Abstract
The importance of cyclic di-GMP (c-di-GMP) and its control of biofilm matrix assembly and production has been a focal point of researchers in recent history. In this issue, Cooley et al. (Cooley RB, Smith TJ, Leung W, Tierney V, Borlee BR, O’Toole GA, Sondermann H, J Bacteriol 198:66–77, http://dx.doi.org/10.1128/JB.00369-15) demonstrate that two c-di-GMP controlled features of Pseudomonas aeruginosa, the periplasmic protease LapG and the surface adhesin CdrA, are linked. CdrA is shown to be a substrate of LapG, with LapG activity controlled by intracellular c-di-GMP levels. This commentary discusses the significance of this finding.
TEXT
The last decade has seen much research directed toward understanding the mechanisms by which microbes form and maintain communities called biofilms (1, 2). A common feature of these communities is an extracellular matrix that holds individual bacteria together. The composition of the matrix is usually complex, consisting of exopolysaccharides, proteins, and nucleic acids (2, 3). Regulation of matrix production and assembly is an area of key focus in the field.
Several common themes have emerged spanning a range of organisms that have been studied in the laboratory. In many Gram-negative species, one prevalent theme involves the intracellular signaling molecule cyclic di-GMP (c-di-GMP), which is known to promote biofilm formation by inducing the production of secreted biofilm matrix polysaccharides and proteins (4–6). In addition, c-di-GMP negatively impacts motility, which has led to c-di-GMP being called the “switch” that controls the transition between sessile and motile lifestyles. Another emerging commonality is the importance of secreted adhesins that provide a variety of functions such as mediating attachment of cells to a surface as well as providing structural integrity to the biofilm matrix itself (7–9). In the model species Pseudomonas aeruginosa, c-di-GMP is known to promote the transcription and synthesis of two matrix polysaccharides, Pel and Psl, as well as the biofilm matrix adhesin CdrA (7, 10). CdrA specifically binds to Psl in the matrix, which can stabilize biofilm structure. All three have been shown to contribute to biofilm formation in this species.
In the current issue of the Journal of Bacteriology, Cooley et al. demonstrate that a periplasmic protease, LapG, controls proteolysis of CdrA in response to low cellular c-di-GMP levels (11). LapD is the protein directly responsive to c-di-GMP and, in turn, controls LapG activity. By cleaving CdrA, LapG releases this adhesin from the outer membrane, presumably disrupting association of the cell with the matrix polysaccharide Psl. This is a significant finding for a few reasons. First, LapG was initially characterized in the related species Pseudomonas fluorescens. In P. fluorescens, LapG was found to act on a large surface-associated adhesin called LapA (12). This adhesin bears no structural similarity to CdrA; thus, this appears to be a conserved but modular mechanism utilized by some of the pseudomonads (i.e., this protease that responds indirectly to c-di-GMP can act on variable targets). This study highlights the presence of LapG and LapD-like orthologs in a number of proteobacteria known to form biofilms, indicating a common mechanism regulating biofilm development. Second, it demonstrates that c-di-GMP can impact CdrA functionality posttranscriptionally, providing an additional level of control. Conceptually, this is a key advance. Biofilm formation is not a dead end. Under the right environmental conditions, we now see that cells have a mechanism by which they sever their association with the biofilm matrix and reenter the planktonic state.
The Lap system was first characterized in Pseudomonas fluorescens (for key references, please see references 12 to 18). In this system, LapD is a cell membrane protein that acts as an intracellular c-di-GMP sensor. Under conditions of high c-di-GMP, LapD is activated, recruiting the periplasmic protease LapG, constraining its reach. When c-di-GMP levels drop, LapG is released from LapD and acts by cleaving the N terminus of the large cell surface-associated adhesin LapA. Intact, uncleaved LapA contributes to cell-surface interactions at the early stages of biofilm formation by promoting irreversible attachment to a surface. LapA is very large (∼520 kDa), has a type I secretion signal sequence, and belongs to the RTX family of adhesins/toxins. Three other proteins encoded by genes adjacent to lapA, LapB, LapC, and LapE, are homologous to ABC-type transporters. Thus, release of the cell from LapA through this process inhibits biofilm formation. An initial search of the P. aeruginosa genome identified LapG and LapD orthologs but failed to identify any LapA-like candidates. The current study begins by identifying a LapG target substrate in P. aeruginosa, CdrA.
The cdrAB genes were initially identified in P. aeruginosa as a bicistronic operon that was induced under conditions of elevated c-di-GMP (7, 19). Subsequent studies linked their expression to biofilm formation (20). Borlee et al. demonstrated that these genes likely belonged to a two-partner secretion system (TPSS; type Vb secretion system) (7). CdrB is a predicted barrel-forming protein that forms a pore in the outer membrane serving as the conduit through which the large adhesin CdrA is secreted. CdrA also harbors a pair of C-terminal cysteine residues that have been implicated in other TPSS adhesins in linking to the outer membrane. Borlee et al. (7) went on to show that CdrA specifically interacts with the matrix exopolysaccharide Psl. This interaction was key for stabilizing matrix integrity, with cdrA mutant strains displaying aggregates with loosely associated Psl and showing biofilms with compromised integrity. CdrA initially was predicted to be translated as an ∼220-kDa protein that was found to be cell associated and also as a processed form (∼150 kDa) in the supernatant of cultures overexpressing it. The resulting model was that under conditions of high c-di-GMP, P. aeruginosa induced biofilm formation through the simultaneous expression of matrix polysaccharides and matrix-binding adhesins.
There are some obvious questions raised by this study. The first relates to the context of this mechanism as it relates to the biology of the organism. When and where is LapG-mediated proteolysis of CdrA important during biofilm formation? This work surely implies that a primary feature of the mechanism would allow the detachment of cells from an existing biofilm. This process has been termed dispersion and has been previously linked to c-di-GMP signaling (21). Dispersion has been described as cells within the interior of a biofilm aggregate exhibiting increased motility until the cells eventually escape the aggregate by active swimming. Two implied prerequisites of this process would be the liberation of cells firmly embedded in the biofilm matrix and activation of swimming motility. LapG cleavage of CdrA and lowered c-di-GMP would enable both to occur. There are some obvious experiments that could test this hypothesis, involving select mutant strains and conditional depletion/synthesis of c-di-GMP.
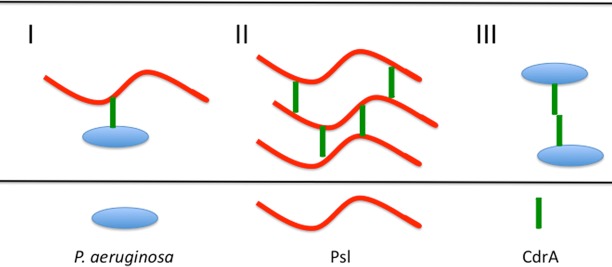
Another question relates to the functionality of CdrA. The presented data, viewed with P. fluorescens LapA in mind, might indicate this as a mechanism that primarily influences attachment/surface association. There is a possibility that the cell-associated and released forms of CdrA both serve important purposes. Ma et al. demonstrated that Psl forms a discrete “shell” around a growing aggregate in a biofilm (22). Could proteolyzed CdrA still function extracellularly to cross-link polymer strands of Psl—independently of the cells themselves? Can CdrA-CdrA interactions mediate intercellular interactions? Or is proteolyzed CdrA nonfunctional? Potential models for CdrA function are depicted in Fig. 1. Note that these models are not mutually exclusive and that LapG activity could potentially have a profound effect on these different scenarios. Finally, it will be interesting to see if there are specific environmental sensors involved in c-di-GMP signaling that specifically interface with LapD.
FIG 1.
Models for how CdrA and Psl may interact within the biofilm matrix.
Another interesting implication of this research is the potential that it represents a common mechanism used by several different species to control surface-associated adhesins. Some of these organisms (listed in Table S1 of the study by Cooley et al. [11]) are verified to utilize c-di-GMP signaling. Additionally, there are several other two-partner secretion system adhesins (such as Bordetella pertussis FHA and Haemophilus influenzae MW1) that might be potentially regulated posttranscriptionally through proteolysis. This, of course, might involve proteases distinct from LapG; however, the general theme/concept could be much more widespread than we appreciate. Much work is needed to address this important question.
In summary, the work of Cooley et al. has made a number of important contributions as well as raising a number of additional questions. Perhaps the most important finding is the implied widespread importance of this mechanism in a range of species. This may not be surprising, as it mirrors the ubiquity of organisms that utilize c-di-GMP signaling. Another exciting aspect of the work pertains to filling in a gap in our understanding of how organisms transition between free-swimming and biofilm lifestyles. That planktonic bacteria are capable of initiating biofilms and that biofilm bacteria can return to the planktonic mode of growth are widely accepted. Perhaps the most poorly understood aspect of this is the mechanism underlying how biofilm bacteria can mobilize into planktonic cells. The use of LapG to cleave CdrA and release a cell tethered to the matrix may be a key step in defining this process.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Costerton JW, Stewart PS. 2001. Battling biofilms. Sci Am 285:74–81. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U. 2012. Cyclic di-GMP, an established secondary messenger still speeding up. Environ Microbiol 14:1817–1829. doi: 10.1111/j.1462-2920.2011.02617.x. [DOI] [PubMed] [Google Scholar]
- 6.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 7.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 9.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooley RB, Smith TJ, Leung W, Tierney V, Borlee BR, O'Toole GA, Sondermann H. 2016. Cyclic di-GMP-regulated periplasmic proteolysis of a Pseudomonas aeruginosa type Vb secretion system substrate. J Bacteriol 198:66–77. doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinsa SM, O'Toole GA. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 14.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd CD, Chatterjee D, Sondermann H, O'Toole GA. 2012. LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J Bacteriol 194:4406–4414. doi: 10.1128/JB.00642-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O'Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife 3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 19.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]