Abstract
The first International Symposium on c-Di-GMP Signaling in Bacteria (22 to 25 March 2015, Harnack-Haus, Berlin, Germany) brought together 131 molecular microbiologists from 17 countries to discuss recent progress in our knowledge of bacterial nucleotide second messenger signaling. While the focus was on signal input, synthesis, degradation, and the striking diversity of the modes of action of the current second messenger paradigm, i.e., cyclic di-GMP (c-di-GMP), “classics” like cAMP and (p)ppGpp were also presented, in novel facets, and more recent “newcomers,” such as c-di-AMP and c-AMP-GMP, made an impressive appearance. A number of clear trends emerged during the 30 talks, on the 71 posters, and in the lively discussions, including (i) c-di-GMP control of the activities of various ATPases and phosphorylation cascades, (ii) extensive cross talk between c-di-GMP and other nucleotide second messenger signaling pathways, and (iii) a stunning number of novel effectors for nucleotide second messengers that surprisingly include some long-known master regulators of developmental pathways. Overall, the conference made it amply clear that second messenger signaling is currently one of the most dynamic fields within molecular microbiology, with major impacts in research fields ranging from human health to microbial ecology.
INTRODUCTION
The first signaling nucleotide discovered was cyclic 3′-5′-adenosine phosphate (cAMP), which was shown to mediate hormone-induced changes in the metabolism of eukaryotic cells (1, 2). For his pionieering work on cellular signaling processes and the action of hormones performed in the late 1950s, Earl W. Sutherland was awarded the Nobel Prize in Physiology or Medicine in 1971. He and his colleague Richard S. Makman were also the first to report the production of cAMP in Escherichia coli in 1965 (3). Shortly afterwards, Agnes Ullmann and Jacques Monod linked cAMP signaling to catabolite repression in E. coli (4), i.e., a phenomenon which allows E. coli and other bacteria to utilize glucose preferentially before other sugar substrates (5). Almost in parallel, “magic spot” or guanosine-(penta)tetraphosphate [(p)ppGpp] was discovered, an alarmone which accumulates as growth rate decreases in E. coli (6) and that gained lasting attention as a small molecule that directly modulates the activities of the bacterial RNA polymerase (RNAP) (7, 8). Both cAMP and (p)ppGpp globally affect transcription, with cAMP in enteric bacteria having emerged as the textbook paradigm for second messenger signaling in bacteria, which for quite some time was considered a (nearly) closed case.
In 1987, bis-(3′,5′)-cyclic diguanosine monophosphate (c-di-GMP) was reported as an allosteric activator for cellulose synthase by Moshe Benziman and his coworkers (9), who had already found a connection to proteins with GGDEF/EAL domains. However, c-di-GMP remained a topic for the adept connoisseur until the first decade of this century, when GGDEF domain proteins were unequivocally demonstrated to have diguanylate cyclase (DGC) activity (10) and EAL and HD-GYP domain proteins were recognized as two distinct families of c-di-GMP-specific phosphodiesterases (PDEs) (11–13). In parallel, bacterial genome sequences began to fill up databases, and it was realized that nearly all bacteria have GGDEF, EAL, and HD-GYP domain proteins, with many encoding multiples of these enzymes (14, 15). It was, above all, the ubiquitous occurrence and this striking multiplicity of c-di-GMP signaling-related enzymes—which raised puzzling questions of specificity of signaling in cells that might contain dozens of different enzymes that make and break c-di-GMP (16)—that brought second messenger signaling back onto the research agenda of molecular microbiology.
Since then, the number of publications on c-di-GMP signaling has literally exploded. After several years of exponential growth, the publication pace has recently slowed down a bit; the “low-hanging fruit” seem to have been picked, with recent studies now going into striking mechanistic detail of signaling complexes and pathways. In parallel, reports on novel second messengers such as c-di-AMP are now coming up at a high rate. There may well be several hundred research groups worldwide who are now engaged in bacterial second messenger research, but this community had never exchanged their findings and views at a dedicated conference. This specific opportunity was provided by the International Symposium on c-Di-GMP Signaling in Bacteria, which was organized by Regine Hengge (Humboldt-Universität zu Berlin) in Berlin in March 2015. The meeting covered the entire range from signaling input via sensory domains as well as structures and functions of DGCs, PDEs, and c-di-GMP binding effectors, to the molecular and physiological functions regulated by c-di-GMP in diverse bacteria (Fig. 1). Moreover, the symposium's focus on c-di-GMP was intended to also provide a blueprint to revisit the classics [cAMP and (p)ppGpp] and to push ahead research on recently discovered novel second messengers, such as c-di-AMP and others.
FIG 1.
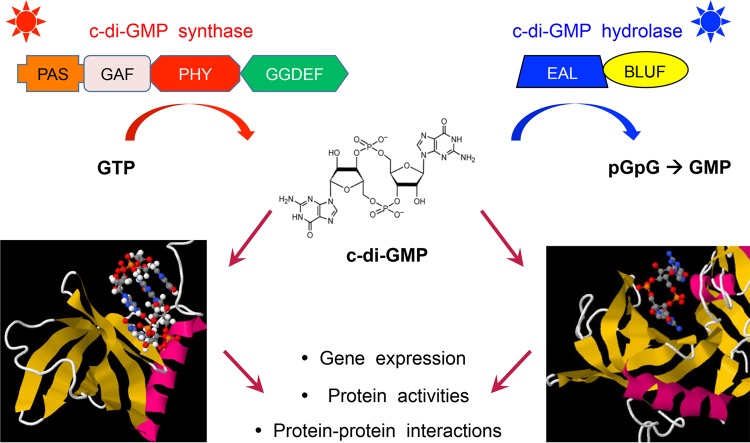
General scheme of production, degradation, mechanism of action, and physiological target processes of the second messenger c-di-GMP. The figure introduces current concepts of c-di-GMP signaling and serves as a guideline, rather than reflecting all the complexities of the many systems currently under study. For instance, while many DGCs and PDEs are membrane attached via their N-terminal sensory domains, others are soluble in the cytoplasm. For further details, see the text.
This report focuses on oral presentations given during the conference, with only a few of the 71 poster presentations mentioned. We attempted to provide here the relevant published literature wherever possible (articles published up to 15 May 2015). Unreferenced statements refer to unpublished results presented at the conference.
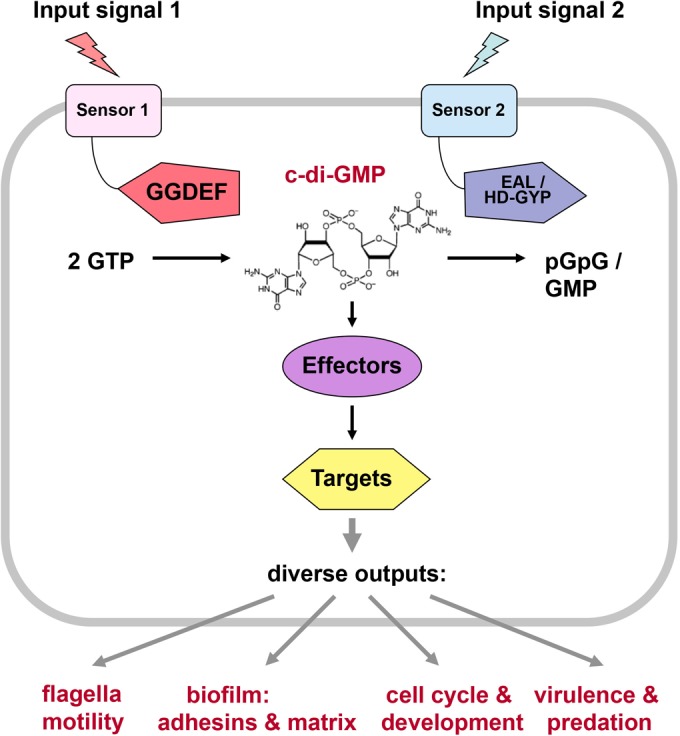
SENSORY INPUT INTO THE CONTROL OF DGC AND PDE ACTIVITIES
The activities of a majority of GGDEF/EAL/HD-GYP domain proteins are controlled by N-terminal sensor domains, which are often integrated in the cytoplasmic membrane and may contain periplasmic loops that bind small ligands or other periplasmic proteins. While some of these domains seem to be specific for c-di-GMP signaling, others are more widespread and also occur in other types of signaling proteins, such as histidine sensor kinases (e.g., PAS, GAF, CHASE, MASE, HAMP, or the light-sensing BLUF, LOV, and bacteriophytochrome domains).
For many of these proteins, the nature of the sensory input signal has remained unknown. Based on work with a fluorescence resonance energy transfer-based c-di-GMP sensor in live cells, Sam Miller (University of Washington, Seattle) reported that not only are c-di-GMP levels heterogeneous in populations of Salmonella enterica serovar Typhimurium (in liquid culture as well as inside host cells) but also many small molecules exert a positive or negative effect on c-di-GMP levels. In particular, micromolar concentrations of arginine were found to activate the Salmonella sp. strain DGC STM1987 via a periplasmic arginine-binding protein that may interact with the large periplasmic domain of the DGC. Andrew Lovering (University of Birmingham) presented a 2.45-Å resolution crystal structure of Bd1971, an EAL domain protein with an N-terminal cAMP-binding domain from Bdellovibrio bacteriovorus, a predator that replicates in the periplasm of prey bacteria such as E. coli. The N-terminal cNMP domain is a homolog of the cAMP-binding domain of the archetypical E. coli cAMP receptor protein (CRP). Mutants lacking this cNMP-EAL protein overproduce extracellular matrix material and produce biofilm-like clumps, a reaction that the wild type may suppress when Bdellovibrio grows inside the host periplasm (Liz Sockett, University of Nottingham) (see below).
A novel membrane-integrated sensory domain with a periplasmic loop region that is specifically found only in EAL domain proteins, the redox-regulated CSS domain, was introduced by Regine Hengge (Humboldt-Universität zu Berlin). E. coli has five PDEs of this CSS-EAL type, which are all expressed but inactive due to disulfide bond formation (between the cysteine of the CSS motif and a second highly conserved cysteine) under standard aerobic growth conditions. Reduction of the CSS domain or mutations that eliminate the relevant cysteine residues result in high PDE activity that can interfere with the production of curli fibers and cellulose, i.e., biofilm matrix components, which in E. coli are a major target of c-di-GMP signaling. As reported by Nicole Frankenberg-Dinkel (Universität Kaiserslautern), biofilm dispersal in Pseudomonas aeruginosa is induced by nitric oxide (NO) via two PDEs, MucR and NbdA, that both feature N-terminal MHYT domains followed by GGDEF and EAL domains (17). Biofilm dispersal-inducing NO concentrations not only stimulate the PDE activities of MucR and NbdA but also activate transcription of nbdA. Intriguingly, very high NO concentrations promote adhesion, i.e., generate the opposite response, possibly by activating the DGC activity of the GGDEF domain(s) of one or both of these composite GGDEF/EAL proteins.
A variety of GGDEF/EAL domain proteins have been shown to be controlled by light, ranging from blue to far red (sensed, e.g., by BLUF, LOV, bacterial phytochrome, or other light receptors) (18–21). As an example of a multidomain light-sensitive GGDEF/EAL protein, Veronika Angerer (from the group of Annegret Wilde, Universität Freiburg) presented Cph2, a Synechocystis sp. protein with a GAF-GAF-GGDEF-EAL-CBCR-GGDEF domain arrangement that, via its blue light-sensing CBCR domain, which stimulates DGC activity of the C-terminal GGDEF domain, suppresses motility and phototaxis while stimulating sessility (22). In addition, the Cph2 EAL domain shows PDE activity, which may be controlled by the degenerate first GGDEF domain and/or the N-terminal GAF domains. As illustrated by Mark Gomelsky (University of Wyoming), light-activated DGCs and PDEs also have considerable potential for synthetic biology, in particular for generating orthogonal optogenetic switches (23). Thus, c-di-GMP accumulation can be triggered by a light-controlled system in engineered cells and can then induce a desired output under the control of a c-di-GMP-activated protein, e.g., a transcription factor, with all these steps based on a modular assembly of bacterial components. As examples, Mark presented a far-red light-sensitive synthetic DGC composed of the phytochrome domain of Rhodobacter sphaeroides BphG1 and a GGDEF domain of Synechocystis sp. Slr1143, as well as a blue light-responsive PDE derived from a multidomain Magnetococcus marinus protein (Fig. 2). Combining these with a c-di-GMP-responsive transcription factor that controls expression of a reporter gene yielded adjustable robust c-di-GMP-mediated switches, which function in bacterial cells as well as in otherwise c-di-GMP-free animal cells. Bacterial phytochromes seem especially well suited for this purpose, since animal cells provide the necessary cofactor biliverdin and far-red light penetrates deeply into mammalian tissues.
FIG 2.
Light-controlled regulation of c-di-GMP levels and c-di-GMP-dependent processes by light-activated enzymes for synthetic biology. For details, see the text. The figure was kindly provided by M. Gomelsky.
NOVEL c-DI-GMP-BINDING EFFECTORS
One of the most astonishing facets of c-di-GMP signaling is the unprecedented diversity of c-di-GMP-sensing effectors or receptors, which then directly affect output reactions of numerous target molecules or larger cellular structures. In principle, either RNA (riboswitches) or proteins can bind c-di-GMP, with protein families that are capable of doing so including several types of transcription factors and PilZ domains, as well as enzymatically inactive degenerate GGDEF (with intact I-site) and EAL domain proteins (24–26). Michael Galperin (NCBI, NIH) provided an overview of the diversity of these c-di-GMP-binding proteins and pointed out that these can bind c-di-GMP monomers in an extended conformation (EAL domain) or as bent/stacked c-di-GMP monomers/dimers and that only a limited number of residues in these effector proteins is critical for c-di-GMP binding, e.g., an arginine in a flexible loop that is part of a binding pocket for bent c-di-GMP.
Holger Sondermann (Cornell University) reported the latest news on an inside-out transmembrane signaling module, which in Pseudomonas fluorescens and Pseudomonas putida regulates biofilm-related cell adhesion posttranscriptionally (27, 28). Here, a transmembrane c-di-GMP receptor (LapD) binds c-di-GMP through its cytoplasmic, enzymatically inactive EAL domain and transmits the signal via a HAMP domain through the inner membrane to its periplasmic PAS domain. In the c-di-GMP-bound state, the receptor inhibits the activity of a periplasmic protease (LapG), which results in retention of an adhesin (LapA) in the outer membrane and thus biofilm formation (29, 30). While the core signaling components LapD and LapG are conserved in several proteobacteria, including Pseudomonas aeruginosa, no substrate or physiological relevance has been previously reported for this species. Based on recent work from his collaboration with the O'Toole group (Dartmouth University), Holger reported that the c-di-GMP-regulated adhesin CdrA, which enhances biofilm formation, is the target of the protease in P. aeruginosa.
The identification of protein partners for the PilZ proteins in P. aeruginosa was reported by Zhao-Xun Liang (Nanyang Technological University Singapore). Studies by the Liang group have revealed that two of the PilZ proteins are likely to bind to a histidine kinase and a chemotaxis methyltransferase, with the interaction between the PilZs and their partners being enhanced by high c-di-GMP concentrations. The two PilZ proteins control motility and biofilm formation through distinct phosphorelay pathways. These studies expand our understanding of the mechanisms of action of PilZ domain c-di-GMP receptors and suggest that, as versatile adaptor proteins, they target different mechanisms (i.e., the flagellar basal body complex, enzymatic activities, phosphorelays) to control diverse outputs.
In P. aeruginosa, c-di-GMP also regulates biofilm formation by binding to the transcriptional regulator FleQ, a member of the large family of AAA+ ATPase proteins, which then activates expression of biofilm-related genes, including genes for Pel and Psl exopolysaccharides, and represses flagellum gene expression (31, 32). Caroline Harwood and coworkers (University of Washington, Seattle) have investigated the FleQ–c-di-GMP interaction and reported the FleQ structure and conformational and functional consequences of c-di-GMP binding to FleQ. It was found that c-di-GMP binds at a site distinct from the ATP-binding site, which induces global conformational changes in FleQ, thereby impacting interactions of FleQ with promoters of biofilm and flagellar genes. Intriguingly, c-di-GMP-mediated repression of flagellar genes depends on the inhibition of the AAA+ ATPase activity of FleQ, which is required at the corresponding σ54-activated flagellar promoters, whereas the c-di-GMP-bound form of FleQ can activate expression of biofilm-related genes in conjunction with vegetative σ70-RNAP.
Vincent Lee (University of Maryland, College Park) described the many applications of the differential radial capillary action of ligand assay (DRaCALA) (33), which allows investigators to directly visualize the binding of c-di-GMP to a protein in a manner that can also be used for quantitative analysis. For instance, this assay allowed Vincent and his group to identify ebselen as a small-molecule inhibitor that binds to the I-site of diguanylate cyclases (34). It was also used to identify novel c-di-GMP-binding proteins from a Vibrio cholerae open reading frame (ORF) collection. One of these novel c-di-GMP effectors is MshE, which is predicted to be a polymerizing ATPase involved in production of the mannose-sensitive hemagglutinin (MSHA), a surface pilus of V. cholerae. He reported that the N-terminal domain of the protein and not the AAA+ ATPase domain is involved in binding c-di-GMP. Fitnat Yildiz (University of California, Santa Cruz) also reported the identification of MshE as a c-di-GMP receptor and showed that MSHA production is enhanced at high c-di-GMP levels (see below).
Novel c-di-GMP effectors have also been identified in several groups of bacteria by affinity pulldown assays using cyclic di-GMP-coupled magnetic beads or a c-di-GMP-specific capture compound (35). Robert Ryan (University of Dundee) reported the identification of c-di-GMP-binding proteins in the plant pathogen Xanthomonas campestris pv. campestris. Of particular interest was XC_3703, which encodes a member of the YajQ family of bacterial proteins that has nucleotide-binding motifs (36). XC_3703 was shown to interact with a transcriptional regulator of the LysR family, thereby impacting its DNA-binding properties and expression of virulence genes. YajQ homologs from P. aeruginosa and Stenotrophomonas maltophilia were also found to bind c-di-GMP (and to be involved in virulence regulation), whereas the YajQ proteins of E. coli and several Gram-positive organisms bind ATP and GTP rather than any cyclic mono- or dinucleotides (37). On a poster that won one of two poster prizes (sponsored by Nature Review Microbiology), Kathrin Sprecher from the Jenal group presented a novel c-di-GMP-binding protein in Caulobacter crescentus that belongs to a family of acetyltransferases. This protein is required for surface adhesion and seems to influence the chemical properties of the holdfast adhesin in a c-di-GMP-controlled manner. Surprisingly, the protein dynamically repositions between the cell membrane and the cytoplasm in response to c-di-GMP binding, arguing that its localization contributes to holdfast control.
Some bacteria make extensive use of c-di-GMP-responsive riboswitches that seem to come in two types (38–41). An example presented by Olga Soutourina (Institut Pasteur) is the emergent enteric pathogen Clostridium difficile, which features 16 riboswitches that are all expressed and respond to c-di-GMP, controlled by 37 c-di-GMP turnover enzymes (42). One of the 12 type I riboswitches shuts down the expression of the major flagellar operon, whereas four activating type II riboswitches promote adhesin synthesis, colonization, and biofilm formation.
c-DI-GMP CONTROL OF MOTILITY, ADHERENCE, EXTRACELLULAR MATRIX PRODUCTION, AND BIOFILM ARCHITECTURE
Perhaps the most prominent function of c-di-GMP signaling is often referred to as an “inverse control of bacterial lifestyles” (43, 44). More precisely, c-di-GMP generally downregulates either the production or activity of flagella and activates the synthesis of adhesins and extracellular matrix components that are important for biofilm formation, i.e., a sessile multicellular lifestyle on surfaces. However, the handy formula that “c-di-GMP inhibits motility and promotes biofilm formation” is a misleading oversimplification, since flagella also play distinct roles during biofilm formation, which can be of a structural nature rather than motility related (45–47), and certain species produce extracellular matrix components also in liquid cultures (e.g., during stationary phase), where these components can induce clumping and sedimentation of cells (48). A directly related c-di-GMP-controlled feature that only recently has gained attention is the high structural organization of biofilms, which becomes especially apparent in macrocolonies. These are large bacterial colonies (grown for extended times on agar plates with complex media) that represent biofilms at the surface of decaying organic material. As a consequence of cellular crowding in combination with a matrix-dependent tissue-like cohesiveness and elasticity, these macrocolonies can buckle into complex three-dimensional structures consisting of intertwined wrinkles, long and high ridges, and/or concentric rings (46, 49–51). Regine Hengge (Humboldt-Universität zu Berlin) showed that this type of biofilm of E. coli contains matrix-producing and matrix-free, yet flagellum-producing, cells in distinct zones and layers, with this architecture in principle translating the temporal succession from growth into stationary phase as observed in a planktonic culture into a spatial pattern in the biofilm (in E. coli, the synthesis of a matrix consisting of amyloid curli fibers and cellulose depends on the stationary-phase sigma factor RpoS) (52). In transition zones, short-range heterogeneity of matrix production can be observed, i.e., cells generating matrix components, or not, are located right next to each other (Fig. 3). The molecular basis of heterogeneity is a switch mechanism that uses the trigger enzyme and PDE YciR (PdeR), which acts not only as a directly binding antagonist of the DGC YdaM (DgcM) and the transcription factor MlrA but also as a sensor for c-di-GMP (a novel systematic nomenclature system for the DGCs and PDEs and their genes in E. coli is introduced in an accompanying article by Hengge et al. in this Journal of Bacteriology issue [96]; since the Y designations, e.g., YdaM, were still used in the presentations at the conference, we also use them in this meeting report but mention the new designations here in parentheses). Under standard conditions, the cellular c-di-GMP level is determined by the balance of the PDE YhjH (PdeH) and the DGC YegE (DgcE), which are shut down and induced, respectively, during entry into stationary phase (44). At high c-di-GMP levels, YciR binds and degrades c-di-GMP, resulting in a release and activation of YdaM, which then also synthesizes c-di-GMP, thereby providing for positive feedback. In parallel, YdaM acts as a coactivator for MlrA, with the YdaM/MlrA complex then activating the expression of CsgD, i.e., the transcription factor required for curli and cellulose production (53). That this complex switch mechanism has the potential to generate bistability was shown on the accompanying posters by Diego O. Serra (from the Hengge group at Humboldt-Universität zu Berlin) and Kaveh P. Yousef (from the group of Max von Kleist at the Freie Universität Berlin), who presented a genetic/microscopic analysis and a mathematical model, respectively.
FIG 3.
Heterogeneous extracellular matrix production at the surface of a macrocolony biofilm of E. coli K-12. The scanning electron micrograph shows an area behind the outer growth zone of the macrocolony of strain W3110. Only a subset of cells (arranged in small chains) are surrounded by a matrix of amyloid curli fibers, whose synthesis depends on high cellular levels of c-di-GMP. The image was previously published in this journal, as part of a larger figure (51).
While the control of curli and cellulose synthesis by CsgD is essentially the same in E. coli and Salmonella, c-di-GMP signaling input is different, since the complements of GGDEF/EAL domain proteins only partially overlap. In her talk, Ute Römling (Karolinska Institutet) focused on two degenerate stand-alone EAL domain proteins in Salmonella, STM1344 and STM1697 (E. coli has only one of this type of proteins, the STM1344 homolog YdiV). By inhibiting the activity and promoting the proteolysis of the flagellar master regulator FlhDC and thereby also reducing the expression of the PDE STM3611 (YhjH in E. coli), which is expressed from a flagellar class 3 promoter, both STM1344 and STM1697 indirectly contribute to c-di-GMP signaling (54, 55). As a consequence, and rather unusual for EAL domain proteins, they in fact promote CsgD expression and thereby biofilm formation. Moreover, these two degenerate EAL domain proteins provide a link between biofilm formation and virulence, with mutants being affected in spleen colonization. STM1697 was also previously shown to suppress acute virulence phenotypes, such as invasion into epithelial cells and secretion of the type III secretion system effector protein SipA (55).
Fitnat Yildiz (University of California, Santa Cruz) discussed how c-di-GMP mechanistically controls the planktonic-to-biofilm switch in Vibrio cholerae, the causative agent of cholera. Mutations in genes involved in MSHA biogenesis were identified as suppressors of the decreased motility phenotype of a V. cholerae mutant that lacked the phosphodiesterase CdgJ, showing that the ATPase proteins MshE and PilT, responsible for polymerizing and depolymerizing MshA pili, impair near-surface motility. Intriguingly, Fitnat showed that MshE, but not PilT, binds c-di-GMP directly, establishing a mechanism for positive c-di-GMP signaling input in MshA pilus production. She also explained her recent collaborative work with Gerard Wong (University of California, Los Angeles) in developing a hydrodynamic model to visually examine different types of motility behavior of V. cholerae, which indicates that MshA pili are crucial for surface selection, irreversible attachment, and ultimately microcolony formation (56).
Cellular aggregation and biofilm formation can also be stimulated if pGpG, the primary product of c-di-GMP cleavage by PDEs, is not further degraded and therefore accumulates and inhibits, via feedback, PDE activity. This was shown by Mona Orr (from Vincent Lee's group), who reported the identification of the oligoribonuclease Orn as the primary enzyme that further degrades pGpG to GMP on a poster that was awarded the second poster prize.
c-DI-GMP CONTROL OF VIRULENCE AND PREDATION
Several presentations at the meeting touched on the role of c-di-GMP in modulating virulence and persistence behaviors in bacteria. Susanne Häussler (Helmholtz Center for Infection Research, Braunschweig, Germany) pointed out the lack of knowledge surrounding the general mechanisms of surface perception and signal transduction to initiate bacterial adherence during host infection. She explained how her group has begun to use a genomics approach to examine clinical small-colony variant (SCV) strains of Pseudomonas aeruginosa to identify adaptive mutations that correlate autoaggregative growth behavior and an enhanced capacity to form biofilms. This led to the identification of a point mutation in the 5′ untranslated region of the accBC gene cluster (encoding subunits of acetyl coenzyme A carboxylase) that is responsible for a stabilized mRNA structure and that results in decreased fatty acid chain lengths. Susanne provided genetic evidence to suggest that changes of the membrane fatty acid composition may serve as a signal for a chemosensory-like surface sensing system (the Wsp system) to produce constitutively elevated levels of c-di-GMP and thus to play a key role in the regulation of adhesion-induced bacterial responses. This may be the first example of a wider range of genetic mechanisms by which bacteria adapt to the mammalian host environment through an influence on c-di-GMP signaling. A poster presentation by Yi-Cheng Sun (Peking Union Medical College) provided another example of how c-di-GMP controls bacterial behavior during infection. Yi-Cheng described how Yersinia pestis, the causal agent of plague, uses the Rcs phosphorelay to regulate biofilm development during flea vector transmission (57). This occurs through tight control of the expression of hmsT and hmsD, which encode diguanylate cyclases. Differential regulation of hmsT and hmsD by Rcs may allow the organism to adapt to different host environments.
Since conventional antimicrobials cannot efficiently eradicate biofilms, such as in the examples above, there is an urgent need to develop alternative measures to combat biofilm-related infections. On her poster, Julie Groizeleau from the laboratory of Tim Tolker-Nielsen (University of Copenhagen) provided an elegant proof of concept in a study showing that modulation of the intracellular c-di-GMP level by using chemical entities in P. aeruginosa is a viable strategy for biofilm control (58). Although still at an early stage, this may provide the basis for the development of new drugs that can inhibit biofilm formation or facilitate eradication of existing biofilms.
Liz Sockett (University of Nottingham) described her group's work on the pathways by which the predatory bacterium Bdellovibrio bacteriovorus uses c-di-GMP to control prey invasion. c-di-GMP is known to control the switch between predatory and nonpredatory life cycles by controlling several enzymes required for prey invasion and subsequent degradation. Interestingly, Bdellovibrio possesses 7 proteins predicted to be involved in c-di-GMP turnover but more than 15 proteins involved in binding the dinucleotide (Fig. 4). Liz highlighted some previously published work showing that CdgA, a degenerate inactive GGDEF domain protein, localizes at the interaction pole, which initiates contact with the prey and is required for prey invasion (59). Her group recently extended this study to demonstrate that CdgA forms complexes with other proteins, including the Ras-GTPase-like proteins MglA and RomR, which are required for prey invasion. Similar complexes are seen in Myxococcus where, by contrast, they act to control cell polarity and type IV pilus-mediated gliding motility (60).
FIG 4.
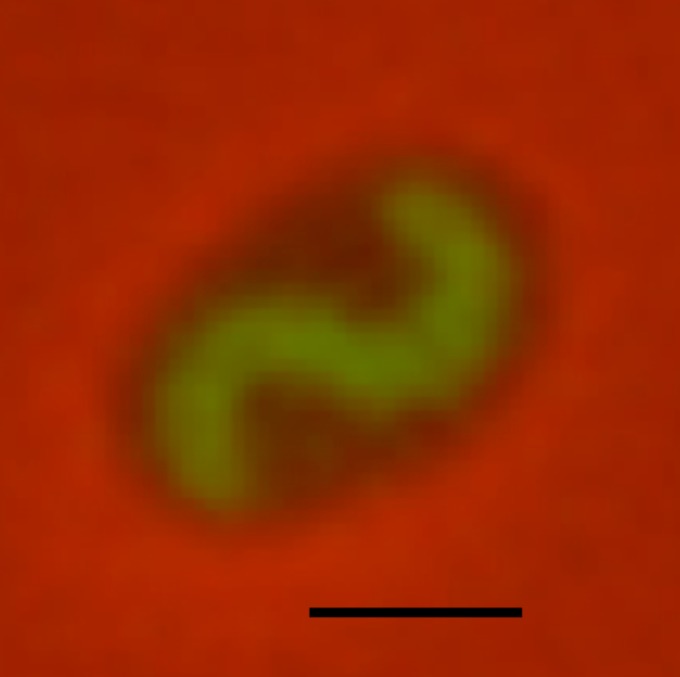
c-di-GMP signaling in Bdellovibrio bacteriovorus. A fluorescently tagged cytoplasmic c-di-GMP-binding protein was expressed in B. bacteriovorus (green) during predatory growth inside an E. coli prey cell (black). Bar, 1 μm. The image was kindly provided by Liz Sockett.
c-DI-GMP CONTROL OF CELL POLARITY AND DEVELOPMENT
c-di-GMP is known to play key roles in cell differentiation and polarity in several model systems of bacterial development. For instance, oscillating levels of c-di-GMP determine much of the bimodal developmental program in Caulobacter crescentus by driving the motile-sessile transition and by organizing cell polarity during asymmetric cell division (61). However, the pivotal role of c-di-GMP appears to go beyond development. In his talk, Urs Jenal (Biozentrum, University of Basel) presented evidence that c-di-GMP is also at the heart of the machinery driving the C. crescentus cell cycle (Fig. 4). He and his coworkers identified the cell cycle kinase CckA as the first member of the large family of sensor histidine kinases that are directly controlled by c-di-GMP. Binding of c-di-GMP to its CA domain switches CckA from its default kinase into the phosphatase mode, thereby licensing S-phase entry through inactivating the replication initiation inhibitor CtrA (62). Through this simple integration, oscillating levels of c-di-GMP effectively coordinate C. crescentus cell cycle progression, with the program determining cell fate. Similar to C. crescentus, P. aeruginosa establishes an asymmetric c-di-GMP distribution during cell division. While work from the Miller lab (University of Washington, Seattle) has suggested that this asymmetry is important for the regulation of cell motility (63), its overall significance is still enigmatic. Benoit Laventie from the Jenal lab introduced FimA, a novel c-di-GMP effector/receptor protein involved in type IV pilus (T4P) function in P. aeruginosa. While FimA and its activation by c-di-GMP is dispensable for T4P-based twitching motility (which requires low c-di-GMP levels and is controlled by the PDE FimX [64]), it is critical for T4P-based surface attachment of P. aeruginosa under conditions of high internal c-di-GMP and biofilm formation. FimA binds c-di-GMP with high affinity and as a result, localizes to poles of the cells, where it presumably regulates some aspects of T4P function.
T4Ps are also required for the development of Myxococcus xanthus. Upon starvation, M. xanthus undergoes a major lifestyle change that culminates in the formation of a spore-filled multicellular fruiting body (65). Lotte Søgaard-Andersen (MPI Marburg) showed that c-di-GMP accumulates in M. xanthus and regulates T4P-based motility and fruiting body formation. During development, cells with reduced c-di-GMP levels failed to aggregate, did not form fruiting bodies, and showed poor sporulation efficiency. Intriguingly, of a plethora of c-di-GMP turnover components encoded by the M. xanthus chromosome, only a few of these proteins seem responsible for the physiological increase of c-di-GMP during development. These findings not only indicate that T4P regulation by c-di-GMP might be a conserved principle but also suggest a fundamental role for c-di-GMP in orchestrating the intricate multicellular developmental program of this organism.
Similarly, c-di-GMP was recently shown to mastermind the development of Streptomyces spp. mycelial growth, sporulation, and production of secondary metabolites. But while c-di-GMP promotes sporulation in M. xanthus, it has the opposite effect in streptomycetes. Increased levels of c-di-GMP delay morphological differentiation until aerial hyphae have been formed, whereas low levels of the second messenger favor premature spore formation in Streptomyces venezuelae. Natalia Tschowri (Humboldt University, Berlin, Germany) presented data showing that c-di-GMP controls Streptomyces development by directly targeting the transcription factor BldD (Fig. 5), one of the key developmental regulators in this group of organisms (66). Initial studies revealed candidate DGCs and PDEs involved in this elementary regulatory node, indicating that c-di-GMP levels are carefully controlled during the developmental cycle.
FIG 5.
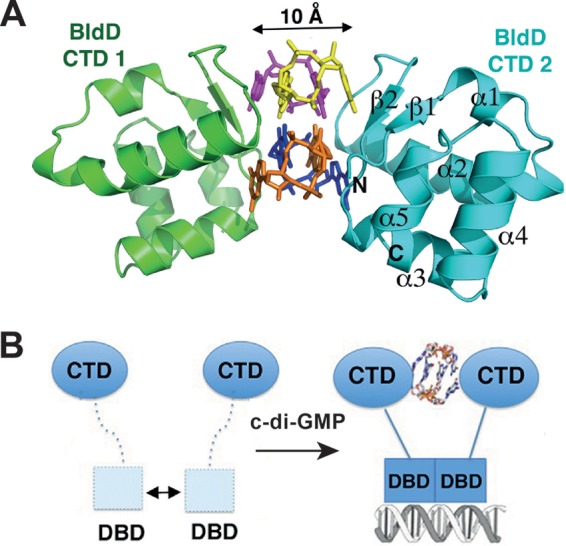
BldD in Streptomyces. A key developmental regulator is regulated by c-di-GMP and reveals a novel mode of c-di-GMP binding. (A) Ribbon diagram of the C-terminal domain (CTD) of BldD in complex with c-di-GMP. The BldD-CTD complex uses a previously unseen mode of c-di-GMP binding in which two noninteracting CTDs separated by 10 Å are glued together by a c-di-GMP tetramer composed of two intercalated c-di-GMP dimers. The four c-di-GMP molecules are shown in different colors. (B) Schematic representation of c-di-GMP-activated BldD binding to DNA. The formation of a c-di-GMP-linked BldD-CTD dimer enables the N-terminal DNA-binding domain (DBD) to optimally dimerize and to effectively bind to target DNA, leading to repression of the BldD regulon. A weak interaction between DBDs in the absence of c-di-GMP is shown by the double-headed arrow. The figure was adapted from reference 66 and is used here with permission from the publisher.
Finally, Pauline Schaap (University of Dundee) expanded the c-di-GMP signaling repertoire to the regulation of development in eukaryotes. Building on their earlier observation that c-di-GMP is required for stalk formation during Dictyostelium discoideum fruiting body formation (Fig. 6) (67), the Schaap group showed that a number of stalk-specific genes are expressed in a c-di-GMP-dependent manner, thereby contributing to the differentiation of precursor cells into highly vacuolated cells that form the stalk structure. c-di-GMP lines up as yet another second messenger involved in this complex social behavior of grazing amoebas. In agreement with cell-cell communication playing a major role in fruiting body formation and sporulation in this organism, c-di-GMP, unlike in the prokaryotic counterparts, seems to have an extracellular role in social amoebas. Intriguingly, the role of c-di-GMP is specifically limited to the differentiation of stalk cells, as earlier and later steps in development are regulated independently of this second messenger. It will be exciting to follow the dissection of this interesting pathway, including its surface-exposed receptor(s) and mechanisms used to limit the c-di-GMP response to a specific cell type of this complex multicellular structure.
FIG 6.
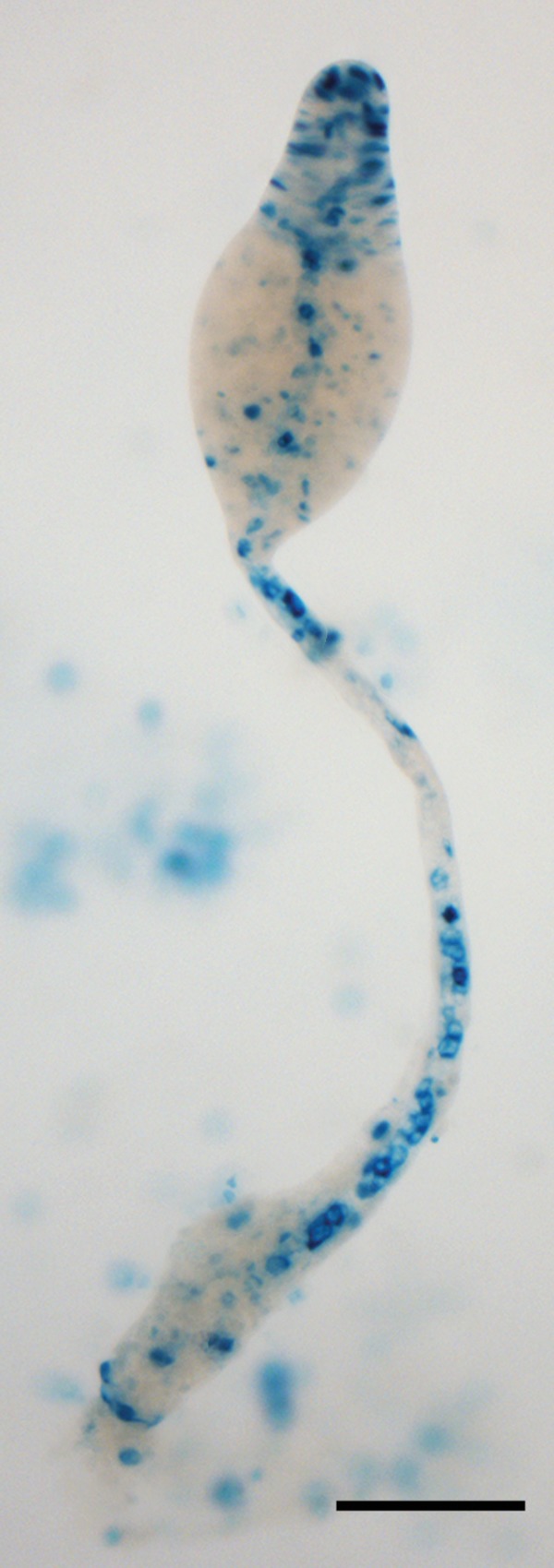
Diguanylate cyclase expression in Dictyostelium. The promoter of the gene encoding the Dictyostelium DgcA was fused to the E. coli lacZ reporter gene and transformed into Dictyostelium cells. Staining of fruiting structures with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) shows that DgcA is expressed in prestalk and stalk cells. Bar, 100 μm. The image was previously published (67) and is reprinted with permission of the publisher.
All of these examples emphasize the role of c-di-GMP in orchestrating multicellularity in bacteria and simple eukaryotes and expand its role to specific developmental processes that exploit complex multicellular interactions. This is reminiscent of the prominent role of c-di-GMP in biofilm formation and points to regulatory links between processes employed by bacteria to colonize surfaces and specific developmental programs that evolved in bacteria to survive harsh environmental conditions.
CLASSICAL BACTERIAL SECOND MESSENGERS REVISITED: cAMP AND ppGpp
cAMP is produced in E. coli by the adenylate cyclase CyaA when glucose is depleted from the medium. Accumulating cAMP will bind to CRP (also known as catabolite gene activator protein, or CAP), which allows this dimeric transcription factor to bind to specific promoter regions, thereby stimulating transcription of a large number of genes, which include many operons encoding enzymes involved in the catabolism of alternative sugars (68). Interestingly, CyaA and CRP are also present in P. putida, a soil bacterium that does not primarily feast on sugars in the environment. Victor de Lorenzo (CNB-CSIC, Madrid, Spain) showed that the cAMP signaling system is functional in P. putida but that the system has been coopted for a new use and instead seems to control the expression of functions involved in cell envelope biogenesis and a range of amino acid transport systems (69). Strikingly, cAMP levels are extremely low in P. putida. This is compensated by a tight binding of cAMP to the corresponding CRP, with a Kd in the low nanomolar range, instead of the micromolar range that is usually observed. Hence, even at a very low cAMP concentration, CRP can bind cAMP in P. putida (70). However, as presented by Victor, the dimeric P. putida CRP binds only one cAMP molecule with high affinity. This anticooperative binding can lead to an activated but “asymmetric” transcription factor that recognizes binding sites in promoter regions which are distinct from the classical CRP-binding sites, leading to the activation of a different set of target genes.
The other “old” signaling nucleotide covered during the conference was (p)ppGpp, which is induced upon amino acid starvation in Gram-negative and Gram-positive bacteria, resulting in specific global transcriptional responses, although the underlying mechanisms are very different (8). In Gram-negative bacteria such as E. coli, regulation of cellular processes by (p)ppGpp is mainly mediated by direct binding to RNAP, whereas, as presented by Jue D. (Jade) Wang (University of Wisconsin, Madison), in Gram-positive bacteria like Bacillus subtilis (p)ppGpp-mediated regulation acts through cellular GTP levels (71). One protein directly targeted by (p)ppGpp in B. subtilis is guanylate kinase (GMK), which is required for the conversion of GMP to GDP, which is subsequently used for GTP production. (p)ppGpp is a competitive inhibitor of GMK, resulting in a decrease in GTP levels under stringent response conditions, when the levels of (p)ppGpp are high. Through a combination of structural, biochemical, and phylogenetic analyses, Jade and her coworkers discovered that the (p)ppGpp-GMK interaction is conserved among Firmicutes, Actinobacteria, and Deinococcus thermus but is not present in Proteobacteria, leading her to postulate that GMK is an ancestral target of (p)ppGpp, with the (p)ppGpp-RNAP interaction having evolved more recently (72). (p)ppGpp not only controls the survival of bacteria under amino acid starvation conditions but also plays a central role in the coordination of cellular processes under a wide range of stress conditions (73). In addition, (p)ppGpp is also involved, along with toxin-antitoxin (TA) modules, in the formation of persister cells in E. coli (74, 75). Kenn Gerdes (University of Copenhagen, Denmark) presented data indicating that the production of (p)ppGpp and slow growth per se are not sufficient to induce E. coli persister cell formation. Thus, when the HipA toxin, which inhibits the glutamyl tRNA synthetase GltX and, via an inhibition of translation, leads to the production of (p)ppGpp and a growth arrest, is induced in the absence of the 10 mRNA endonuclease toxins, this does not lead to persister cell formation. Rather, true persister cells are only formed when increased (p)ppGpp levels inhibit exopolyphosphatase (PPX), which results in polyphosphate-activated Lon protease degrading the antitoxins of the 10 mRNA endonuclease toxins (76).
NEWCOMERS IN BACTERIAL SECOND MESSENGER SIGNALING: c-DI-AMP AND cAMP-GMP
In the final session of the meeting, more-recently discovered newcomer signaling nucleotides were discussed. This area of research started with the discovery of c-di-AMP in a crystallographic study of the DNA integrity scanning protein DisA (77). Both c-di-AMP-binding proteins, which include components of transport systems and enzymes (78–80) as well as a c-di-AMP-binding riboswitch (81, 82), have been identified. During its intracellular existence, Listeria monocytogenes even secretes c-di-AMP into the host cell cytosol, where it activates the type I interferon response (83). Novel insights into the role of c-di-AMP in Staphylococcus aureus were presented by Angelika Gründling (Imperial College, London, United Kingdom). High cellular levels of c-di-AMP in S. aureus can compensate for the lack of the essential cell wall polymer lipoteichoic acid (LTA) (84), although cell wall polymer synthesis on the outside of the cell and nucleotide signaling on the inside of the cell have at first sight not much in common. However, the discovery of the c-di-AMP target proteins KtrA, the cytoplasmic gating component of a potassium transport system (likely corresponding to the KtrC protein in Bacillus subtilis), KdpD, the sensor histidine kinase required under osmotic stress conditions for the expression of the high-affinity potassium transport system Kdp, and CpaA, a predicted cation proton antiporter (79), suggested that the connection between these two processes has a function in osmoregulation. The hypothesis was put forward that an increase in the cellular c-di-AMP concentration leads to a reduction in the cellular turgor pressure, allowing S. aureus cells to survive in the absence of the cell wall polymer LTA, which in Gram-positive bacteria may function in a manner similar to that of the membrane-derived oligosaccharides (MDOs; also called osmoregulated periplasmic glucans) in the periplasm of E. coli, which help to balance the osmolarity within the cell and the cell wall. Jörg Stülke (University of Göttingen) showed the latest development on the c-di-AMP signaling network in B. subtilis. In this organism, c-di-AMP is produced by three diadenylate cyclase (DAC) enzymes, CdaA (or DacA), DisA, and CdaS, and is degraded by the DHH/DHHA1 domain-containing phosphodiesterase GdpP (YybT) and a second predicted phosphodiesterase containing a DH domain with homology to the Listeria monocytogenes enzyme PgpH (85, 86). Jörg highlighted the finding that c-di-AMP is the first signaling nucleotide that directly controls both expression and activity of a target protein, KtrA in B. subtilis, by binding to the ydaO riboswitch that is linked to the ktrA gene as well as to the KtrA protein itself. He also reported the crystal structure of a trimeric c-di-AMP-binding effector protein, DarA, a homolog of the ubiquitous PII signaling proteins which was found in pulldown assays with a biotin-tagged nucleotide (87). Furthermore, it was recognized early on that, in contrast to c-di-GMP, the c-di-AMP signaling network is essential in many bacteria (88, 89), which suggests that DACs may represent a promising drug target.
Ming C. Hammond (University of California, Berkeley) reported on c-di-nucleotide-specific riboswitches linked to the dye-binding “spinach” RNA molecule which act as fluorescent biosensors. These allowed her to investigate cellular c-di-nucleotide levels in bacterial cells and to provide experimental evidence that c-di-AMP is produced by Archaea (90). Next, an artificial riboswitch binding to c-GMP-AMP, a signaling molecule initially discovered in V. cholerae (91), was constructed by introducing a single base change in a naturally occurring c-di-GMP-specific riboswitch. This led to the realization that a large number of such c-GMP-AMP-specific riboswitches (GEMM-I) are present naturally in energy-harvesting Geobacter spp. (92). Moreover, this work led to the discovery of a novel class of c-GMP-AMP cyclase enzymes. Notably, the same riboswitch was discovered independently and presented on a poster by James W. Nelson (from the laboratory of Ronald R. Breaker, Yale University) (93). The meeting was closed by a presentation by Volkhard Kaever, who spoke on the detection and quantification of signaling nucleotides by high-performance liquid chromatography (HPLC)-coupled tandem mass spectrometry (MS/MS) methods. He highlighted the importance of proper sample preparation, in particular for short-lived signaling molecules and discussed the importance of internal isotope-labeled standards for the accurate quantification of signaling nucleotides as well as the use of multiple mass qualifiers to unequivocally identify signaling molecules. He ended on the note that certainly additional nucleotides can be detected in bacterial and eukaryotic cells, and the future will tell if some of these act as novel signaling molecules.
PERSPECTIVES AND TRENDS
The overarching trend that became apparent in a majority of presentations of this conference is the high connectivity of c-di-GMP and other nucleotide second messengers to other regulatory pathways and networks; we are clearly approaching a systems-level perspective on nucleotide second messenger signaling in bacteria, in which global cellular functions such as growth control and the cell cycle become tightly interlinked with differentiation and development.
At the molecular level, diverse ATPases are emerging as being targeted by c-di-GMP, a link that reflects the central roles of both components in regulatory switches controlling many key cellular processes. Several examples were presented at the conference, including the transcription regulator FleQ from P. aeruginosa, a member of the NtrC family harboring an AAA+ ATPase domain. Binding of c-di-GMP to this domain induces a conformational change that interferes with ATP binding and thus controls overall activity of FleQ (Caroline Harwood and Holger Sondermann). Another example is MshE, an AAA+ domain ATPase involved in the assembly of the adhesive MshA pilus at the surface of V. cholerae that was shown to bind c-di-GMP (Fitnat Yildiz and Vincent Lee). Furthermore, histidine sensor kinases of two-component phosphorylation cascades can be subject to direct control by c-di-GMP. For instance, in the case of the bifunctional histidine kinase CckA from C. crescentus, c-di-GMP binds to the CA domain (which binds ATP to promote autophosphorylation) and switches CckA from its default kinase into the phosphatase mode, which is a key event in cell cycle control (Urs Jenal). Another variation of cross talk between c-di-GMP and two-component signaling is realized in the SagS histidine sensor kinase of P. aeruginosa, whose activity is modulated by a c-di-GMP-binding PilZ effector protein (Zhao-Xun Liang).
Another regulatory network-building trend is the emerging cross talk between different nucleotide second messengers. Striking examples are a Bdellovibrio c-di-GMP-degrading PDE controlled by an N-terminal cAMP-binding sensory input domain (Andrew Lovering), or the recent finding that in P. aeruginosa, c-di-GMP and cAMP levels, and therefore biofilm formation and virulence gene expression, are inversely coordinated (94). When S. aureus enters into stationary phase, two signaling nucleotides seem engaged in a positive feedback cycle: c-di-AMP may help to activate the stringent response through an RSH (RelA/SpoT homolog), i.e., lead to an increase in (p)ppGpp, which in turn inhibits a c-di-AMP-degrading phosphodiesterase (Angelika Gründling) (95).
High specificity within these signaling networks is often generated by the formation of large multicomponent complexes containing DGCs, PDEs, and c-di-GMP-binding effector proteins, such as those observed at the different poles of C. crescentus (reported by Urs Jenal) or Bdellovibrio (reported by Liz Sockett). Besides a scaffolding function, protein-protein interactions in such complexes can also assume a direct regulatory role, which, as exemplified by the PDE and “trigger enzyme” YciR (PdeR) in E. coli, may even become the major function that is modulated by binding and degradation of c-di-GMP (Regine Hengge). Within larger complexes, c-di-GMP-binding effectors, such as PilZ domain proteins (Zhao-Xun Liang) or the newly discovered YajQ proteins (Robert Ryan), can act as versatile adaptors that link c-di-GMP signal input to the activity of enzymes, structural components, or transcription factors.
Another theme that became apparent during the symposium is the versatile and rapid evolution of second messenger binding components. In particular, riboswitches apparently can switch specificity rather easily, a property that paves the way for yet another, more technical trend, i.e., the production of new tools, such as fluorescent sensors for the quantification of nucleotide second messengers in live cells and the identification of novel signaling proteins (Ming Hammond). Finally, both riboswitches and also proteins associated with second messenger signaling, in particular those that respond to light, can provide promising novel orthogonal switches for synthetic biology (Mark Gomelsky).
Just a few years ago, nobody would have imagined this complexity and central role of second messengers in the regulatory networks of bacterial cells, but we are quite sure that c-di-GMP as well as its classical and more recently emerging cousins will continue to surprise us. To facilitate this, the Deutsche Forschungsgemeinschaft announced—just 2 days before the beginning of this conference—the establishment of a novel Priority Programme (Nucleotide Second Messenger Signaling in Bacteria; SPP 1879), which will not only provide funding for research over the next 6 years to German research groups but will also allow continued organization of biannual international symposia on bacterial nucleotide second messenger signaling in the future.
ACKNOWLEDGMENTS
We thank all participants of the conference for their contributions, those participants whose presentations are mentioned here for their feedback, and our colleagues Mark Gomelsky, Liz Sockett, Natalia Tschowri, and Pauline Schaap for providing figures for this meeting report.
Financial support for the symposium was provided by the European Research Council under the European Union's Seventh Framework Programme (ERC-AdG 249780 to R.H.) and the Deutsche Forschungsgemeinschaft.
We declare that we do not have any conflicts of interest.
REFERENCES
- 1.Rall TW, Sutherland EW. 1958. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem 232:1065–1076. [PubMed] [Google Scholar]
- 2.Sutherland EW, Rall TW. 1958. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem 232:1077–1091. [PubMed] [Google Scholar]
- 3.Makman RS, Sutherland EW. 1965. Adenosine 3′,5′-phosphate in Escherichia coli. J Biol Chem 240:1309–1314. [PubMed] [Google Scholar]
- 4.Ullmann A, Monod J. 1968. Cyclic AMP as an antagonist of catabolite repression in Escherichia coli. FEBS Lett 2:57–60. doi: 10.1016/0014-5793(68)80100-0. [DOI] [PubMed] [Google Scholar]
- 5.Brückner R, Titgemeyer F. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett 209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 6.Cashel M, Gallant J. 1969. Two compounds implicated in the funciton of the RC gene of Escherichia coli. Nature 221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 7.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 8.Liu K, Bittner AN, Wang JD. 2015. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylate. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 10.Paul R, Weiser S, Amiot N, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 12.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He Y-W, Zhang L-H, Heeb S, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 15.Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 16.Hengge R. 2009. Principles of cyclic-di-GMP signaling. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 18:1015–1018. [DOI] [PubMed] [Google Scholar]
- 19.Gomelsky M, Hoff WD. 2011. Light helps bacteria make important lifestyle decisions. Trends Microbiol 19:441–448. doi: 10.1016/j.tim.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 21.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue light response of E. coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savakis P, de Causmaecker S, Angerer V, Ruppert U, Anders K, Essen L-O, Wilde A. 2012. Light-induced alteration of c-di-GMP levels controls motility of Synechocystis sp. PCC 6803. Mol Microbiol 85:239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 23.Ryu MH, Gomelsky M. 2014. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol 3:802–810. doi: 10.1021/sb400182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengge R. 2010. Cyclic-di-GMP reaches out into the bacterial RNA world. Sci Signal 3:pe44. doi: 10.1126/scisignal.3149pe44. [DOI] [PubMed] [Google Scholar]
- 25.Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci 21:929–948. doi: 10.1002/pro.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan RP, Tolker-Nielsen T, Dow JM. 2012. When the PilZ don't work: effectors for c-di-GMP action in bacteria. Trends Microbiol 20:235–242. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 29.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O'Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife 3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newell PD, Boyd CD, Sondermann H, O′Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A 108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebermann OJ, Orr MW, Wang Y, Lee VT. 2014. High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem Biol 9:183–192. doi: 10.1021/cb400485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. 2012. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics 75:4874–4878. doi: 10.1016/j.jprot.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 36.Saveanu C, Miron S, Borza T, Craescu CT, Labesse G, Gagyi C, Popescu A, Schaeffer F, Namane A, Laurent-Winter C, Barzu O, Gilles AM. 2002. Structural and nucleotide-binding properties of YajQ and YnaF, two Escherichia coli proteins of unknown function. Protein Sci 11:2551–2560. doi: 10.1110/ps.0217502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An S-Q, Caly DL, McCarthy Y, Murdoch SL, Ward J, Febrer M, Dow JM, Ryan RP. 2014. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog 10:e1004429. doi: 10.1371/journal.ppat.1004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulshina N, Baird NJ, Ferré-D'Amaré AR. 2009. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol 16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soutourina O, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguénec C, Coppée J-Y, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficila. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 44.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 46.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4(2):e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signaling within the σS network of Escherichia coli. Mol Microbiol 62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 49.Römling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci 62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okegbe C, Price-Whelan A, Dietrich LE. 2014. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol 18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serra DO, Hengge R. 2014. Stress responses go three-dimensional: the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ Microbiol 16:1455–1471. doi: 10.1111/1462-2920.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain phosphodiesterase YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simm R, Remminghorst U, Ahmad I, Zakikhany K, Römling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol 191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, Anwar N, Chuah ML, Lünsdorf H, Frank R, Rhen M, Liang ZX, Lindqvist Y, Römling U. 2013. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella typhimurium. Mol Microbiol 90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- 56.Utada AS, Bennett RR, Fong JC, Gibiansky ML, Yildiz FH, Golestanian R, Wong GC. 2014. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun 5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo XP, Ren GX, Zhu H, Mao XJ, Sun YC. 2015. Differential regulation of the hmsCDE operon in Yersinia pestis and Yersinia pseudotuberculosis by the Rcs phosphorely system. Sci Rep 5:8412. doi: 10.1038/srep08412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christensen LD, van Gennip M, Rybtke MT, Wu HY, Chiang WC, Alhede M, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic-di-GMP level in the bacteria. Infect Immun 81:2705–2713. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milner DS, Till R, Cadby I, Lovering AL, Basford SM, Saxon EB, Liddell S, Williams LE, Sockett RE. 2014. Ras GTPase-like protein MglA, a controller of bacterial social motility in myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Genet 10:e1004253. doi: 10.1371/journal.pgen.1004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 63.Kulasekara BR, Kamischke C, Kulasakara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain R, Behrens AJ, Kaever V, Kazmierczak BI. 2012. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J Bacteriol 194:4285–4294. doi: 10.1128/JB.00803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 66.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen ZH, Schaap P. 2012. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci U S A 102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milanesio P, Arce-Rodríguez A, Munoz A, Calles B, de Lorenzo V. 2011. Regulatory exaptation of the catabolite repression protein (Crp)-cAMP system in Pseudomonas putida. Environ Microbiol 13:324–339. doi: 10.1111/j.1462-2920.2010.02331.x. [DOI] [PubMed] [Google Scholar]
- 70.Arce-Rodríguez A, Durante-Rodríguez G, Platero R, Krell T, Calles B, de Lorenzo V. 2012. The Crp regulator of Pseudomonas putida: evidence of an unusually high affinity for its physiological effector, cAMP. Environ Microbiol 14:702–713. doi: 10.1111/j.1462-2920.2011.02622.x. [DOI] [PubMed] [Google Scholar]
- 71.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. 2015. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell 57:735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaca AO, Colomer-Winter C, Lemos JA. 2015. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 75.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 24:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 76.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Witte G, Hartung S, Büttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 78.Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 79.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. 2014. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao A, Serganov A. 2014. Sructural insights into the recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol 10:787–792. doi: 10.1038/nchembio.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren A, Patel DJ. 2014. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol 10:780–786. doi: 10.1038/nchembio.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. 2015. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A 112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mehne FM, Schröder-Tittmann K, Eijlander RT, Herzberg C, Hewitt L, Kaever V, Lewis RJ, Kuipers OP, Tittmann K, Stülke J. 2014. Control of the diadenylate cyclase CdaS in Bacillus subtilis: an autoinhibitory domain limits cyclic di-AMP production. J Biol Chem 289:21098–21107. doi: 10.1074/jbc.M114.562066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gundlach J, Dickmanns A, Schröder-Tittmann K, Neumann P, Kaesler J, Kampf J, Herzberg C, Hammer E, Schwede F, Kaever V, Tittmann K, Stülke J, Ficner R. 2015. Identification, characterization, and structure analysis of the cyclic di-AMP-binding PII-like signal transduction protein DarA. J Biol Chem 290:3069–3080. doi: 10.1074/jbc.M114.619619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. 2013. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high-level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Commichau FM, Pietack N, Stülke J. 2013. Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol Biosyst 9:1068–1075. doi: 10.1039/c3mb25595f. [DOI] [PubMed] [Google Scholar]
- 90.Kellenberger CA, Chen C, Whiteley AT, Portnoy DA, Hammond MC. 2015. RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP. J Am Chem Soc 137:6432–6435. doi: 10.1021/jacs.5b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kellenberger CA, Wilson SC, Hickey SF, Gonzalez TL, Su Y, Hallberg ZF, Brewer TF, Iavarone AT, Carlson HK, Hsieh YF, Hammond MC. 2015. GEMM-1 riboswitches from Geobacter sense the bacterial second messenger c-AMP-GMP. Proc Natl Acad Sci U S A 112:5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nelson JW, Sudarsan N, Phillips GE, Stav S, Lünse CE, McCown PJ, Breaker RR. 2015. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci U S A 112:5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Almblad H, Harrison JJ, Rybtke M, Groizeleau J, Givskov M, Parsek MR, Tolker-Nielsen T. 2015. The cAMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by c-di-GMP. J Bacteriol 197:2190–2200. doi: 10.1128/JB.00193-15 (Erratum, 197: 2731, doi:.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corrigan RM, Bowman L, Willis AR, Kaever V, Gründling A. 2015. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J Biol Chem 290:5826–5839. doi: 10.1074/jbc.M114.598300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hengge R, Galperin MY, Ghigo J-M, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2016. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J Bacteriol 198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]