ABSTRACT
The transcription factor FleQ from Pseudomonas aeruginosa derepresses expression of genes involved in biofilm formation when intracellular levels of the second messenger cyclic diguanosine monophosphate (c-di-GMP) are high. FleQ also activates transcription of flagellar genes, and the expression of these genes is highest at low intracellular c-di-GMP. FleQ thus plays a central role in mediating the transition between planktonic and biofilm lifestyles of P. aeruginosa. Previous work showed that FleQ controls expression of the pel operon for Pel exopolysaccharide biosynthesis by converting from a repressor to an activator upon binding c-di-GMP. To explore the activity of FleQ further, we carried out DNase I footprinting at three additional biofilm gene promoters, those of psl, cdrAB, and PA2440. The expression of cdrAB, encoding a cell surface adhesin, was sufficiently responsive to FleQ to allow us to carry out in vivo promoter assays. The results showed that, similarly to our observations with the pel operon, FleQ switches from a repressor to an activator of cdrAB gene expression in response to c-di-GMP. From the footprinting data, we identified a FleQ DNA binding consensus sequence. A search for this conserved sequence in bacterial genome sequences led to the identification of FleQ binding sites in the promoters of the siaABCD operon, important for cell aggregation, and the bdlA gene, important for biofilm dispersal, in P. aeruginosa. We also identified FleQ binding sites upstream of lapA-like adhesin genes in other Pseudomonas species.
IMPORTANCE The transcription factor FleQ is widely distributed in Pseudomonas species. In all species examined, it is a master regulator of flagellar gene expression. It also regulates diverse genes involved in biofilm formation in P. aeruginosa when intracellular levels of the second messenger c-di-GMP are high. Unlike flagellar genes, biofilm-associated genes are not always easy to recognize in genome sequences. Here, we identified a consensus DNA binding sequence for FleQ. This allowed us to survey Pseudomonas strains and find new genes that are likely regulated by FleQ and possibly involved in biofilm formation.
INTRODUCTION
Cyclic diguanosine monophosphate (c-di-GMP) is an intracellular second messenger that is produced by bacteria and has diverse effects on bacterial physiology. c-di-GMP binds to riboswitches and effector proteins to modulate transcription, translation, and protein activities (1, 2). In the opportunistic pathogen Pseudomonas aeruginosa, the transcription factor FleQ binds c-di-GMP to derepress expression of genes for biofilm components (3, 4). Biofilms are surface-attached communities of bacteria embedded in a matrix made of exopolysaccharides, proteins, and DNA. Biofilm infections are particularly difficult to treat because bacteria are protected by the biofilm matrix, making them resistant to antimicrobial treatment compared to planktonic cells (5). Genes controlled by FleQ include pel genes, coding for Pel exopolysaccharide; psl genes, coding for Psl exopolysaccharide; cdrAB, coding for an adhesin; and PA2440, coding for a polysaccharide deacetylase (4, 6–10). In a previous study of pel operon expression, we found that FleQ undergoes a conformational change when it binds c-di-GMP to switch it from a repressor to an activator of pel gene expression (3). FleQ also activates flagellar gene expression. In conjunction with σ54, FleQ activates the expression of the two-component regulatory genes fleSR, as well as genes required for flagellar export apparatus assembly and flagellar basal body assembly (6, 11). c-di-GMP inhibits FleQ ATPase activity (12), which explains the modest (1.5- to 2-fold) downregulation of flagellar gene expression that is seen in the presence of c-di-GMP (4, 8). By inversely regulating biofilm and flagellar gene expression in response to c-di-GMP, FleQ helps to control the planktonic-to-biofilm lifestyle transition of P. aeruginosa.
To extend our understanding of FleQ as a transcriptional regulator of psl, cdrAB, and PA2440 genes involved in biofilm formation, we carried out DNA footprinting at high and low c-di-GMP concentrations. We also looked at the effects of FleQ and c-di-GMP on the in vivo promoter activities of the cdrAB operon, which encodes an adhesin. We determined that FleQ binds at two to three sites near the psl, cdrAB, and PA2440 operon promoters. This allowed us to define a common FleQ consensus binding sequence motif. A search for this conserved sequence in the genome sequence of strain PAO1 and several other Pseudomonas species led to the identification of FleQ binding sites in the promoter regions of a number of new genes.
MATERIALS AND METHODS
Strains and growth media.
P. aeruginosa strain PAO1 and Escherichia coli strains were routinely grown on LB medium at 37°C. Antibiotics were added at the following concentrations: 60 μg/ml gentamicin or tetracycline for P. aeruginosa and 10 μg/ml gentamicin or tetracycline for E. coli. c-di-GMP was purchased from Biolog Life Science Institute (Bremen, Germany).
For all primer sequences, lowercase letters refer to restriction endonuclease recognition sequences. A cdrAB promoter deletion was constructed by sequence overlap extension PCR with primers cdrA-del-E1 (gaagaattcTCCTGGCAACTGACCGAGCAGCAC), cdrA-del-I1 (GCTGGCTGATCTTGATCTCGTTGGGTTTTCCTCGACCATGCCTCCCTC), cdrA-del-I2 (GAGGGAGGCATGGTCGAGGAAAACCCAACGAGATCAAGATCAGCCAGC), and cdrA-del-E2 (gaaggatccGCATCCTGCGGCTTGCGGGTAACC). The cdrAB promoter deletion starts 341 bases upstream and ends 151 bases downstream of the translation start site. Upstream and downstream flanking regions of about 500 bases were cloned into the EcoRI and BamHI sites of suicide vector pEX19Gm and introduced into PAO1 Δpel Δpsl or PAO1 Δpel Δpsl ΔfleQ by conjugation with E. coli S17-1 (13). A similar procedure was followed to construct a PAO1 strain with rpoN deleted. Mutations in the fleQ gene (fleQD245A and fleQT224S) were generated by overlapping PCR with oligonucleotides containing the mutations and flanking primers with the following sequences: FleQmutup, gacgaattcAGGCCGGCAACTGGGATGCCATCG, and FleQmutdown, gacaagcttAGGGAAATCTCGTGACCGTCATCC. The EcoRI-HindIII fragments were then cloned into pEX19-Gm and introduced into PAO1 ΔfleQ by conjugation with E. coli S17-1.
Construction of chromosomal cdrA-lacZ fusions and measurement of promoter activities in vivo.
Chromosomal cdrA-lacZ reporter fusions were constructed using the mini-CTX-lacZ vector (14). A 441-bp DNA fragment starting 331 bases upstream of the translation start site and ending 110 bases downstream of the translation start site was amplified by PCR using primers PcdrA-up (gaaggatccATGCTGGCGATGGCGAACATGAGG) and PcdrA-down (gaagaattcGCATGGCTCATGGAAGGTTCCTTG). The fragment was cloned between the BamHI and EcoRI sites of mini-CTX-lacZ vector. Overlapping PCR was used to generate mutations in the cdrA promoter. Mini-CTX-lacZ-cdrAmut1 was constructed by replacing the ACTGAC sequence with CAGTCA centered at position −206 relative to the cdrA translation start site. Mini-CTX-lacZ-cdrAmut2 was constructed by replacing the ACTGAC sequence with CAGTCA centered at position −54 relative to the cdrAB translation start site. Mini-CTX-lacZ-cdrAmut3 was constructed by replacing the TTTGAC sequence with GGGTCA centered at position −30 relative to the cdrA translation start site. The plasmids mini-CTX-lacZ-cdrAWT, -mut1, -mut2, and -mut3 were transformed into the E. coli S17-1 strain and then introduced at the attB site of the PAO1 Δpel Δpsl ΔcdrAprom or PAO1 Δpel Δpsl ΔfleQ ΔcdrAprom chromosome by mating. Tetracycline-resistant colonies were selected and confirmed by PCR analysis. Plasmids pJN105 (vector control, gentamicin resistant) and pJN1120 (allowing the overexpression of the diguanylate cyclase encoded by PA1120) were introduced by electroporation (4). Strains were grown aerobically overnight in LB with gentamicin (60 μg/ml) with shaking. β-Galactosidase activities of whole cells were measured by the Miller method (15). Results are the averages from three independent experiments.
Protein purification.
FleQ and FleN were purified from E. coli strains with plasmid pFleQ-Int1 or pFleN-Int1, as previously described (4).
DNase I footprinting.
DNase I footprinting experiments were performed using a nonradiochemical capillary electrophoresis method as previously described (3, 16). The fragments of DNA containing the cdrAB, psl, or PA2440 promoter were generated by PCR using 6-FAM (6-carboxyfluorescein phosphoramidate) primers labeled at the 5′ end with P. aeruginosa PAO1 chromosomal DNA as the template or mini-CTX-lacZ-cdrAB mutated versions when needed. The cdrAB promoter DNA fragment was generated using primers CdrA-FAM-up (5′-FAM-CTGGCGATGGCGAACATGAG), which paired 107 bases downstream of the cdrA translation start site, and PcdrA-down (gaagaattcGCATGGCTCATGGAAGGTTCCTTG), which paired 331 bases upstream of the cdrAB translational start site, leading to a DNA fragment of 438 bases. The psl fragment was obtained with primer Psl-FAM-down (5′-FAM-TCGATGAATCCAGCCCGTGTCAG), annealing 53 bases downstream of the pslA translational start site, and primer TR0 (cggcgaattcGGCTGGTTCTGGTAGAGCCGTCCG), annealing 541 bases upstream of the pslA translational start site. The PA2440 DNA fragment was generated by PCR using primers PA2440-FAM-down (5′-FAM-TTCAGGAACGTCATCCCGATCAG), which paired 32 bases downstream of the PA2440 translational start site, and PA2440-up (ACCACCGGACATCCGTGCAAAC), which paired 490 bases upstream of the PA2440 translational start site. The DNA fragments (0.45 pmol) were mixed with purified FleQ and/or FleN in 50-μl reaction mixtures containing ATP (10 μM), c-di-GMP (500 μM), and binding buffer (10 mM Tris-HCl, pH 7.8, 50 mM KCl, 8 mM magnesium acetate, 50 ng/μl bovine serum albumin [BSA], and 5% glycerol). After incubation for 30 min at room temperature, DNase I (0.3 units; Promega) was added for 2 min. The samples were phenol extracted and ethanol precipitated. Fragments were analyzed (Genewiz, Inc.), and fragment sizes were determined using ABI Peak Scanner software.
Gel shift assays.
To assess FleQ binding in gel shift assays, the probe DNA fragments (0.30 pmol) were mixed with 4 pmol of FleQ and/or 4 pmol of FleN in the presence of 50 μM ATP and in the presence or absence of 100 or 500 μM c-di-GMP in a 15-μl reaction mixture containing the same binding buffer as that used in footprinting experiments. After incubation for 25 min at room temperature, the samples were separated by electrophoresis on a 5% nondenaturing acrylamide Tris-glycine-EDTA (10 mM Tris [pH 8.0], 380 mM glycine, and 1 mM EDTA) gel in Tris-glycine-EDTA buffer at 4°C. The gel was soaked in 10,000-fold-diluted SYBR green I nucleic acid stain (Lonza, Walkersville, MD), and DNA was visualized under UV light. The DNA fragments containing the cdrAB or the PA2440 promoter were the same as the cdrAB or PA2440 fragment used for footprinting experiments (438 bases and 522 bases, respectively). The DNA fragment containing the psl operon promoter was generated by PCR using primer Psl-FAM-up (5′-FAM-CCCAGGCCAGAGCGCTCGCGG), which paired 297 bases upstream of the pslA translational start site, and primer DN1 (gatcggatccCTCGATGGAATCCACCCGTGTCA), which paired 54 bases downstream of the translation start site of pslA and led to a fragment of 361 bp.
RNA extraction, RT-PCR, and primer extensions.
Cell growth and RNA extraction were performed as previously described (3). Reverse transcription was performed using SuperScript II reverse transcriptase (Invitrogen) for reverse transcription-PCR (RT-PCR) analysis or SuperScript III reverse transcriptase for primer extension experiments. RT-PCRs were performed using a Sybr Green master mix with 1 ng of cDNA as the template and 200 nM each set of primers (pelA-F, CCTTCAGCCATCCGTTCTTCT, and pelA-R, TCGCGTACGAAGTCGACCTT, or PA4625-F, CCAGGTGATCATCTACAACTCCAA, and PA4625-R, GGAAGTGTTCGTCGCTGATGT). Cycling parameters were previously described (4). Genomic DNA was used as a standard, and the transcript levels of all genes were normalized to total cDNA. Data presented are the averages from at least two biological replicates. Error bars represent the standard deviation between samples. Primer extension analysis was performed on 15 μg of total RNA using the primer CdrA-FAM-up (also used for footprinting experiments) and following a preestablished protocol (3).
Bioinformatics.
MEME (Multiple Em for Motif Elicitation) software was used to identify a putative FleQ binding site motif (17). The 13 FleQ binding sites identified by footprinting were used as input into the MEME algorithm, and one occurrence per sequence was searched. MEME builds a position-specific scoring matrix wherein there is a probability associated with the occurrence of each base at each position. The MEME output also contains a sequence logo of the conserved motif as well as an E value, which is the probability of finding an equally well conserved pattern in random sequences. Position-dependent scoring matrices, as defined by MEME, were used as input to the MAST algorithm for searching in genome databases (18). Multiple alignments were performed using ClustalW from the EMBL-EBI website. The Pseudomonas Genome Database was used to retrieve pel, psl, PA2440, and cdrA orthologs (19).
RESULTS
Identification of the cdrAB operon transcription start site.
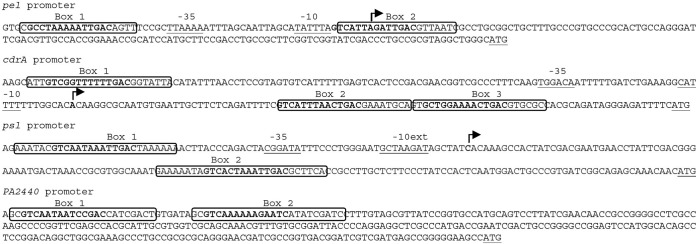
The cdrAB operon encodes an adhesin essential for Psl exopolysaccharide structure (9). To determine the cdrAB transcription start site, we isolated RNA from P. aeruginosa strains PAO1 Δpel Δpsl ΔfleQ and PAO1 Δpel Δpsl ΔwspF and performed primer extension analysis. The expression of cdrA (PA4625) has been shown to be high in both fleQ and wspF mutants (4). The wspF mutation causes an increase in the intracellular concentration of c-di-GMP (7), and FleQ is an apparent transcriptional repressor of cdrA expression (4). As a control, we isolated RNA from a strain (PAO1 Δpel Δpsl ΔcdrAprom) with a cdrAB promoter deletion. The deletion starts 341 bases upstream, and ends 151 bases downstream, of the cdrA translation start site. One major transcript was detected with RNA isolated from the ΔwspF and the ΔfleQ strains but not detected with RNA isolated from the strain with the cdrAB promoter deletion (Fig. 1). This fragment was 206 bp long, allowing us to map the cdrAB transcription start site to a location 99 bp upstream of the ATG start codon. FleQ has been shown to work with σ54 at flagellar promoters (6). These promoters are characterized by a GC doublet located between 11 and 14 bases upstream of the transcription start site (−12 site) and a conserved GG doublet that lies 10 bases farther upstream (−24 site) (20). No typical −24/−12 σ54 RNA polymerase binding site was found upstream of the transcription start site of cdrAB (Fig. 2). Instead, there are putative −10 (CATATT) and −35 (TTAAAA) boxes indicative of control by σ70 RNA polymerase in the promoter region of cdrAB.
FIG 1.
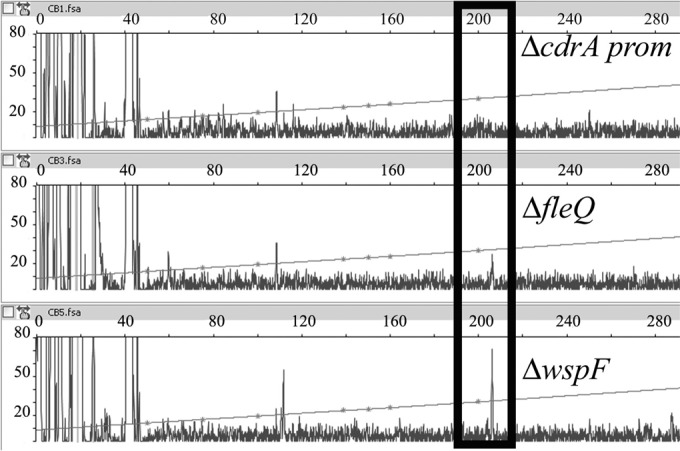
Identification of the cdrAB transcription start site. Primer extension analysis of the cdrAB transcript was done using a 6-FAM primer, which annealed to the mRNA 84 to 104 nucleotides downstream from the cdrAB translation start site. Total RNA was extracted from strains PAO1 Δpel Δpsl ΔcdrAprom (ΔcdrAprom), PAO1 Δpel Δpsl ΔwspF (ΔwspF), and PAO1 Δpel Δpsl ΔfleQ (ΔfleQ). The fluorescence intensity is plotted against the sequence length of the DNA fragment. The horizontal lines designate an internal size ladder. The major transcript with a size of 206 nucleotides is boxed.
FIG 2.
Nucleotide sequences of the pel, cdrAB, psl, and PA2440 promoter regions. The FleQ binding sites are boxed, and repeat sequences are in bold. The arrows indicating the transcription start sites were determined in this paper or previously. The translation start sites and the putative −10 and −35 elements are underlined.
We attempted but failed to identify the transcription start site of PA2440 using either an S1 nuclease protection assay or 5′ primer extension analysis. Our own transcriptome data and published transcriptome data (21) also failed to reveal a transcription start site for PA2440, possibly because this gene is typically expressed at very low levels. Thus, for footprinting and gel shift assays, we used a DNA fragment of 522 bp that spanned most of the intergenic region between PA2440 and its neighboring divergently transcribed gene, starting 32 bp downstream of the PA2440 translational start site.
Identification of FleQ binding sites in cdrAB, psl, and PA2440 promoter regions.
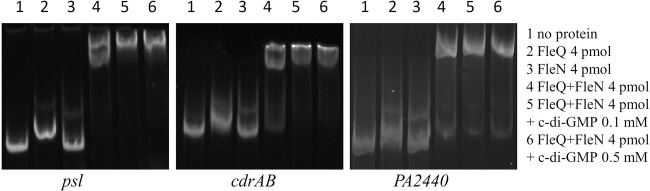
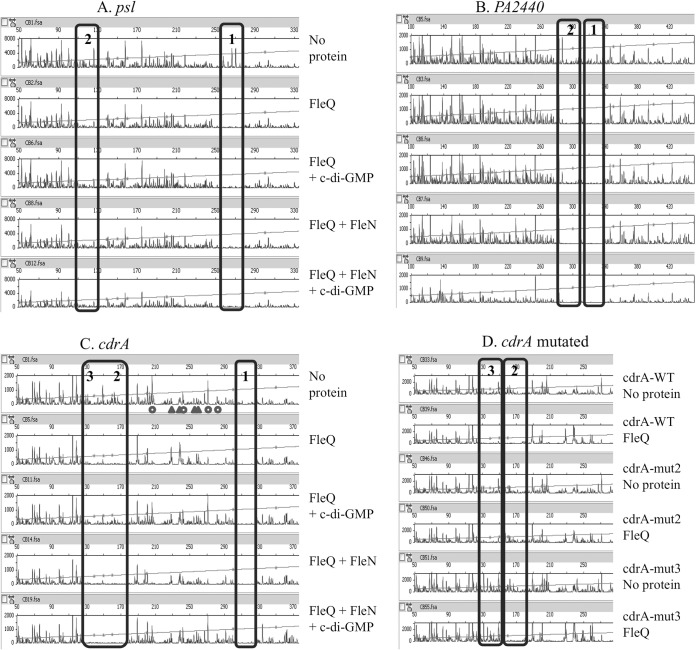
We performed gel shift assays to test if FleQ binds directly to the promoter regions of cdrAB, PA2440, and the psl operon. As shown in Fig. 3, FleQ caused a modest mobility shift of psl and cdrAB promoter DNA fragments when added alone to reaction mixtures. Addition of FleQ alone did not convincingly shift the mobility of the PA2440 DNA fragment. However, as described below in more detail, there was a strong shift in the mobility of all the DNA fragments tested, including the PA2440 fragment, when both FleQ and the protein FleN were present (Fig. 3). FleN has been shown to bind to FleQ but does not bind to DNA on its own (3, 22). To map the location of FleQ binding sites on these promoters, we performed footprinting experiments using 5′-FAM-labeled probes (Fig. 4). The assays shown in Fig. 4 were performed in the presence of 10 μM ATP and in the presence or absence of 500 μM c-di-GMP to allow for comparison to previously published results (3). In other experiments, we found that the properties of binding of FleQ to the DNA fragments shown in Fig. 4 were unchanged regardless of the presence or absence of ATP. ATP addition to a final concentration of 500 μM also had no effect on the DNA binding characteristics of FleQ. We tested c-di-GMP concentrations ranging from 1 μM to 1 mM in FleQ footprinting assays and found that the DNA binding properties of FleQ did not differ over this c-di-GMP concentration range. Footprinting of the psl operon promoter revealed two FleQ binding sites of 26 and 28 bases, centered at positions −62 and +76 relative to the previously determined transcription start site of the psl operon (Fig. 4A, rows 1 and 2, and Fig. 2) (23). Two FleQ binding sites were also identified upstream of the PA2440 coding region. These were centered at −294 and −263 relative to the translation start site of this gene (Fig. 4B, rows 1 and 2, and Fig. 2). DNase I footprinting with a fragment of DNA containing the cdrAB promoter revealed two protected regions separated by 131 bases: one of these is 24 bases and the second is 44 bases in length (Fig. 4C, rows 1 and 2, and Fig. 2). A closer look at the 44-base region revealed a repeated motif, ACTGAC, similar to the ATTGAC motif found in the FleQ binding sites in the pel promoter (3). Independent mutation of each of these motifs suggested that the 44-base box actually includes two FleQ binding sites, each about 22 bases in size (Fig. 4D, compare rows 1 with 2, 3 with 4, and 5 with 6). The cdrAB promoter thus contains three FleQ boxes centered at positions −109, +45, and +67 relative to its transcription start site (Fig. 2). The DNase I digestion profile observed between the boxes differed depending on the presence or absence of FleQ. Hyper- and hyposensitive sites appeared between FleQ boxes 1 and 2/3 when FleQ was bound. The positions of these sites are indicated by circles and triangles in the first row of Fig. 4C. This result suggests that FleQ binding induces a rearrangement of DNA, possibly DNA bending or wrapping of DNA around the FleQ protein.
FIG 3.
FleQ binds to the psl, cdrA, and PA2440 promoters. Fragments of DNA containing the psl (361-bp), cdrAB (438-bp), and PA2440 (522-bp) promoters (0.3 pmol) were incubated with or without FleQ (4 pmol), FleN (4 pmol), or c-di-GMP (0.1 or 0.5 mM). All reaction mixtures contained 50 μM ATP.
FIG 4.
FleQ binding does not require FleN or c-di-GMP. FAM-labeled DNA (0.45 pmol) was incubated with or without FleQ or FleN in the presence of ATP (10 μM) and in the presence or absence of c-di-GMP (500 μM) and then submitted to DNase I treatment (0.3 U) and analyzed by capillary electrophoresis. The fluorescence intensity (arbitrary units, ordinate) is plotted against the sequence length (bases, abscissa) of the fragment relative to the first base of the FAM-labeled primer. The horizontal line indicates the GS-500 internal size standard. The FleQ binding sites are boxed. (A) psl DNA (594 bp) was tested with 10 pmol of FleQ or FleN. (B) PA2440 DNA (522 bp) was tested with 50 pmol of FleQ or FleN. (C) cdrA fragment DNA (438 bp) was tested with 50 pmol of FleQ or FleN, and the locations of major putative DNA rearrangements are indicated by triangles (hypersensitive sites) or circles (hyposensitive sites). (D) The wild-type (WT) cdrA DNA fragment or cdrA fragments mutated in box 2 or 3 were tested with 50 pmol of FleQ.
FleQ stays bound to the cdrAB, psl operon, and PA2440 promoters in the presence of c-di-GMP or FleN.
FleQ works in concert with another protein, FleN, which is an ATPase with a deviant Walker A motif (24, 25). FleN is required for full expression of biofilm-related genes in the presence of c-di-GMP (4), but its exact biological role in biofilm gene expression is not clear. In studies of pel operon transcriptional control, we found that FleN interacts with FleQ but does not bind to DNA (3). We previously showed that FleN, in conjunction with FleQ, induced a bending of pel operon promoter DNA (3). We hypothesized that the bending increases repression of pel transcription. To test if a similar phenomenon occurs at the psl, cdrAB, and PA2440 promoter regions, we performed footprinting experiments and gel shift assays with FleQ and FleN. Consistent with previous results, FleN alone did not bind to the promoter DNA (Fig. 3). When FleQ and FleN were both present, a band of much lower mobility than that observed with FleQ alone was observed in gel shifts (Fig. 3), suggesting that FleN binds to FleQ to give higher-molecular-weight complexes. When we footprinted the different promoter regions, the electropherograms were not modified when FleQ and FleN were incubated together prior to DNase I treatment (Fig. 4, compare rows 2 and 4). From this, we conclude that the FleQ-FleN complex binds to the same DNA sequences as does FleQ alone. No DNA rearrangements were observed to result from the presence of FleN at the psl, cdrAB, or PA2440 promoter region DNA. This is in contrast to what we observed at the pel operon promoter (3).
Addition of c-di-GMP did not dramatically change FleQ footprinting patterns at the cdrAB, psl operon, or PA2440 promoters in the presence or absence of FleN. c-di-GMP also did not change the promoter DNA mobility shifts observed in the presence of FleQ and FleN (Fig. 3 and 4, compare rows 2 with 3 and 4 with 5). From this, it appears that the FleQ-FleN complex remains on the promoters even in the presence of c-di-GMP, similarly to what we have observed at the pelA promoter (3). It is noteworthy that the DNA rearrangement observed when FleQ is bound to the cdrAB promoter is relieved in the presence of c-di-GMP, irrespective of the presence of FleN (Fig. 4C, compare rows 2 with 3 and 4 with 5).
FleQ both activates and represses cdrAB expression.
To determine if c-di-GMP changes FleQ from a repressor to an activator of cdrAB expression in a manner similar to that observed for pel operon expression, we analyzed the cdrAB promoter region in more detail. We constructed transcriptional fusions of lacZ to cdrAB promoter fragments carrying wild-type or mutated versions of the FleQ boxes. We then introduced these constructs into the wild-type or fleQ-deletion strains with high (strains carrying pJN1120, expressing the diguanylate cyclase PA1120) or low (strains carrying pJN105, vector control) intracellular concentrations of c-di-GMP. The wild-type strain carrying a wild-type cdrA promoter (PcdrA-wt) had low β-galactosidase activity when c-di-GMP was low, while β-galactosidase activity was much higher when c-di-GMP was high (Fig. 5). In a strain with fleQ deleted, the same PcdrA-wt fusion had an intermediate level of expression in the presence or absence of c-di-GMP. These results suggest that FleQ represses cdrAB expression in the absence of c-di-GMP and activates cdrAB expression in the presence of c-di-GMP. Promoter fusions carrying mutations inside FleQ box 2 or 3 had low levels of expression with or without c-di-GMP, indicating that intact box 2 or 3 is not required for repression but that the binding of FleQ to both boxes 2 and 3 is required for cdrAB activation. The activity of PcdrA-mut1 was high whatever the concentration of c-di-GMP, suggesting that the binding of FleQ to this box is required for cdrAB repression.
FIG 5.
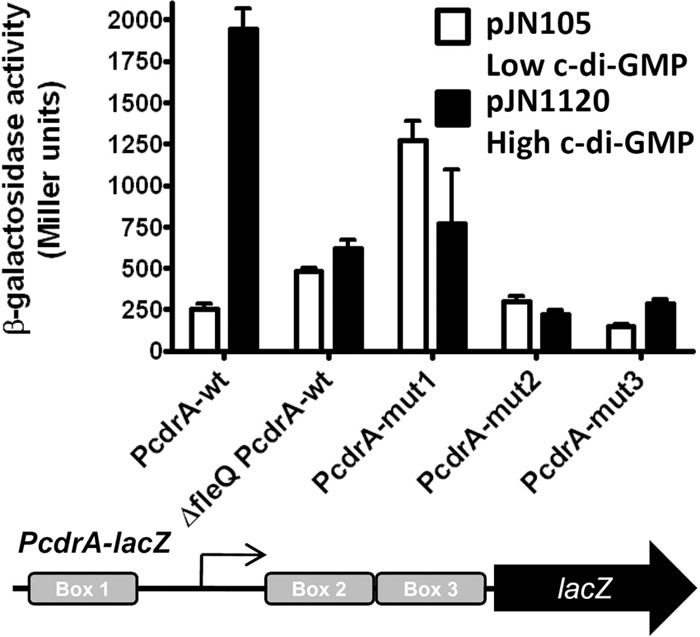
FleQ acts as a repressor in the absence of c-di-GMP and as an activator in the presence of c-di-GMP at the cdrAB promoter. β-Galactosidase activities of wild-type (wt) or mutated PcdrA-lacZ fusions were measured in PAO1 Δpel Δpsl ΔcdrAprom (except when ΔfleQ is indicated, which corresponds to the PAO1 Δpel Δpsl ΔcdrAprom ΔfleQ strain) carrying pJN105 (vector control) or pJN1120 (allowing the overexpression of the diguanylate cyclase encoded by PA1120). PcdrA-mut1 contains a mutation in FleQ box 1, PcdrA-mut2 contains a mutation in FleQ box 2, and PcdrA-mut3 contains a mutation in FleQ box 3, as indicated below the graph. The arrow indicates the position of the transcription start site of cdrA. Transcription of PcdrA-wt is significantly different in the fleQ-deleted strain than in the wild type under low c-di-GMP (t test, P value of 0.01).
ATPase activity and RpoN are not required for biofilm gene expression.
FleQ is an enhancer binding protein (EBP). Typically, EBPs interact with σ54 (RpoN) and open complex formation is driven by ATP hydrolysis catalyzed by the AAA ATPase domain of this class of protein. Expression of the pel or cdrAB operon was not affected in a strain with rpoN deleted (6). Also, the cdrAB and pel operon promoters contain −10 and −35 boxes instead of the −12/−24 boxes required for binding of σ54. We found that the level of pel or cdrAB transcripts in wild-type or rpoN strains was identical regardless of the concentration of c-di-GMP (Fig. 6). A mutation in the σ54 binding domain of FleQ (FleQT224S) did not induce a change in pel expression, confirming that the regulation of biofilm gene expression does not depend on σ54 RNA polymerase (Fig. 6) (6).
FIG 6.
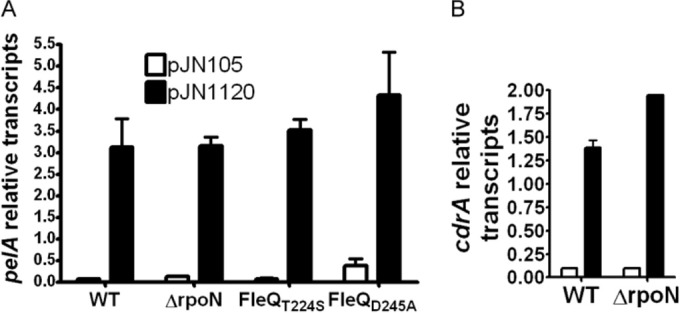
Neither FleQ ATPase activity nor σ54 is required for pel or cdrAB expression. Relative transcript levels of pel (A) or cdrAB (B) were measured by RT-PCR in the wild-type (WT) strain, a strain deleted for rpoN, or strains with fleQ mutations at the native chromosomal location. fleQT224S has a mutation in the σ54 binding domain of FleQ. fleQD245A has a mutation in the Walker B site of FleQ required for ATPase activity.
We also assessed the role of the ATPase activity of FleQ at the pel promoter, by introducing at the native location a mutation in the Walker B site of fleQ (fleQD245A). This mutation has previously been shown to abolish FleQ ATPase activity (12). pel expression was low in this mutant strain in the absence of c-di-GMP and high in the presence of c-di-GMP (Fig. 6). This result indicates that FleQ ATPase activity is not required for either pel repression or pel activation. Thus, FleQ is an unusual EBP able to both repress and activate gene expression independently of σ54 and ATPase activity.
Identification of a FleQ binding motif.
From the FleQ footprinting data at different promoters, we were able to identify a conserved FleQ binding motif of 14 bases, GTCAATaaATTGAC (Fig. 7). We also found this motif, although in less conserved form, in previously identified FleQ binding sites in the promoter regions of several FleQ-controlled flagellar genes (Fig. 7) (11).
FIG 7.
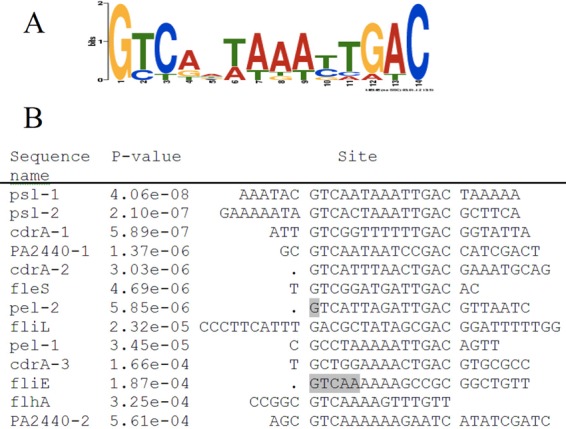
FleQ binding consensus sequence. (A) MEME logo of the conserved sequence of FleQ binding sites. According to the MEME software, the E value of this motif is 7.9e−018. (B) Alignment and P values of the different FleQ boxes. The consensus sequences are centered. Gray bases were not initially identified as part of FleQ binding sites.
Conservation of the FleQ binding sites in other Pseudomonas species and identification of new FleQ binding sites.
We examined the promoter regions of pel, psl, cdrAB, and PA2440 genes in other P. aeruginosa strains and other Pseudomonas species for the presence of FleQ consensus binding sequences. We found that the promoter sequences upstream of these genes are well conserved in all P. aeruginosa strains examined, with particularly high conservation in the FleQ consensus binding sequences (see Fig. S1A in the supplemental material). In looking at other species, we found that the region upstream of pelA in Pseudomonas protegens strains CHA0 and Pf-5 does not contain FleQ boxes. However, both strains have a gene coding for a putative epimerase (PFL_2971 and PFLCHAO_c30140) upstream of the pelA ortholog (see Fig. S1B). There are two putative FleQ boxes spaced 36 bases apart upstream of these genes, suggesting that the epimerase genes are the first genes in the pel operons in strains CHA0 and Pf-5 (see Fig. S1C). Pseudomonas putida, Pseudomonas syringae, and Pseudomonas stutzeri have orthologs of psl genes, but their promoter regions are not conserved and they do not have FleQ binding sites (data not shown).
From these observations, we can conclude that the FleQ binding sites that we determined experimentally are highly conserved in P. aeruginosa and can also be found in other Pseudomonas species, suggesting a common mechanism of regulation. We next looked for possible new FleQ target genes. Position-dependent scoring matrices, as defined by MEME, were used as input to the MAST algorithm for searching the FleQ binding sites in the upstream regions of genes. First, we examined the PAO1 genome. We found two FleQ binding consensus sequences in the promoter region of bdlA (PA1423) centered at positions −173 and −223 bases upstream of the bdlA translation start site (Fig. 8). bdlA codes for a chemosensory protein involved in bacterial dispersal. The phosphodiesterase DipA is activated where BldA is proteolytically processed. This then results in a net reduction of c-di-GMP and biofilm dispersal (26). We also identified two FleQ consensus sequences in the promoter region of siaA (PA0172), which is the first gene of the siaABCD operon involved in cell aggregation (27). The two FleQ boxes are spaced 58 bases apart and centered at positions −66 and −138 bases upstream of siaA ATG (Fig. 8).
FIG 8.
Promoter regions of putative FleQ-regulated genes. For each promoter region, the best possible match to the FleQ consensus binding motif and the position P value from the MAST algorithm are shown. Bases whose match to the motif has a positive score are indicated by a plus sign. The putative match sequences are indicated in gray, and the location of the sequences is indicated below the sequence and is relative to the translation start site.
When we looked for other potential FleQ binding sites among other Pseudomonas species, a common class of genes that emerged was lapA-like genes (Fig. 8). LapA is an outer membrane adhesin protein responsible for keeping cells attached to surfaces. When c-di-GMP is low, the LapD protein, which is a c-di-GMP receptor, allows LapG, a periplasmic protease, to interact with LapA and cleave the LapA N-terminal domain, inducing the release of LapA from the cell surface and promoting biofilm detachment (28–30). To illustrate the presence of putative FleQ binding sites in the promoter regions of a few lapA-like adhesins, an alignment of the promoter regions of these genes is shown in Fig. S2 in the supplemental material. In Pseudomonas mendocina NK01, the promoter region of MDS_0805 contains two putative FleQ binding sites located at positions centered at −80 and −106 bases upstream of the translation start site (Fig. 8). MDS_0805 codes for a response regulator-GGDEF domain protein, homologous to GcbA, which was shown to facilitate P. aeruginosa biofilm dispersion by activating BdlA (31). Finally, in Pseudomonas brassicacearum and Pseudomonas fluorescens F113 strains, we found homologous operons that have in their promoter regions (upstream of PSEBR_a3747 and PSF113_1970) two sequences (GTCAAGGACTTGAT and GTCATTTTGCTGAC) that resemble the FleQ consensus binding motif (see Fig. S3 in the supplemental material). These operons appear to encode an exopolysaccharide that is distinct from Pel or Psl.
DISCUSSION
Here, we identified FleQ binding sites in the promoter regions of cdrAB, PA2440, and the psl operon. We found that FleQ and FleN bind as a complex at these sites irrespective of the presence of c-di-GMP. We also found that FleQ both represses and activates cdrAB expression in response to c-di-GMP, similar to our results describing pel operon transcription in response to FleQ (3). However, the two promoters are organized differently. In the case of cdrAB, FleQ binds to three sites, one site located upstream of the transcription start site and two sites located side by side downstream of the transcription start site. The upstream box is required for FleQ-mediated cdrAB repression in the absence of c-di-GMP, whereas the downstream boxes are required for FleQ-mediated cdrAB activation in the presence of c-di-GMP. In the pel operon, the box upstream of the transcription start site is required for activation and the FleQ box that overlaps the transcription start site is required for repression. Our model for cdrAB expression is that in the absence of c-di-GMP, FleQ binds to the cdrAB promoter and induces a distortion of DNA. This could impair RNA polymerase binding and repress cdrAB expression. We know that binding of c-di-GMP induces a conformational change in FleQ (3). This serves to relieve the DNA distortion even though FleQ remains bound to the cdrAB promoter. This may improve RNA polymerase binding, leading to activation of cdrAB expression. FleQ may also both repress and activate psl and PA2440 gene expression. However, we did not analyze the expression of these genes in response to c-di-GMP in detail. lacZ transcriptional fusions that we prepared to the promoter regions of these genes showed a 3-fold or less induction of expression in response to c-di-GMP.
It is unusual for transcription activator binding sites to be located downstream of transcription start sites in bacteria. A few transcription activator proteins have been shown to bind downstream of the transcription start site, but little is known concerning their mechanism of activation (32–34). In the case of regulation of bacteriophage lambda pR promoter activity by DnaA, the location of the binding site downstream of the transcription start site allows a direct interaction between DnaA and the RNA polymerase β subunit (32). Weak binding of DnaA to DNA may contribute to removal of this activator after transcription initiation (32).
FleQ interacts with σ54 RNA polymerase to activate flagellar gene expression. In contrast, neither σ54 nor the FleQ σ54 binding domain was required for biofilm gene expression (Fig. 6). This is in good agreement with the presence of −10/−35 boxes and the absence of −12/−24 boxes in the promoter regions of biofilm-related genes. Together, these data indicate that biofilm-related genes are probably regulated by σ70. We also note, however, that P. aeruginosa PAO1 encodes 24 putative RNA polymerase sigma factors, 19 of which are classified as σ70-like extracytoplasmic function sigma factors (35). We cannot exclude that one of these other sigma factors participates in FleQ-mediated regulation of biofilm-related genes. A few EBPs regulate gene expression in conjunction with σ70. However, in these proteins, the σ54 binding domain is often mutated, as in HupR and NtrC from Rhodobacter capsulatus, or deleted, as is the case for VpsR from Vibrio cholerae and TyrR from E. coli (36–41). As with other EBPs, FleQ has an ATPase activity (12), and this activity is required for flagellar gene expression. Here, we determined that ATP hydrolysis was not required for either FleQ-mediated repression or activation of pel operon expression (Fig. 6). We do not know if a mechanism exists to inhibit FleQ ATPase activity at biofilm gene promoters.
At this point, FleQ is probably the best-characterized c-di-GMP-responsive transcription factor in terms of its mechanism of action. Its homologue FlrA from Vibrio cholerae is also a master regulator of flagellar synthesis, and c-di-GMP inhibits its ability to activate transcription (42). FlrA and FleQ may respond to c-di-GMP similarly at flagellar gene promoters. However, FlrA does not appear to directly regulate biofilm biosynthesis genes. Transcription factors belonging to several other protein families bind c-di-GMP as an effector, and it will be interesting to explore their mechanisms of action in depth. These include VpsT from Vibrio cholerae, which inversely regulates biofilm matrix production and motility in response to c-di-GMP and is a member of the LuxR family of regulators (43, 44). In Xanthomonas campestris, c-di-GMP acts as a negative regulator of the protein Clp, impairing the binding of Clp to DNA and the induction of genes important for virulence (45). A homologous protein, Bcam1349, from Burkholderia cenocepacia regulates cellulose synthase (46). Both Clp and Bcam1349 are homologous to the well-studied cyclic AMP (c-AMP)-responsive regulator Crp. LtmA, an apparently new class of regulator from Mycobacterium smegmatis, binds c-di-GMP to positively regulate the expression of lipid transport and metabolism genes (47). Finally, BrlR, a MerR family member from P. aeruginosa, was recently shown to bind c-di-GMP to control gene expression (48).
The data presented here allowed the identification of a consensus sequence among the 13 FleQ binding sites characterized in this study as well as in previous studies (3, 11). This motif contains the inverted repeat GTCAAT and ATTGAC separated by two nucleotides that are mostly A or T rich (Fig. 7). The promoter regions of both biofilm- and flagellum-related genes contain this consensus sequence. Biofilm-related gene promoters contain two to three FleQ binding sites, whereas flagellar genes contain only one poorly conserved FleQ binding consensus sequence, explaining why this motif was not discovered earlier. A search of genomes for FleQ consensus binding sequences led to the identification of several possible new FleQ-regulated genes, including bdlA, siaA, and lapA-like genes, as well as a homologue of gcbA in P. mendocina and a new exopolysaccharide synthesis operon in P. brassicacearum and P. fluorescens F113. The promoter regions of bdlA and siaA contain two putative FleQ binding sites. It has been shown that the expression of bdlA and the siaABCD operon was induced in a wspF mutant (high c-di-GMP) by a factor of 2 or 2 to 5, respectively (7, 8). Expression of bdlA was also shown to be low in planktonic cells compared to biofilm growth conditions (49). Two putative FleQ boxes were also found in the promoter regions of different lapA-like genes. Recent data showing that FleQ modulates lapA expression in response to c-di-GMP in P. putida make us more confident in our prediction (50). The promoter region of the P. mendocina gcbA homologue contains particularly well conserved putative FleQ binding sites; however, gcbA expression does not seem to be regulated by c-di-GMP or fleQ in P. aeruginosa strain PAO1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joe J. Harrison and Matthew Parsek for the PAO1 rpoN strain.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00539-15.
REFERENCES
- 1.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol 15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 7.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jyot J, Dasgupta N, Ramphal R. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184:5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 14.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–952. [DOI] [PubMed] [Google Scholar]
- 15.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 16.Wilson DO, Johnson P, McCord BR. 2001. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis 22:1979–1986. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 18.Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 19.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock REW, Brinkman FSL. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kustu S, Santero E, Keener J, Popham D, Weiss D. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev 53:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin EV. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 25.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dörrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B. 2009. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ Microbiol 11:3073–3086. doi: 10.1111/j.1462-2920.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 28.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro MVAS, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glinkowska M, Majka J, Messer W, Wegrzyn G. 2003. The mechanism of regulation of bacteriophage lambda pR promoter activity by Escherichia coli DnaA protein. J Biol Chem 278:22250–22256. doi: 10.1074/jbc.M212492200. [DOI] [PubMed] [Google Scholar]
- 33.Munson GP, Scott JR. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol Microbiol 36:1391–1402. [DOI] [PubMed] [Google Scholar]
- 34.Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. 2013. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9:e1003839. doi: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 36.Pittard AJ, Davidson BE. 1991. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol 5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 37.Herrera MC, Ramos JL. 2007. Catabolism of phenylalanine by Pseudomonas putida: the NtrC-family PhhR regulator binds to two sites upstream from the phhA gene and stimulates transcription with sigma70. J Mol Biol 366:1374–1386. doi: 10.1016/j.jmb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Bowman WC, Kranz RG. 1998. A bacterial ATP-dependent, enhancer binding protein that activates the housekeeping RNA polymerase. Genes Dev 12:1884–1893. doi: 10.1101/gad.12.12.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dischert W, Vignais PM, Colbeau A. 1999. The synthesis of Rhodobacter capsulatus HupSL hydrogenase is regulated by the two-component HupT/HupR system. Mol Microbiol 34:995–1006. doi: 10.1046/j.1365-2958.1999.01660.x. [DOI] [PubMed] [Google Scholar]
- 40.Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol 183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wozniak DJ, Ohman DE. 1991. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol 173:1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava D, Hsieh M-L, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamorano-Sánchez D, Fong JCN, Kilic S, Erill I, Yildiz FH. 2015. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J Bacteriol 197:1221–1235. doi: 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro. MVAS, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao F, He Y-W, Wu D-H, Swarup S, Zhang L-H. 2010. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol 192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol 82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 47.Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res 40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers JR, Liao J, Schurr MJ, Sauer K. 2014. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol Microbiol 92:471–487. doi: 10.1111/mmi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez-Gil M, Ramos-González MI, Espinosa-Urgel M. 2014. Roles of cyclic di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol 196:1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.