Abstract
Werner syndrome (WS) is an autosomal recessive segmental progeroid syndrome caused by null mutations at the WRN locus, which codes for a member of the RecQ family of DNA helicases. Since 1988, the International Registry of Werner syndrome had enrolled 130 molecularly confirmed WS cases from among 110 worldwide pedigrees. We now report 18 new mutations, including two genomic rearrangements, a deep intronic mutation resulting in a novel exon, a splice consensus mutation leading to utilization of the nearby splice site, and two rare missense mutations. We also review evidence for founder mutations among various ethnic/geographic groups. Founder WRN mutations had been previously reported in Japan and Northern Sardinia. Our Registry now suggests characteristic mutations originated in Morocco, Turkey, The Netherlands and elsewhere.
Introduction
Progeroid syndromes can be defined as diseases that present features suggestive of accelerated aging. Segmental progeroid syndromes affect multiple organs and tissues, whereas unimodal syndromes predominantly impact a single organ (e.g., dementias of the Alzheimer type) (Martin 1978; Martin and Oshima 2000). The prototypical example of a segmental progeroid syndrome is the Werner syndrome, caused by the mutations at the WRN locus (Yu et al. 1996).
The clinical phenotype of WS (WS; MIM# 277700) has been succinctly summarized as a “caricature of aging” (Epstein et al. 1966; Goto 1997). WS patients usually develop normally until they reach the second decade of life. The first clinical sign is a lack of the pubertal growth spurt during the teen years. In their 20s and 30s, patients begin to suffer from skin atrophy, regional loss of subcutaneous fat, and loss and graying of hair. The loss of fat is progressive and is often associated with characteristic ulcerations around the elbows, and, ankles. The lower limb ulcers are associated with neuropathy and vascular insufficiency; they may eventually require amputation. Other complications include type 2 diabetes mellitus, osteoporosis, bilateral ocular cataracts, gonadal atrophy, several forms of arteriosclerosis (atherosclerosis, arteriolosclerosis and medial calcinosis), and peripheral neuropathy (Leistritz et al. 2007). Multiple cancers have been observed by middle age; in contrast to usual aging, however, these include a disproportionate number of sarcomas (Goto et al. 1996). The osteoporosis is also unusual in that there is disproportionate involvement of the lower limbs. Bilateral ocular cataracts are the most penetrant phenotype. Our recent survey of WS patients with a molecularly confirmed diagnosis revealed a prevalence of 100% (87/87) (Huang et al. 2006); these cataracts were surgically removed at a median age of 30 years. At the time of diagnosis, prevalence of osteoporosis was 91%, hypogonadism 80%, diabetes mellitus 71%, and atherosclerosis 40%. Surprisingly, the median age of death in the most recent study was 54 years, a significant increase over what had been observed several decades ago (Epstein et al. 1966; Goto 1997), perhaps the result of improved medical management. A similar trend has been observed among Japanese patients (Goto and Matsuura 2008). The most common cause of death was myocardial infarction (Huang et al. 2006).
Classical WS is caused by mutations of the WRN gene on chromosome 8. The locus spans approximately 250 kb and consists of 35 exons, 34 of which are protein coding (Yu et al. 1996). WRN encodes a 180-kd multifunctional nuclear protein that belongs to the RecQ family of helicases (Gray et al. 1997) (Fig. 1). Recent structural study revealed a unique interaction between the RecQC-terminal (RQC) domain of WRN protein and the DNA substrates during the base-separation (Kitano et al. 2010). In contrast to other members of the RecQ family, WRN includes an N-terminal domain that codes for exonuclease activity (Huang et al. 1998). A domain for single strand-DNA annealing activity is in the C-terminal region (Muftuoglu et al. 2008a). Its preferred substrates resemble various DNA metabolic intermediates, substrates for which its helicase and exonuclease activities function in a coordinated manner, suggestive of roles in DNA repair, recombination, and replication (Brosh et al. 2006). The WRN protein is also involved in telomere maintenance. Proliferating fibroblasts from WS patients exhibit an acceleration of telomere shortening (Opresko 2008).
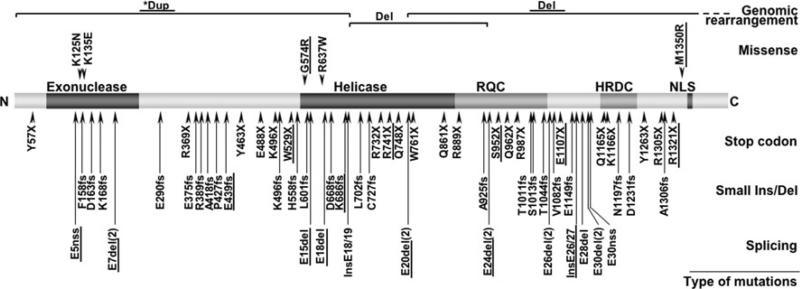
Fig. 1.
WRN mutations in WS patients. A diagram of the full-length wild-type WRN protein is shown, with its N-terminus on the left (N) and C-terminus on the right (C). Known functional domains are marked with darker shades; the exonuclease domain, the helicase domain, the RecQ helicase conserved region (RQC), the helicase RNaseD C-terminal conserved region (HDRC), and the nuclear localization signal (NLS). Mutations are grouped according to canonical classes and further identified by their amino acid changes using the reference sequence, NP_000544.2, and the nomenclature guidelines of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/). Splicing mutations are indicated by the affected exons. Splicing mutations that result in identical exon skipping are combined and indicated by the number of unique mutations as in (2). Deep intron mutations that create new exons are indicated as “Ins” along with the flanking exons. Splice mutations that create a new splice site (“nss”) are indicated as such. Genomic rearrangements, either deletion (Del) or triplication (Trip), are shown at corresponding protein locations, with regions extending beyond this figure in dotted lines. Newly identified mutations and those requiring further study are also underlined. Asterisk indicates uncertainty in the interpretation of array CGH results (see text)
There have been approximately 60 WRN disease mutations reported from all over the world (Huang et al. 2006; Matsumoto et al. 1997; Oshima et al. 1996; Uhrhammer et al. 2006). The majority of these mutations result in truncations of the WRN protein, thus eliminating the C-terminal nuclear localization signal (Suzuki et al. 2001). Regardless of the site of the mutations, these truncated WRN proteins that lack nuclear localization signals would be unable to enter the nuclei, thus becoming functionally null. This seemed to be satisfactory explanation of why we did not observe noticeable phenotype differences among the various common WRN mutations thus far (Huang et al. 2006).
We now report 18 new mutations, including two genomic rearrangements, two missense mutations, a deep intronic mutation that creates a new exon, and a splice consensus mutation that creates a new splice site. Given the availability of a total of 71 distinct mutations, we have initiated efforts to associate specific mutations with specific ethnic/geographical groups around the world. Some mutations appear to be specific to certain regions of Europe, the Middle East, and other parts of the world.
Materials and methods
Werner syndrome patients and samples
The International Registry of Werner Syndrome (Department of Pathology, University of Washington, Seattle, WA, USA; http:\www.wernersyndrome.org) has been recruiting patients suspected of having WS from all over the world since 1988. Referrals were based upon the clinical interpretations of referring physicians and guided by published descriptions of the characteristic signs and symptoms, including a set of criteria developed for the research project that led to the positional cloning of the WRN locus (Nakura et al. 1994; Yu et al. 1996). These clinical criteria have been recently reviewed (Leistritz et al. 2007; Nakura et al. 1994). The study has ongoing approval from the University of Washington Institutional Review Board.
Sample processing and preparation
The details of sample processing were described previously (Huang et al. 2006). In brief, peripheral blood samples from patients and, when feasible, from members of their nuclear pedigrees, were processed immediately upon arrival for (1) cryopreservation of aliquots of primary cells (buffy coats) and plasma, (2) isolation of DNA and RNA, and (3) establishment of lymphoblastoid cell lines using Epstein-Barr virus. Biopsies of skin were occasionally submitted and were used for the establishment of strains of fibroblasts. A definitive molecular diagnosis of WS was determined based on the results of genomic sequencing of the WRN coding exons and Western analysis of WRN protein (Muftuoglu et al. 2008b). RT-PCR was employed when the Western analysis revealed an abnormality but the genomic sequence appeared to be wild type or contained polymorphic variants. RT-PCR was also performed when we identified mutations at the splice junctions that required confirmations at the mRNA level (Muftuoglu et al. 2008b).
Genomic DNA was isolated from blood leukocytes or skin fibroblasts using a Puregene DNA isolation kit (Gentra Systems). Total RNA was isolated from whole blood using Trizol LS (Invitrogen, Carlsbad, CA, USA) or from the cultured cell pellet using Trizol (Invitrogen), as previously described (Huang et al. 2006).
PCR, RT-PCR and sequencing of the WRN gene
Genomic PCR primes of WRN were designed based on the GenBank reference sequence NG_008870.1 These were designed to amplify all coding exons together with flanking intronic sequences (50–100 bp) (Muftuoglu et al. 2008b). RT-PCR was performed with primer sets based upon Genbank reference sequence NM_000553.4 to amplify the WRN cDNA in three overlapping sections (Oshima et al. 1996). When we suspected large deletions or intronic mutations outside of the regions covered by our standard protocol, additional PCR reactions and sequencing, using additional primers were performed.
PCR and RT-PCR products were sequenced using Big Dye terminator version 3.1 chemistry on an ABI 3730 XL or ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed with commercially available Mutation Survey software (SoftGenetics, State College, PA, USA).
Western analysis of WRN protein
Total proteins derived from 4 × 105 of LCLs or 2 × 105 fibroblasts were separated on NuPAGE 4-12% Bis–Tris gel (InVitrogen) and transferred to nylon membranes. WRN proteins were detected with a mouse monoclonal antibody against amino acids 1,074–1,432 of human WRN protein (clone 195C, 1:2,000 dilution; catalog# W0393, Sigma–Aldrich, St. Louis, MO, USA) and a biotinylated anti-mouse antibody (1:500 dilution; catalog# BA9200, Vector Laboratories, Burlingame, CA, USA). The reactions were visualized with Western Lightening Chemiluminescence Reagent (catalog# NEL100, Perkin Elmer, Waltham, MA, USA) according to the manufacturer's instruction.
Affymetrix genome-wide SNP array 6.0 comparative genomic hybridization analysis
The Affymetrix Genome-Wide Human SNP Array 6.0 utilizing more than 906,600 SNPs and 946,000 probes for the detection of copy number variations was used for the detection of possible deletions or duplications at the WRN locus. Quantitative data analyses were performed with GTC 3.0.1 (Affymetrix Genotyping Console) using Hap-Map270 (Affymetrix, Inc., Santa Clara, CA, USA) as reference file. The Segment Reporting Tool (SRT) was used to locate segments with copy number changes in the copy number data with the assumption of a minimum of 10 kb per segment and minimum genomic size of five markers of a segment.
Determination of rearrangement breakpoints
The rearrangement breakpoints were ascertained by PCR primer walking using the Expand Long Template PCR System (Roche Diagnostics, Indianapolis, IN, USA). PCR primers were designed based on the GenBank reference sequence NC_000008.10 (NCBI Build 36.1; UCSC Genome Browser on Human Mar. 2006 Assembly (hg18), http://www.genome.ucsc.edu/). PCR products were sequenced as described above.
Results
WRN mutations in WS patients
In our previous review of WRN mutations (Huang et al. 2006), we reported on a total of 99 WS patients with confirmed WRN mutations, including 32 new cases. As of January 2010, the Registry had enrolled 130 cases with confirmed mutations from among 110 pedigrees. We also diagnosed five clinically atypical cases shown to be wild type at the WRN locus, all of which exhibited LMNA mutations (Chen et al. 2003). Forty-one cases submitted to our Registry as possible cases of WS were wild type at both the WRN and LMNA loci. These cases remain of considerable interest, as they may reflect new and novel segmental progeroid syndromes.
Figure 1 shows 18 newly identified WRN mutations (underlined) along with previously reported mutations (see Supplementary Table 2 for a description). Newly identified mutations included two genomic rearrangements, five stop codon mutations, two small deletions, four splicing mutations that resulted in exon skipping, one mutation that created a new splice acceptor site, one deep intron mutations that created novel exons, and two missense mutations. We also identified a mutation that resulted in the absence of exon 20, but it was not associated with any detectible mutation at the splice junction. While stop codon mutations, small deletions and insertions, and splice junction mutations that cause the skipping of the exons were common among the WRN mutations in WS patients, others are relatively rare.
Genomic rearrangements involving WRN locus
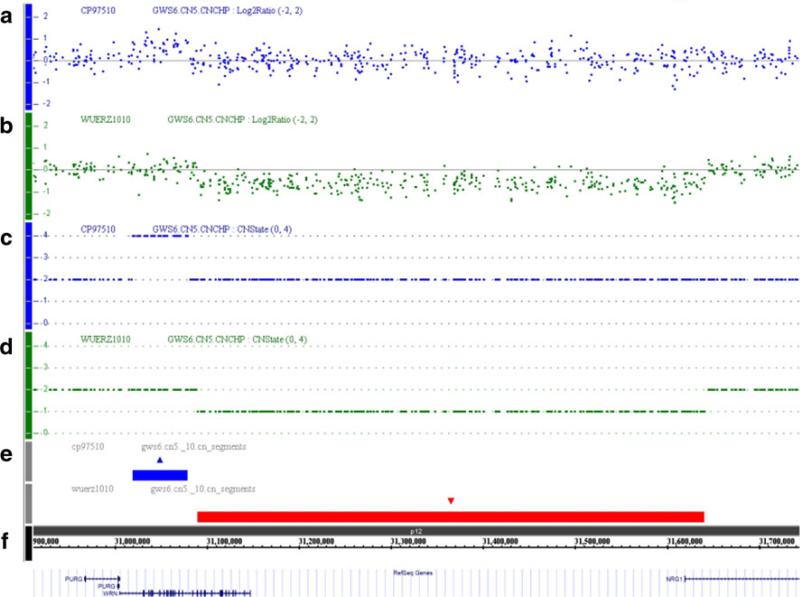
A large genomic deletion was identified in a female German patient, Registry# WU1010 (Fig. 2). The patient was referred to us at age 50 with a definite clinical diagnosis of WS. The sequencing of the coding exons showed the heterozygous stop codon mutation, c.171C>G (p.Y57X) in exon 3, but we did not identify any other mutation. Array CGH study revealed a heterozygous gross deletion (>500 kb) encompassing intron 19 of the WRN gene to intron 1 of the NRG1, is located at centromeric (or 3′) relative to WRN. Junction fragment analysis confirmed the array CGH data and revealed an approximately 553-kb deletion, chr8:g.31,089,233-31,642,239del, with a 32-bp insertion of a SINE repeat derived sequence (InsCTCACT GCAGCCTCAAACTCCTGGGTTCAAC) at the site of deletion. The 5′ breakpoint was within the FRAME sequence that belongs to the Alu repeat at chr8:31,089,153–31,089,298 and the 3′ breakpoint was within the MIRb sequence at chr8:31,642,175–31,642,390 (UCSC Genome Browser on Human Mar. 2006 Assembly (hg18)).
Fig. 2.
Array CGH analyses of the WS cases involving WRN locus. Panels show result of the quantitative SNP-analysis for patients, Registry# WU1010 and CP97510. The X-axis indicates the physical position of the chromosome's 8p12 region with the telomeric region on the left and the centromeric on the right. The Y-axis of a–d indicates the copy number calculated by log2 (test/control) ratios of the probes. a and d A scattered plot (a) and copy number plot (c) of CP97510 show an amplification (CN-State 4) with a size of approximately 60 kb in the N-terminal part of WRN. b and d A scattered plot (b) and copy number plot (d) of WU1010 show a large heterozygous deletion (CN-State 1) encompassing the C-terminal part of WRN and the first exon of NRG1 with a size of about 550 kb. e The region of copy number gain in CP97510 is indicated by a blue rectangle. The region of copy number loss in WU1010 is indicated by a red rectangle. f The chromosome band (8p12), nucleotide coordinates and the annotated loci in this region are shown with stripes that are scaled to the exact size of the deletion/gain regions on the chromosome
We also identified an increase in copy number of the N-terminal region of the WRN gene in a French patient, Registry# CP97510 (Fig. 2). In the initial RT-PCR sequencing, we were able to amplify WRN mRNA in three sections but we did not identify any mutations. However, Western analysis showed no WRN protein, which confirmed the diagnosis of WS. Array CGH revealed an increased copy number (4 copies) of the genomic regions encompassing WRN introns 1 through 19. This could be a homozygous duplication of this region in both homologues of chromosome 8. Alternatively, it could represent a triplication in one allele together with the single copy represented by the other allele.
Unusual intron mutations identified in WS patients
The deep intron mutation noted above was homozygous. It was identified in intron 26 from a Turkish patient, the offspring of a first cousin mating (Registry# CO1010). Genomic sequencing of the coding exons and flanking introns did not reveal the site of the mutation, but Western analysis of proteins derived from an LCL from this subject failed to reveal any WRN protein. RT-PCR sequencing of the coding region of WRN revealed an insertion of 69 nucleotides between exons 25 and 26 (r.3233_3234ins69). The inserted sequence aligned with the region in intron 26, c.3234-159 through c.3234-91 that was flanked by ttcaaACATC and AAAGGgtaat in the reference sequence. Genomic sequencing targeting to the region revealed an A-to-G substitution at c.3234-160, one that created a new splicing acceptor site, ttcagACATC, consistent with the insertion of the novel exon observed by RT-PCR.
A mutation that alters a splice acceptor site was found in intron 4 of a French Caucasian Werner patient, Registry# AB10010. The subject was a compound heterozygote. The initial RT-PCR sequencing showed a deletion of 11 nucleotides at the 5′ end of exon 5, r.356_366del11, which would cause a frame shift, p.V119 fs (Huang et al. 2006). Genomic PCR revealed a splicing consensus mutation, c.356-2A>C. This mutation was reported in another French patient by another group (Uhrhammer et al. 2006).
We also identified a heterozygous exon 20 skipping mutation in a US WS patient, Registry# SI1010, by cDNA analysis. We did not find alterations in exon 20 or in adjacent intronic regions by genomic sequencing in DNA from this subject, however, although there was a common polymorphism, c.2361G>T, p.L787L within exon 20. This patient was also heterozygous for an exon 22 stop codon mutation, p.889X, which would result in the loss of the nuclear localization signal and, presumably, nonsense-mediated decay. The Western analysis of LCL from this patient revealed a trace of WRN protein of the normal size. The steady-state-level of WRN protein in SI1010 was estimated to be approximately 1.1% of the control. These findings raise the possibility of a mutation that affects the splicing efficiency of exon 20. We sequenced parts of intron 19 (4,554 bp in size) and intron 20 (3,414 bp in size), the two introns that surround exon 20. However, we were unable to identify the cause of this presumably leaky mutation.
Novel missense mutations of WRN gene
We also identified two missense alterations that are likely pathogenic. One was found in a compound heterozygous patient of German origin (Registry# HN2010). He carried a heterozygous splice site mutation at the junction of intron 17 and exon 18, c.1982-1G>A. That mutation is expected to cause exon 18 skipping at the mRNA level, although we were unable to demonstrate this by RT-PCR because cell materials were not available. He also carried a heterozygous missense alteration in exon 14, c.1720G>A, p.Gly574Arg. We did not find any other nucleotide changes aside from the known polymorphisms. Gly574 is a consensus amino acid within the ATPase motif of the RecQ helicase domain I (Yu et al. 1996). Mutation here could be expected to result in the abolishment of ATPase activity.
The PolyPhen program (accessed at http://www.genetics. bwh.harvard.edu/pph/) (Ramensky et al. 2002) predicted that p.Gly574Arg change would be “probably damaging”. The SIFT program (accessed at http://www.sift.jcvi.org/) (Kumar et al. 2009) also predicted that this alteration would be deleterious.
Another missense mutation was found in a compound heterozygote of Dutch origin (Registry# UT1010). He carried a heterozygous mutation, c.3590delA, p.Asn1197fs, in exon 31, one that had been found in another Dutch Werner patient (Registry# ER1010). Registry# UT1010 also had a heterozygous substitution, c.4049T>G, p.Met1350Arg, in exon 34. We did not find any other changes except for common polymorphisms. The p.Met1350Arg change was located at N-terminal of the nuclear localization signal, NKRRCF, at residues 1,370–1,375 (Matsumoto et al. 1998) and N-terminal of the nucleolar localization signal, RK, at residues 1,403–1,404 (Suzuki et al. 2001). There are no known overlapping functional domains. The Polyphen program predicted this change would be “probably damaging” and the SIFT program predicted that this change would be deleterious.
Mutations thought to be specific for various ethnic-geographic groups
Exon 26 deletions due to a splice site mutation, c.3139-1G>C, in intron 25, have been widely reported in Japanese WS patients (Goto et al. 1997; Matsumoto et al. 1997; Satoh et al. 1999). These are regarded as reflecting a founder mutation. Similarly, deep intronic mutations, c.2089-3025A>G in intron 19 that create a new exon between exon 18 and 19 (r.2088_2089ins106), have been identified in 18 WS patients from multiple families in Sardinia (Masala et al. 2007). In this study, we observed other mutations that appear to be specific for various other ethnic-geographic groups. These have been summarized in Table 1.
Table 1.
Ethnic specific WRN mutations found in WS patients
| Ethnic origin | No of pedigrees | Mutation type | Nucleotide change | Consequence of mutation | Affected exon | References |
|---|---|---|---|---|---|---|
| Sardinian | 2 | Deep intron mutation | c.2089-3025A>G | New exon | IVS18 | Masala et al. (2007) |
| Moroccan | 2 | Insertion | c.2179dupT | p.C727fs | Exon 19 | Huang et al. (2006) |
| Turk | 3 | Splice site mutation | c.3460-2A>G | Exon 30 deletion | IVS29 | Huang et al. (2006) |
| Japanese | 23 | Splice site mutation | c.3139-1G>C | Exon 26 deletion | IVS25 | Satoh et al. (1999); Yu et al. (1996) |
| Dutch | 2 | Deletion | c.3590delA | pN1197fs | Exon 31 | Huang et al. (2006) |
Nucleotide and amino acid notations are based on the reference sequence, GenBank accession number NM_000553.4
No of Pedigrees are ones in our Registry only
We identified two Moroccan brothers who carry the same one-nucleotide duplication, c.2179dupT in exon 19 previously found in another Moroccan patient. Two additional
Turkish patients were found to have a splicing mutation, c.3460-2A>G in intron 29, that was previously seen in one Turkish patient. Similarly, a single nucleotide deletion, c.3590delA in exon 31, previously identified in a Dutch WS patient was found in a second Dutch patient. While the exon 19 mutation in our Moroccan patients and the mutations resulting in exon 30 skipping in our Turkish patients were all homozygous, c.3590delA in exon 31 was seen as homozygous in one Dutch patient and heterozygous in the other.
While not specific to a particular ethnic group, a p.R732X mutation in exon 19 was seen mostly in Italian WS patients, a p.Q1165X mutation in exon 30 was mainly observed in Turkish patients, and a p.R1305X mutation in exon 33 was predominantly found in Japanese WS patients.
A complete list of mutations is given in Supplementary Table 2.
Discussion
In this mutation update, we report the analysis of 18 mutations that were newly identified or further investigated since our last report (Huang et al. 2006). Combined with the previously published mutations by our group and those of others, a total of 71 WRN disease mutations have now been identified in clinically diagnosed Werner patients. These mutations are summarized in this report. They include 21 stop codon mutations, 25 small deletions/insertions, 17 splicing mutation, 5 missense mutations, 1 intragenic large deletion, and 2 genomic rearrangement extending beyond the WRN locus. Particularly interesting were the genomic rearrangements involving WRN and neighboring loci, deep intron mutations, and missense mutations, all of which are rare among Werner syndrome patients.
In this study, we identified an approximately 553-kb deletion spanning intron 19 of the WRN gene to the intron 1 of the NRG1 gene. To our knowledge, this is the first report of the genomic arrangements of WRN involving another locus. The breakpoints were both located within the SINE sequences, one in the FRAME sequence that belongs to the Alu repeat, and the other in the MIRb sequence. We previously identified approximately 27 kb intragenic deletion in which the 5′ breakpoint, chr8:31,078,664, was located within the AluSq sequence at chr8:31,078,563–31,078,870, and 3′ breakpoint, chr8:31,105,734, within the AluY sequence at chr8:31,105,636–31,105,924. Both deletions are likely facilitated by the presence of the repeat sequence that conferred genomic instability in this region (Gu et al. 2008; Hastings et al. 2009; Oshima et al. 2009).
The NRG1 gene encodes a signaling protein, neuregulin-1, that is involved in cell proliferation particularly in the central and peripheral nervous system (Holmes et al. 1992; Tan et al. 2007). Association studies suggested a link between NRG1 with schizophrenia (Stefansson et al. 2002; Tan et al. 2007). Our patient did not report any neurological or psychological abnormalities in her early 50s at the time of this report. This may be because there are a number of isoforms for neuregulin-1, many of which lack exon 1 (Tan et al. 2007) and the haploinsufficiency of one isoform that contains exon 1 does not present any symptoms. Another possibility is that the patient may be too young to develop symptoms due to the NRG1 mutation.
It has been reported in Japan that approximately 10% of WS patients develop schizophrenia (Goto 1997). Most of these patients carry the typical Japanese mutation at 26 exon. Association studies of NRG1 polymorphisms and haplotypes among the Werner syndrome patients and the appropriate controls might reveal the role of the NRG1 locus that contributes to higher susceptibility to schizophrenia observed among the Japanese WS patients.
We also identified an amplification (4 regions) of the genomic region that includes WRN exons 2 through 18. The copy number change was associated with the loss of normal WRN expression, as evidenced by Western analysis. We think it is equally possible to have a homozygous duplication or triplication in one allele and a single copy in the other allele. However, we have not been able to determine the genomic structure or the breakpoints of this genomic rearrangement; there are too many possibilities, including potential involvements of other chromosomes. Further studies are in progress.
Increasing numbers of pseudoexon inclusions have been seen at different loci associated with human diseases (Buratti et al. 2006). While point mutations in deep intronic regions that create efficient splice donor or splice acceptor sites are more common among deep intronic mutations, mutations of intronic splicing regulatory elements within pseudoexons have also been identified (Buratti et al. 2006; Pagani and Baralle 2004). Deep intronic mutations in the WRN gene have been shown to create a new splice donor site (c.2089–3025A>G in intron 18) in multiple Sardinian pedigrees (Masala et al. 2007). In this study, an intron mutation that creates a new splice acceptor site (c.3234-160A>G in intron 25) was observed in a Turkish pedigree. Because these mutations can be easily missed, genomic sequencing, Western analysis of proteins, and RT-PCR are necessary for an accurate molecular diagnosis. This is especially important for the patients from ethnic/geographic groups that have known deep intronic mutations.
In a previously published genetic investigation of a French WS patient, we identified a splice acceptor consensus mutation, c.356-2A>C, which leads to the utilization of the splice acceptor consensus just at the 3′ of the mutated site, resulting in 11-bp deletion at the mRNA level. Splice consensus mutations generally result in the skipping of exons. However, there are instances in which another splicing consensus nearby is utilized, as seen in our case. Such findings underscore the importance of RT-PCR sequencing for the confirmation of splice site mutations suspected from the results of genomic sequencing.
We identified an instance of heterozygous exon 20 skipping that was not associated with mutations of the splice acceptor or donor sites of exon 20 in a US Caucasian patient. Cell extracts from that subject revealed trace amounts of WRN protein (approximately 1% of those of controls). This is suggestive of a leaky mutation that suppresses the inclusion of exon 20, possibly a mutation in one of the flanking introns. Because of the relatively large sizes of intron 19 and 20, we have not been able to pinpoint these suspected intronic mutations.
Two pathogenic missense mutations have been observed in our studies. A c.1720G>A, p.Gly574Arg missense mutation was found in the ATPase consensus motif of the helicase domain I (Yu et al. 1996). It occurred in a German compound heterozygous patient. Previous biochemical studies using a recombinant WRN protein bearing an experimentally generated p.Lys577Met mutation within the ATPase domain provided evidence that ATPase activity was essential for WRN helicase activity (Gray et al. 1997), although we have not identified a Werner patient carrying the p.Lys577Met mutation. We speculate that p.Gly574Arg also results in the abolishment of helicase activity and is responsible for the Werner syndrome phenotype in the patient.
Unlike other truncation mutants, this p.Gly574Arg mutant retains the nuclear localization signal that allows the entry of mutant proteins into the nuclei. The p.Gly574Arg mutant protein, although likely lacking helicase activity, is expected to retain exonuclease domains and other functional or interacting domains. The presence of the full-length p.Gly574Arg mutant WRN protein in nuclei, either homozygous or heterozygous, may give rise to atypical Werner phenotypes. Further follow-up and studies are needed to clarify the genotype–phenotype correlation for this missense mutation.
In addition, the c.1720G missense mutation occurred at the last nucleotide of exon 14; the underlying G-to-A transition might affect exon 14 splicing if the c.1720G is a part of the splice acceptor consensus. Various splice site prediction programs predicted that this G-to-A change would not grossly affect the splicing of exon 14. We were unable to rule out the possibility that splicing of WRN exon 14 may be affected by this c.1720G>A change, however, as we were unable to obtain suitable cell materials from this patient.
Another missense alteration, p.Met1350Arg in exon 34, was found as a heterozygous change in a Dutch WS patient.
He also carried a heterozygous frame shift mutation in exon 31. There is no known functional domain overlapping Met1350. The closest functional domain is the nuclear localization signal at residues 1,370–1,375. Two programs, Polyphen and SIFT, predicted that this change would be deleterious, as methionine at residue 1,350 is well conserved across species.
Our evidence of mutations that were specific for particular ethnic/geographical groups are consistent with mutational founder effects (Zlotogora 2007). These conclusions are of clinical importance, as they provide guidance for more focused molecular genetic diagnostic studies. They also can guide research on potential vulnerabilities of heterozygous carriers, an important and neglected area of public health research. We note, for example, that canonical mutations in Japan permitted estimates that the prevalence of heterozygote carriers was approximately 1/167 (Satoh et al. 1999). The prevalence of heterozygous carriers in Sardinia was estimated to be of the order of 1/120 (Masala et al. 2007). Our study has provided evidence of additional such examples: an insertion, c.2179dupT in exon 19, in Moroccan pedigrees and a splice site mutation, c.3460-2A>G, that causes exon 30 skipping in Turkish patients. Among the mutations found in Caucasian WS patients, a specific deletion in exon 31 (c.3590delA, p.N1197fs) was seen only in two patients from The Netherlands. We anticipate that additional examples of potential founder mutations effect will be identified among various populations as we continue to expand our International Registry.
Supplementary Material
Acknowledgments
We thank Ms. Mizue Kishida for her assistance in curating the mutations. This work was supported by R24CA78088 (Martin), R21AG033313 (Oshima), the Ellison Medical Foundation Senior Scholar Award (Oshima), and the German Research Foundation (DFG) in the framework of the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (Kubisch).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-010-0832-5) contains supplementary material, which is available to authorized users.
Contributor Information
Katrin Friedrich, Center for Molecular Medicine Cologne, Institute of Human Genetics, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany.
Lin Lee, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA.
Dru F. Leistritz, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA
Gudrun Nürnberg, Center for Molecular Medicine Cologne, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), Cologne Centre For Genomics, University of Cologne, Cologne, Germany.
Bidisha Saha, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA.
Fuki M. Hisama, Department of Medicine, University of Wahsignton, Seattle, WA, USA
Daniel K. Eyman, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA
Davor Lessel, Center for Molecular Medicine Cologne, Institute of Human Genetics, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany.
Peter Nürnberg, Center for Molecular Medicine Cologne, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), Cologne Centre For Genomics, University of Cologne, Cologne, Germany.
Chumei Li, Toronto General Hospital, Toronto, Canada.
María J. Garcia-F-Villalta, Hospital de la Princesa, Madrid, Spain
Carolien M. Kets, Radboud University, Nijmegen, The Netherlands
Joerg Schmidtke, Institut für Humangenetik, Medizinische Hochschule, Hannover, Germany.
Vítor Tedim Cruz, Hospital Sao Sebastiao, Santa Maria da Feira, Portugal.
Peter C. Van den Akker, Department of Genetics, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands
Joseph Boak, Riverview Medical Associates, Red Bank, NJ, USA.
Dincy Peter, Christian Medical College, Vellore, India.
Goli Compoginis, University of Southern California, Los Angeles, CA, USA.
Kivanc Cefle, Istanbul Medical Faculty, Istanbul University, Istanbul, Turkey.
Sukru Ozturk, Istanbul Medical Faculty, Istanbul University, Istanbul, Turkey.
Norberto López, Clinic Hospital Virgen de la Victoria, Malaga, Spain.
Theda Wessel, Department of Pediatric Endocrinology, Charité University Hospital, Berlin, Germany.
Martin Poot, Department of Medical Genetics, University Medical Center Utrecht, Utrecht, The Netherlands.
P. F. Ippel, Department of Medical Genetics, University Medical Center Utrecht, Utrecht, The Netherlands
Birgit Groff-Kellermann, Department of Dermatology and Venerology, Karl Landsteiner Institute for Dermatological Research, General Hospital St. Poelten, St. Poelten, Austria.
Holger Hoehn, Department of Human and Medical Genetics, University of Würzburg, Würzburg, Germany.
George M. Martin, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA
Christian Kubisch, Center for Molecular Medicine Cologne, Institute of Human Genetics, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany.
Junko Oshima, Department of Pathology, University of Washington, Box 357470, 1959 NE Pacific St., Seattle, WA 98195-7470, USA.
References
- Brosh RM, Jr, Opresko PL, Bohr VA. Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol. 2006;409:52–85. doi: 10.1016/S0076-6879(05)09004-X. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lee L, Kudlow BA, Dos Santos HG, Sletvold O, Shafeghati Y, Botha EG, Garg A, Hanson NB, Martin GM, Mian IS, Kennedy BK, Oshima J. LMNA mutations in atypical Werner's syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Goto M, Matsuura M. Secular trends towards delayed onsets of pathologies and prolonged longevities in Japanese patients with Werner syndrome. Biosci Trends. 2008;2:81–87. [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol Bio-markers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Goto M, Imamura O, Kuromitsu J, Matsumoto T, Yamabe Y, Tokutake Y, Suzuki N, Mason B, Drayna D, Sugawara M, Sugimoto M, Furuichi Y. Analysis of helicase gene mutations in Japanese Werner's syndrome patients. Hum Genet. 1997;99:191–193. doi: 10.1007/s004390050336. [DOI] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WE, Sliwkowski MX, Akita RW, Henzel WJ, Lee J, Park JW, Yansura D, Abadi N, Raab H, Lewis GD, et al. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256:1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonu clease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von Herbay A, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Leistritz DF, Hanson NB, Martin GM, Oshima J. Werner syndrome. In: GeneReviews at Genetests: Medical Genetics Information Resource (database online) Copyright, University of Washington; Seattle: 2007. pp. 1997–2010. http://www.genetests.org. [Google Scholar]
- Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- Masala MV, Scapaticci S, Olivieri C, Pirodda C, Montesu MA, Cuccuru MA, Pruneddu S, Danesino C, Cerimele D. Epidemiology and clinical aspects of Werner's syndrome in North Sardinia: description of a cluster. Eur J Dermatol. 2007;17:213–216. doi: 10.1684/ejd.2007.0155. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Imamura O, Yamabe Y, Kuromitsu J, Tokutake Y, Shimamoto A, Suzuki N, Satoh M, Kitao S, Ichikawa K, Kataoka H, Sugawara K, Thomas W, Mason B, Tsuchihashi Z, Drayna D, Sugawara M, Sugimoto M, Furuichi Y, Goto M. Mutation and haplotype analyses of the Werner's syndrome gene based on its genomic structure: genetic epidemiology in the Japanese population. Hum Genet. 1997;100:123–130. doi: 10.1007/s004390050477. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Imamura O, Goto M, Furuichi Y. Characterization of the nuclear localization signal in the DNA helicase involved in Werner's syndrome. Int J Mol Med. 1998;1:71–76. doi: 10.3892/ijmm.1.1.71. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008a;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu M, Oshima J, von Kobbe C, Cheng WH, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008b;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakura J, Wijsman EM, Miki T, Kamino K, Yu CE, Oshima J, Fukuchi K, Weber JL, Piussan C, Melaragno MI, et al. Homozygosity mapping of the Werner syndrome locus (WRN). Genomics. 1994;23:600–608. doi: 10.1006/geno.1994.1548. [DOI] [PubMed] [Google Scholar]
- Opresko PL. Telomere ResQue and preservation—roles for the Werner syndrome protein and other RecQ helicases. Mech Ageing Dev. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Oshima J, Yu CE, Piussan C, Klein G, Jabkowski J, Balci S, Miki T, Nakura J, Ogihara T, Ells J, Smith M, Melaragno MI, Fraccaro M, Scappaticci S, Matthews J, Ouais S, Jarzebowicz A, Schellenberg GD, Martin GM. Homozygous and compound heterozygous mutations at the Werner syndrome locus. Hum Mol Genet. 1996;5:1909–1913. doi: 10.1093/hmg/5.12.1909. [DOI] [PubMed] [Google Scholar]
- Oshima J, Magner DB, Lee JA, Breman AM, Schmitt ES, White LD, Crowe CA, Merrill M, Jayakar P, Rajadhyaksha A, Eng CM, del Gaudio D. Regional genomic instability predisposes to complex dystrophin gene rearrangements. Hum Genet. 2009;126:411–423. doi: 10.1007/s00439-009-0679-9. [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Imai M, Sugimoto M, Goto M, Furuichi Y. Prevalence of Werner's syndrome heterozygotes in Japan. Lancet. 1999;353:1766. doi: 10.1016/S0140-6736(98)05869-3. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shiratori M, Furuichi Y, Matsumoto T. Diverged nuclear localization of Werner helicase in human and mouse cells. Oncogene. 2001;20:2551–2558. doi: 10.1038/sj.onc.1204344. [DOI] [PubMed] [Google Scholar]
- Tan W, Wang Y, Gold B, Chen J, Dean M, Harrison PJ, Weinberger DR, Law AJ. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007;282:24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- Uhrhammer NA, Lafarge L, Dos Santos L, Domaszewska A, Lange M, Yang Y, Aractingi S, Bessis D, Bignon YJ. Werner syndrome and mutations of the WRN and LMNA genes in France. Hum Mutat. 2006;27:718–719. doi: 10.1002/humu.9435. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zlotogora J. Multiple mutations responsible for frequent genetic diseases in isolated populations. Eur J Hum Genet. 2007;15:272–278. doi: 10.1038/sj.ejhg.5201760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.