergence of one or more germ tubes. Apical hyphal extension and branching directed by the coiled-coil protein DivIVA (44) lead to the formation of the vegetative (or substrate) mycelium, which is a dense network of filamentous vegetative hyphae (Fig. 1). The reproductive stage begins with the emergence of aerial hyphae when stress (e.g., nutrient limitation) is encountered. A fibrous sheath consisting of the amyloid chaplin proteins (45, 46), the rodlins (47), and, on some media, the lantibiotic-like surfactant peptide SapB (48) enables aerial hyphae to grow out of the aqueous environment of the substrate mycelium into the air to form the aerial mycelium, which gives the developing colonies their characteristic fuzzy appearance (Fig. 1). The onset of sporulation begins with the synchronous septation of each multigenomic aerial hypha into ca. 50 to 100 unigenomic prespore compartments and ends with spore maturation. Mature spores are pigmented, resulting in gray colonies in S. coelicolor or green colonies in S. venezuelae (Fig. 1).
FIG 1.
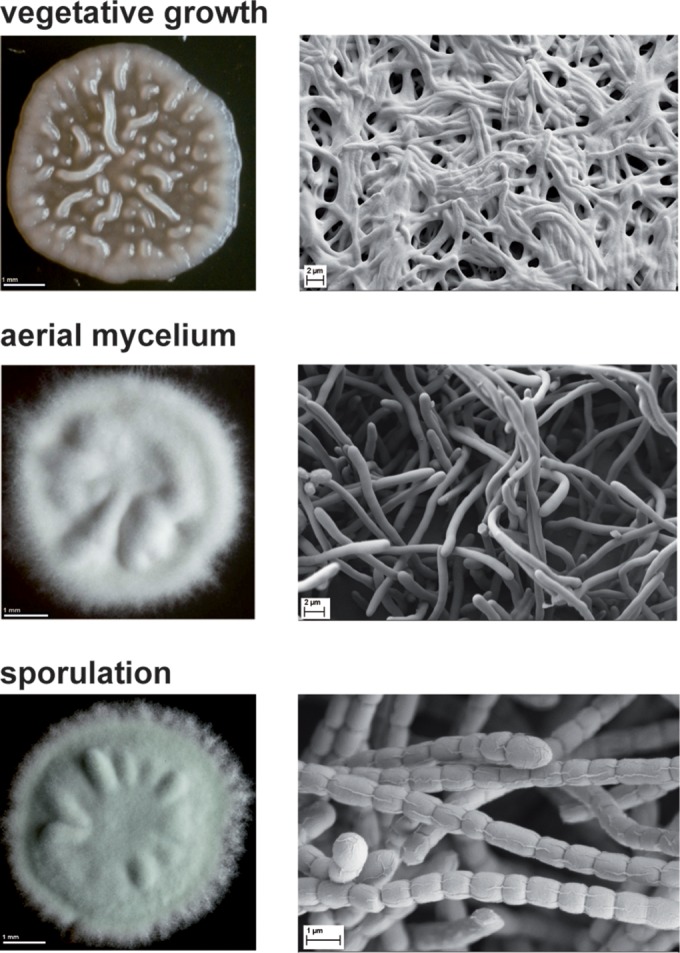
S. venezuelae developmental stages. Colony morphology (left) and scanning electron micrographs (right) are shown. During the vegetative growth phase, hyphae form a dense mat of filaments on and in the substrate and the colony appears bald and shiny. The emergence of aerial hyphae marks the beginning of morphological and physiological differentiation and results in a fuzzy and white colony phenotype. In the course of sporulation, multigenomic aerial hyphae differentiate into 50 to 100 unigenomic prespore compartments, which mature into dormant exospores. The synthesis of a polyketide pigment during spore maturation gives the final S. venezuelae colonies a characteristic green color.
Differentiation is controlled by the Bld and Whi developmental regulators. Mutations in bld loci block aerial mycelium formation and therefore cause a “bald” and shiny colony phenotype, whereas whi mutants make aerial hyphae but are blocked in spore formation, resulting in white colonies due to the absence of the polyketide pigment associated with mature spores (2).
A key recent study demonstrated that intracellular c-di-GMP levels dictate the speed at which the Streptomyces life cycle is completed (12). By overexpressing an active DGC in S. venezuelae, the authors showed that elevated levels of c-di-GMP arrest development in the vegetative growth phase. Conversely, low levels of the c-dinucleotide, generated by overexpressing a PDE, accelerate development, causing the vegetative hyphae to sporulate without the need to form an aerial mycelium.
SYNTHESIS AND DEGRADATION OF c-DI-GMP IN STREPTOMYCES
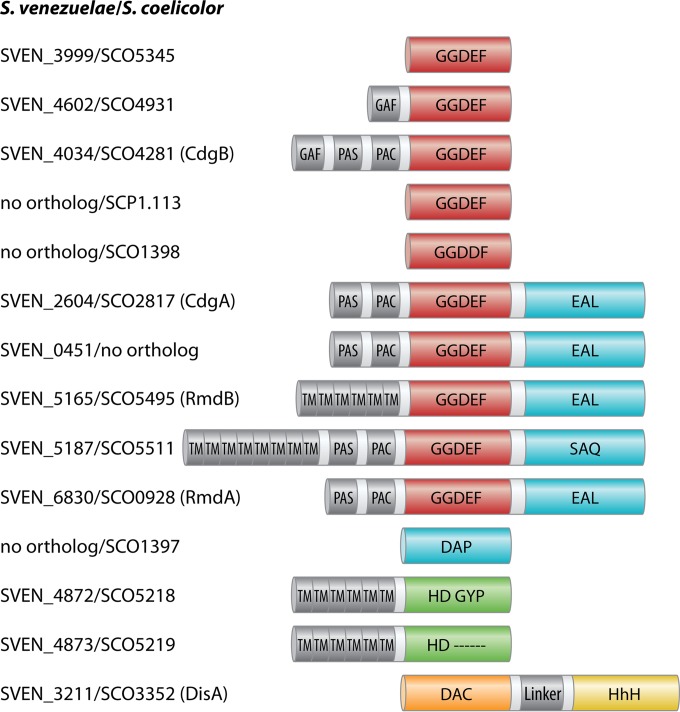
Although most streptomycetes contain overlapping sets of up to 85% identical proteins that make or break c-di-GMP, a few variations exist; for example, S. venezuelae encodes a GGDEF-EAL composite protein, SVEN_0451, no ortholog of which can be found in S. coelicolor. On the other hand, S. coelicolor has one additional conserved GGDEF protein, SCP1.113, a GGDDF domain protein, SCO1398, and an extra degenerate EAL protein, SCO1397, which have no equivalents in S. venezuelae (Fig. 2).
FIG 2.
Domain organization of predicted c-di-GMP and c-di-AMP metabolizing proteins in S. venezuelae and S. coelicolor. S. venezuelae and S. coelicolor encode an overlapping set of GGDEF, EAL, HD-GYP, and DAC domain proteins, with minor modifications. SVEN_0451 is unique to S. venezuelae. SCP1.113, SCO1398, and SCO1397 orthologs are not present in S. venezuelae. Predicted signaling domains (GAF, PAS, and PAC) and transmembrane helices (TM) as well as linker regions are shown in gray. Missing residues in SVEN_4873 are shown as a dashed line, and the helix-hairpin-helix domain (HhH) of DisA is indicated.
The GGDEF-EAL CdgA protein was the first c-di-GMP-metabolizing enzyme studied in Streptomyces. Aiming to characterize the function of CdgA, den Hengst et al. altered the levels of the protein and found that overexpression of cdgA blocks the formation of aerial hyphae and the synthesis of the blue antibiotic actinorhodin in S. coelicolor. These effects could be achieved only with a protein version containing an intact GGDEF motif and not with a mutagenized variant containing AADEF residues in the active site, strongly arguing that c-di-GMP was involved in the observed phenotype and thereby setting the stage for the participation of c-di-GMP in Streptomyces development and secondary metabolite production (4). Nevertheless, a direct biochemical demonstration of the DGC activity of CdgA is still missing and the function of the conserved C-terminal EAL domain remains to be determined.
One year later, in 2011, Tran et al. provided evidence that the GGDEF domain protein CdgB is another DGC involved in S. coelicolor development (49). By using high-performance liquid chromatography (HPLC)-based c-di-GMP detection analysis, they demonstrated that purified CdgB synthesizes c-di-GMP in vitro. Depending on the media used, the overproduction of CdgB showed effects similar to those seen with overproduction of CdgA, i.e., inhibition of aerial mycelium formation and actinorhodin production.
Two GGDEF-EAL proteins, RmdA and RmdB, were shown to function as PDEs and to impact development in S. coelicolor (50). Deletion of either rmdA or rmdB causes a mild defect in development, delaying spore formation. In contrast, deletion of both genes has a dramatic effect, causing a total inhibition of aerial mycelium formation, which suggests functional redundancy. Aiming to identify the signals that activate the enzymatic function of RmdA, the authors found that hemin can bind to this protein. However, evidence that hemin specifically binds to the PAS-PAC sensory domain and that binding of the ligand has an effect on the PDE function is still lacking. In addition, the role of the conserved GGDEF domain in RmdA as well as in RmdB remains unknown.
BldD LINKS c-DI-GMP SIGNALING AND DEVELOPMENT IN STREPTOMYCES
BldD is a DNA binding protein that directly controls key developmental genes in Streptomyces (51–53). With the identification of the complete BldD regulon, comprising ca. 167 genes, including 42 targets encoding regulatory proteins, it became apparent that BldD has a central, pleiotropic role in Streptomyces development and acts as a master regulator that represses sporulation genes during vegetative growth (4). Nevertheless, how the BldD-dependent repression of sporulation genes is relieved during development remained a mystery.
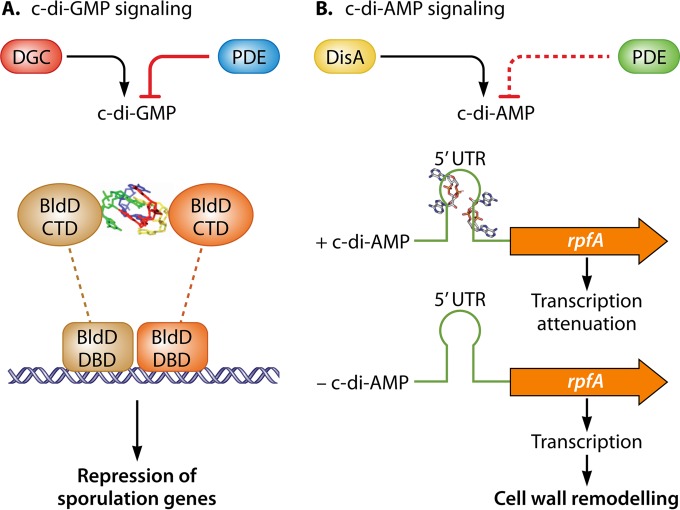
This mystery was solved when BldD was shown to be a c-di-GMP effector protein (12). Tschowri et al. demonstrated that c-di-GMP binds to the RXD-X8-RXXD motif located in the C-terminal domain (CTD) of BldD and enhances its activity as a transcriptional repressor (12). Notably, the crystal structure of the BldD CTD–c-di-GMP complex revealed that four molecules of c-di-GMP assemble into a tetramer that enables the two BldD CTDs to form a stable dimer and thus drive DNA binding. Uniquely, there are no protein-protein contacts between the two CTDs and it is the ligand only that connects the two protein domains (see Fig. 4A). Consequently, high levels of c-di-GMP drive BldD dimerization, leading to DNA binding through the N-terminal domains (NTDs), repression of the sporulation cascade, and a block in development (see Fig. 4A). On the other hand, when c-di-GMP levels are low, BldD is inactive, leading to activation of the developmental cascade and sporulation.
FIG 4.
Graphical summary of the c-di-GMP and c-di-AMP signaling pathways. (A) GGDEF-type diguanylate cyclases (DGCs) and EAL- and HD-GYP-type phosphodiesterases (PDEs) make and break c-di-GMP. A tetrameric c-di-GMP binds to the developmental master regulator BldD and links the two otherwise separate C-terminal domains (CTDs), resulting in protein dimerization. This enables the transcriptional repressor BldD to effectively bind to target DNA via its N-terminal DNA binding domain (DBD), leading to repression of sporulation genes during vegetative growth (4, 12). (B) DisA is the sole DAC protein responsible for synthesis of c-di-AMP. A c-di-AMP degrading PDE has yet to be identified (indicated by a dashed line). Binding of 2 c-di-AMPs to the ydaO-like riboswitch (41) in the 5′ untranslated region (5′ UTR) upstream of rpfA (resuscitation-promoting factor A) results in transcriptional attenuation. The absence of c-di-AMP favors rpfA transcription and accumulation of the RpfA cell wall-remodelling enzyme during spore germination (71).
bldD is an ancient gene that was present in very early actinobacteria but that has been lost several times in the course of evolution of the phylum (54). Orthologs of bldD show high conservation and local synteny and are found in all sporulating actinomycetes (54). Moreover, the c-di-GMP binding signature is present in all sequenced bldD orthologs and c-di-GMP is predicted to be present in all sporulating actinomycetes. This suggests that the BldD–c-di-GMP complex controls developmental processes not only in streptomycetes but also in all sporulating actinobacteria.
c-DI-GMP TURNOVER ENZYMES ARE DIRECT TARGETS OF DEVELOPMENTAL REGULATORS
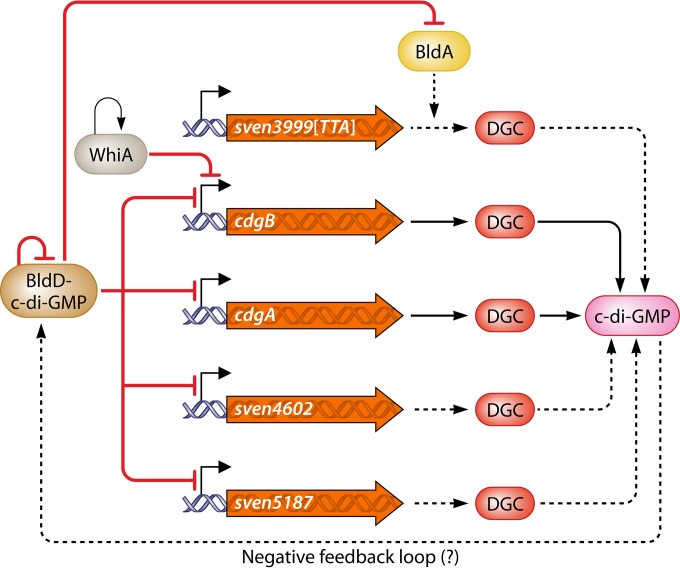
BldD not only senses c-di-GMP levels in the cell but also controls the expression of c-di-GMP-producing enzymes in response to ligand binding. Among the direct BldD targets in S. venezuelae are four genes encoding proven or putative DGCs: cdgA, cdgB, sven4602, and sven5187 (4, 12, 49) (Fig. 3). Thus, 4 of 6 potential DGCs in S. venezuelae are transcriptionally controlled by a c-di-GMP-sensing regulator. This wiring might suggest that there is a crucial need for a carefully fine-tuned c-di-GMP level in the cell. However, evidence for the involvement of a negative feedback loop is lacking, and whether bldD and any of these 4 DGCs are expressed and active in the same time and place during development and do indeed constitute a common c-di-GMP signaling module needs to be clarified.
FIG 3.
Developmental regulators control the levels of c-di-GMP synthesizing diguanylate cyclases (DGCs) in S. venezuelae. The BldD–c-di-GMP complex directly represses the expression of genes encoding proven (cdgB and cdgA) and predicted (sven4602 and sven5187) DGCs (4, 12, 49). This suggests that a negative feedback loop might be involved whereby c-di-GMP made by these DGCs binds to BldD, leading to repression of transcription. At the onset of sporulation, cdgB is also subject to repression by the developmental regulator WhiA (55). sven3999 encoding a putative DGC contains the rare TTA codon, suggesting that the BldA tRNA is required for its translation. Dashed lines indicate predicted wiring.
In addition to BldD, the WhiA transcriptional regulator is also involved in c-di-GMP signaling by directly controlling the expression of cdgB. WhiA is widespread in Gram-positive bacteria and is required for cessation of aerial growth and initiation of sporulation in Streptomyces species. This regulator is predominantly active at the onset of sporulation and moderately represses the transcription of cdgB (55) (Fig. 3). Since BldD and WhiA are active at distinct differentiation stages in Streptomyces species, it is unlikely that these two transcriptional regulators compete for binding to the cdgB promoter. However, the function of WhiA seems to be subject to posttranslational regulation (55) and it will be interesting to identify the conditions that lead to derepression of cdgB by WhiA.
Further, the developmental regulatory BldA tRNA appears to participate in the c-di-GMP signaling cascade by providing a leu-tRNAUUA for translation of TTA codons in sven3999 and the orthologous gene sco5345 (Fig. 3) as well as in rmdB. The TTA codon is rare in GC-rich streptomycetes, and bldA encodes the sole tRNA able to translate the UUA codon into the amino acid leucine (56). A bldA mutant is bald mainly because adpA and amfR contain a TTA codon in their DNA, and the presence of bldA is essential for their efficient translation (57). The AdpA transcriptional regulator not only controls the expression of sporulation genes but also governs chromosome replication by binding to the oriC region (58, 59). AmfR is required for the synthesis of the SapB surfactant peptide, which contributes to the hydrophobic sheath formation to allow aerial hyphae to escape the aqueous substrate environment during aerial mycelium formation. Similarly, sco5345 in S. coelicolor and the orthologous sven3999 gene in S. venezuelae, encoding a conserved GGDEF protein, contain a TTA codon (Fig. 3). Moreover, translation of the PDE RmdB in S. coelicolor also seems to depend on BldA due to the presence of a TTA codon in rmdB (54). The regulation of several c-di-GMP metabolizing enzymes by the developmental regulators BldD, WhiA, and BldA further emphasizes the central role of c-di-GMP signaling in Streptomyces differentiation.
BldD-MEDIATED ROLES OF c-DI-GMP IN ANTIBIOTIC PRODUCTION AND PATHOGENICITY
Antibiotic production in Streptomyces species is linked to morphological differentiation, and the developmental regulators that control aerial mycelium formation often also impact natural product synthesis, directly or indirectly (60). In the case of BldD, the BldD–c-di-GMP-controlled adpA and bldA genes are directly involved in the production of three different antibiotics in S. coelicolor: the blue actinorhodin (ACT), the red undecylprodigiosin (RED), and methylenomycin (MM) (60). The synthesis of actinorhodin is completely dependent on the pathway-specific regulator ActII-ORF4, which activates the expression of genes that encode biosynthetic enzymes within the act gene cluster (61). Among others, AdpA seems to directly bind to the actII-ORF4 promoter region, as shown by DNA-affinity capture assays (62). The same screen identified AdpA as a direct regulator of redD, which requires RedZ for expression and activates undecylprodigiosin biosynthetic genes (62, 63). However, how direct binding of AdpA to the actII-ORF4 as well as to the redD promoter regions affects gene expression awaits further investigation.
Strikingly, several genes involved in antibiotic synthesis in S. coelicolor contain a TTA codon and are therefore dependent on the BldA tRNA for translation. A TTA codon is present in adpA (64), actII-ORF4 (61), and redZ (65) as well as in mmyB and mmfL, encoding an activator and an enzyme involved in methylenomycin biosynthesis, respectively (66). In S. venezuelae, the indirect role of BldD–c-di-GMP extends to the regulation of chloramphenicol biosynthesis. Recently, it was shown that BldD–c-di-GMP-dependent response regulator BldM represses the expression of the chloramphenicol biosynthetic gene cluster (67), but the regulatory mechanism is not understood.
In Saccharopolyspora erythraea, BldD directly regulates the synthesis of the antibiotic erythromycin by binding to the promoters of the erythromycin biosynthetic gene cluster (68). S. erythraea encodes 13 GGDEF proteins, 11 GGDEF-EAL composite proteins, one GGDEF-HD-GYP fusion protein, and one EAL domain protein (28). Since the RXD-X8-RXXD c-di-GMP binding motif is conserved in S. erythraea BldD, it is very likely that c-di-GMP signals through BldD to control erythromycin production in this actinobacterium. The involvement of BldD–c-di-GMP in the production of a commercially valuable secondary metabolite raises the possibility that the manipulation of c-di-GMP levels is a promising tool for the full exploitation of antibiotic synthesis in sporulating actinomycetes.
A recent study systematically investigated the influence of classical bld genes on the pathogenicity of the agronomically important plant pathogen Streptomyces scabies, and the researchers found that bldD, bldC, bldA, adpA, and bldG are required for the full virulence of the bacterium (69). S. scabies is one of the best-characterized plant-pathogenic species and is the causative agent of the scab disease of taproot crops such as potato, carrot, radish, turnip, and beet. The prime source of the ability of S. scabies and related pathogenic streptomycetes to cause the scab disease is the production of the virulence factor thaxtomin A, which is activated by the regulator TxtR and functions as a cellulose synthesis inhibitor (70). Bioinformatic analysis revealed that a putative BldD binding site is present in the upstream region of txtR, suggesting that BldD may directly control the expression of txtR (69). BldD from S. scabies is 98% identical to the BldD ortholog in S. venezuelae, and the c-di-GMP binding signature is well conserved. Moreover, S. scabies encodes 4 GGDEF and 4 GGDEF-EAL domain proteins (28), suggesting that c-di-GMP signaling plays a role in the biology of the organism and that c-di-GMP signals through BldD to influence the virulence of pathogenic streptomycetes.
THE EMERGING ROLE OF c-DI-AMP IN CELL WALL METABOLISM OF STREPTOMYCES SPECIES
Streptomyces species encode one DAC domain-containing protein, which is homologous to DNA integrity-scanning protein A (DisA) in B. subtilis and consists of a N-terminal DAC domain and a C-terminal helix-hairpin-helix domain separated by a short linker region (Fig. 2). The DGA and RHR motifs, crucial for enzymatic activity (29), are conserved in DisA from Streptomyces species, and the protein was recently shown to be an active DAC in vivo in S. venezuelae (71). Streptomycetes do not have any DHH-DHHA1 domain-containing proteins or HD-type PDEs able to cleave c-di-AMP, so the counterpart to DisA remains unknown.
The function of DisA has been thoroughly studied in B. subtilis, and it is generally accepted that DisA is involved in DNA repair mechanisms. It has been shown that DisA scans chromosomal DNA for integrity and reduces c-di-AMP synthesis when DNA lesions are encountered, resulting in a delayed entry into sporulation to allow repair of DNA damage (72, 73). Moreover, it has been reported that DisA is involved in the arrest of DNA replication during B. subtilis spore outgrowth, to ensure that germinating spores are free of damaged DNA (74). How DisA and c-di-AMP contribute to recruitment of DNA repair factors is not well understood, but the diadenylate cyclase has been shown to interact with the RadA recombination protein, which participates in processing of Holliday junctions (75, 76).
The role of c-di-AMP in Streptomyces species has yet to be investigated in any detail; however, in contrast to evidence from B. subtilis and other species (see the introduction), it is already clear that c-di-AMP is not essential in S. venezuelae, since a disA mutant lacks detectable c-di-AMP and is viable under standard growth conditions (71). The first relevant published study provided evidence that c-di-AMP made by DisA may be involved in cell wall remodelling during spore germination in Streptomyces species (71). In actinobacteria, the resuscitation-promoting factor (Rpf) proteins are a class of enzymes that hydrolyze the sugar backbone of peptidoglycan and play an important role in the resuscitation of dormant cells (77–79). A detailed analysis of rpfA regulation revealed that this locus functions as a regulatory node that integrates signals from at least three different second messengers: cAMP-CRP is required for transcription initiation, a c-di-AMP binding ydaO-like riboswitch is involved in transcript elongation, and a ppGpp-responsive protease controls RpfA stability (71).
The ydaO riboswitch is widespread in Gram-positive bacteria and is typically located in the 5′ untranslated (leader) region (UTR) of controlled genes (80). In B. subtilis, the ydaO riboswitch is present in the 5′ UTR of the ydaO gene, the function of which is unknown, and of the ktrAB operon, encoding the two subunits of a potassium transporter. Representing a so-called genetic OFF switch, binding of c-di-AMP to the ydaO switch results in premature transcription termination (41). Similarly, the ydaO-like riboswitch in S. coelicolor is located in the 5′ UTR of rpfA and leads to transcription attenuation upon c-di-AMP binding (Fig. 4B). The ydaO or rpfA riboswitch is conserved in actinobacteria and is mostly associated with genes involved in cell wall metabolism, suggesting that c-di-AMP controls cell wall remodelling in these bacteria (80). However, rpfA is very likely not the only c-di-AMP-regulated gene in Streptomyces since there are at least three additional genes encoding cell wall lytic enzymes that carry an rpfA-like riboswitch in their 5′ UTRs (71, 77). The identification of further c-di-AMP effector molecules in Streptomyces is an important goal for the future.
CONCLUSIONS
Given that c-di-GMP stimulates hyphal vegetative growth and inhibits sporulation in Streptomyces species, it remains faithful to its role as a lifestyle regulator. But, in contrast to its control of the “stick-or-swim” switch in Gram-negative species, it controls the hypha-to-spore transition in Streptomyces species. By controlling the activity of the BldD developmental master regulator, c-di-GMP determines the timing of sporulation. On the basis of recent findings, we now have a detailed understanding of the processes triggered by binding of c-di-GMP to BldD, but we have little knowledge of the signaling events that occur upstream of BldD. To get a more complete picture of c-di-GMP signaling in streptomycetes, it will be essential to identify the DGCs and PDEs that specifically control the c-di-GMP sensed by BldD and also to determine the internal and external stimuli that fine-tune the levels and activities of these enzymes.
To date, very little is known about c-di-AMP signaling in streptomycetes, but low levels seem to favor spore germination by stimulating the accumulation of enzymes involved in cell wall remodelling. Whether c-di-AMP synthesis is also connected to DNA integrity, as is the case in B. subtilis, remains to be determined. In the absence of DHH-DHHA1- and HD domain-containing proteins, how c-di-AMP is degraded in streptomycetes also awaits future investigations.
The most recently discovered c-dinucleotide in the collection of the bacteria is the hybrid molecule c-AMP-GMP, which is synthesized from ATP and GTP by DncV in Vibrio cholerae (81). Streptomyces species do not have a DncV homolog, but it is tempting to speculate that, with c-di-GMP and c-di-AMP, the set of c-dinucleotides employed by these bacteria is not yet complete. Altogether, recent findings indicate that c-dinucleotides are important players in signal transduction networks in streptomycetes and represent an exciting research area for future studies.
ACKNOWLEDGMENTS
I thank Mark Buttner, Susan Schlimpert, and Anja Richter for reading the manuscript and providing constructive comments.
REFERENCES
- 1.Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY. [Google Scholar]
- 2.McCormick JR, Flärdh K. 2012. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev 36:206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 4.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 5.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 6.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem 282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 7.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 12.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Chuah ML, Dong X, Xie K, Luo Z, Tang K, Liang ZX. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J Biol Chem 286:2910–2917. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 18.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Romling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 21.Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res 40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. 2010. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol 396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 23.Leduc JL, Roberts GP. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol 191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol 82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 25.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witte G, Hartung S, Buttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. 2015. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A 112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. 2013. Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. 2013. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4:e00282–13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol Cells 19:365–374. [PubMed] [Google Scholar]
- 36.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Li W, He ZG. 2013. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem 288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller M, Hopfner KP, Witte G. 2015. c-di-AMP recognition by Staphylococcus aureus PstA. FEBS Lett 589:45–51. doi: 10.1016/j.febslet.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. 2014. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corrigan RM, Grundling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 43.Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stulke J. 9 May 2015, posting date. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 44.Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, Parker JL, Richards DM, Jung YG, Buttner MJ, Flardh K. 2012. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci U S A 109:E2371–E2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, Dijkhuizen L, Wosten HA. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev 17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev 17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claessen D, Wosten HA, van Keulen G, Faber OG, Alves AM, Meijer WG, Dijkhuizen L. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol 44:1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 48.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641–650. doi: 10.1016/0092-8674(91)90096-H. [DOI] [PubMed] [Google Scholar]
- 49.Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J Bacteriol 193:3100–3108. doi: 10.1128/JB.01460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hull TD, Ryu MH, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. 2012. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J Bacteriol 194:4642–4651. doi: 10.1128/JB.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elliot M, Damji F, Passantino R, Chater K, Leskiw B. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol 180:1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliot MA, Leskiw BK. 1999. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J Bacteriol 181:6832–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- 54.Chandra G, Chater KF. 2014. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol Rev 38:345–379. doi: 10.1111/1574-6976.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. 2013. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in streptomyces venezuelae. mBio 4:e00684–13. doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leskiw BK, Mah R, Lawlor EJ, Chater KF. 1993. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J Bacteriol 175:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Higo A, Horinouchi S, Ohnishi Y. 2011. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol Microbiol 81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- 58.Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res 19:259–273. doi: 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolański M, Jakimowicz D, Zakrzewska-Czerwińska J. 2012. AdpA, key regulator for morphological differentiation regulates bacterial chromosome replication. Open Biol 2:120097. doi: 10.1098/rsob.120097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769–780. doi: 10.1016/0092-8674(91)90120-N. [DOI] [PubMed] [Google Scholar]
- 62.Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 36:1073–1083. doi: 10.1007/s10295-009-0591-2. [DOI] [PubMed] [Google Scholar]
- 63.Rudd BA, Hopwood DA. 1980. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol 119:333–340. [DOI] [PubMed] [Google Scholar]
- 64.Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- 65.White J, Bibb M. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol 179:627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Rourke S, Wietzorrek A, Fowler K, Corre C, Challis GL, Chater KF. 2009. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol Microbiol 71:763–778. doi: 10.1111/j.1365-2958.2008.06560.x. [DOI] [PubMed] [Google Scholar]
- 67.Fernández-Martínez LT, Borsetto C, Gomez-Escribano JP, Bibb MJ, Al-Bassam MM, Chandra G, Bibb MJ. 2014. New insights into chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712. Antimicrob Agents Chemother 58:7441–7450. doi: 10.1128/AAC.04272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chng C, Lum AM, Vroom JA, Kao CM. 2008. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc Natl Acad Sci U S A 105:11346–11351. doi: 10.1073/pnas.0803622105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bignell DR, Francis IM, Fyans JK, Loria R. 2014. Thaxtomin A production and virulence are controlled by several bld gene global regulators in Streptomyces scabies. Mol Plant Microbe Interact 27:875–885. doi: 10.1094/MPMI-02-14-0037-R. [DOI] [PubMed] [Google Scholar]
- 70.Loria R, Kers J, Joshi M. 2006. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 71.St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA. 16 March 2015, posting date. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol Microbiol doi: 10.1111/mmi.12971. [DOI] [PubMed] [Google Scholar]
- 72.Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. 2006. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 73.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep 12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campos SS, Ibarra-Rodriguez JR, Barajas-Ornelas RC, Ramirez-Guadiana FH, Obregon-Herrera A, Setlow P, Pedraza-Reyes M. 2014. Interaction of apurinic/apyrimidinic endonucleases Nfo and ExoA with the DNA integrity scanning protein DisA in the processing of oxidative DNA damage during Bacillus subtilis spore outgrowth. J Bacteriol 196:568–578. doi: 10.1128/JB.01259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L, He ZG. 2013. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis. J Biol Chem 288:22426–22436. doi: 10.1074/jbc.M113.464883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carrasco B, Cozar MC, Lurz R, Alonso JC, Ayora S. 2004. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J Bacteriol 186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haiser HJ, Yousef MR, Elliot MA. 2009. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J Bacteriol 191:6501–6512. doi: 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Telkov MV, Demina GR, Voloshin SA, Salina EG, Dudik TV, Stekhanova TN, Mukamolova GV, Kazaryan KA, Goncharenko AV, Young M, Kaprelyants AS. 2006. Proteins of the Rpf (resuscitation promoting factor) family are peptidoglycan hydrolases. Biochemistry 71:414–422. doi: 10.1134/S0006297906040092. [DOI] [PubMed] [Google Scholar]
- 79.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. 2006. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol 14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 80.Block KF, Hammond MC, Breaker RR. 2010. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol 192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]