Abstract
Abundant ribonucleotide incorporation in DNA during replication and repair has profound consequences for genome stability, but the global distribution of ribonucleotide incorporation is unknown. We developed Ribose-seq, a method to capture unique products generated by alkaline cleavage of DNA at embedded ribonucleotides. High-throughput sequencing of these fragments from yeast Saccharomyces cerevisiae DNA revealed widespread ribonucleotide distribution with a strong preference for cytidine and guanosine, and identified hotspots of ribonucleotide incorporation in nuclear and mitochondrial DNA. Ribonucleotides were primarily incorporated on the newly-synthesized leading strand of nuclear DNA and were present upstream of G+C-rich tracts in the mitochondrial genome. Ribose-seq is a powerful tool for the systematic profiling of ribonucleotide incorporation in genomic DNA.
Introduction
Genomic DNA contains embedded ribonucleotides (rNMPs) that are incorporated during DNA replication and repair, or formed during DNA damage (reviewed in 1). The modifications have been linked to genome instability and disease, but no method currently exists to profile their locations genome-wide.
rNMPs were initially found at specific DNA loci in mouse and human mitochondrial DNA2 and the mating type locus of fission yeast3, but have since been detected in a variety of cell types4. Many DNA polymerases can incorporate rNMPs into DNA, including the human replicative DNA polymerase (Pol) δ5 and mitochondrial Pol γ6, budding yeast nuclear replicative Pol α, δ, and ɛ7, Escherichia coli polymerase V8, and the polymerase component of bacterial non-homologous end joining ligases9. rNMP incorporation could also be a consequence of incomplete maturation of Okazaki fragments during lagging strand synthesis in DNA replication10. Moreover, generation of hydroxyl radicals during oxidative stress can modify DNA deoxyribose sugars to ribose, forming rNMPs in DNA both in vitro and in vivo11.
Ribonuclease H type 2 (RNase H2/HII) cleaves single rNMPs or longer rNMP tracts incorporated in DNA12 and initiates ribonucleotide excision repair (RER), the major rNMP repair mechanism in bacterial DNA and in eukaryotic nuclear DNA (13 and references therein). By contrast, RNase H1/HI only recognizes rNMP tracts longer than 4 nucleotides. Inactivation of RNase H2/HII leads to the accumulation of high amounts of rNMPs in genomic DNA, enabling >1 million rNMPs to be quantified per mouse embryonic fibroblast genome and suggesting that rNMPs are the most common non-canonical nucleotides in dividing mouse cells14. Similar measurements on genomic DNA derived from RNase H2-deficient (rnh201Δ) budding yeast estimated a few thousand rNMPs incorporated per genome per cell cycle15,16, and RNase HII-null Bacillus subtilis cells have high levels of incorporated rNMPs17. Embedded rNMPs in DNA have highly reactive 2′-hydroxyl groups, altering its properties, structure, and function18,19 and leading to genome instability16,20–22. In humans, mutations in any of the three subunits of RNase H2 are associated with the neurological syndrome Aicardi-Goutieres (AGS)23.
Despite abundant evidence for the frequent incorporation of rNMPs in DNA, a comprehensive and detailed picture of rNMP incorporation throughout a genome is lacking. Here, we introduce Ribose-seq: a technique for mapping rNMPs in genomic DNA.
Results
Ribose-seq strategy to capture rNMPs in DNA
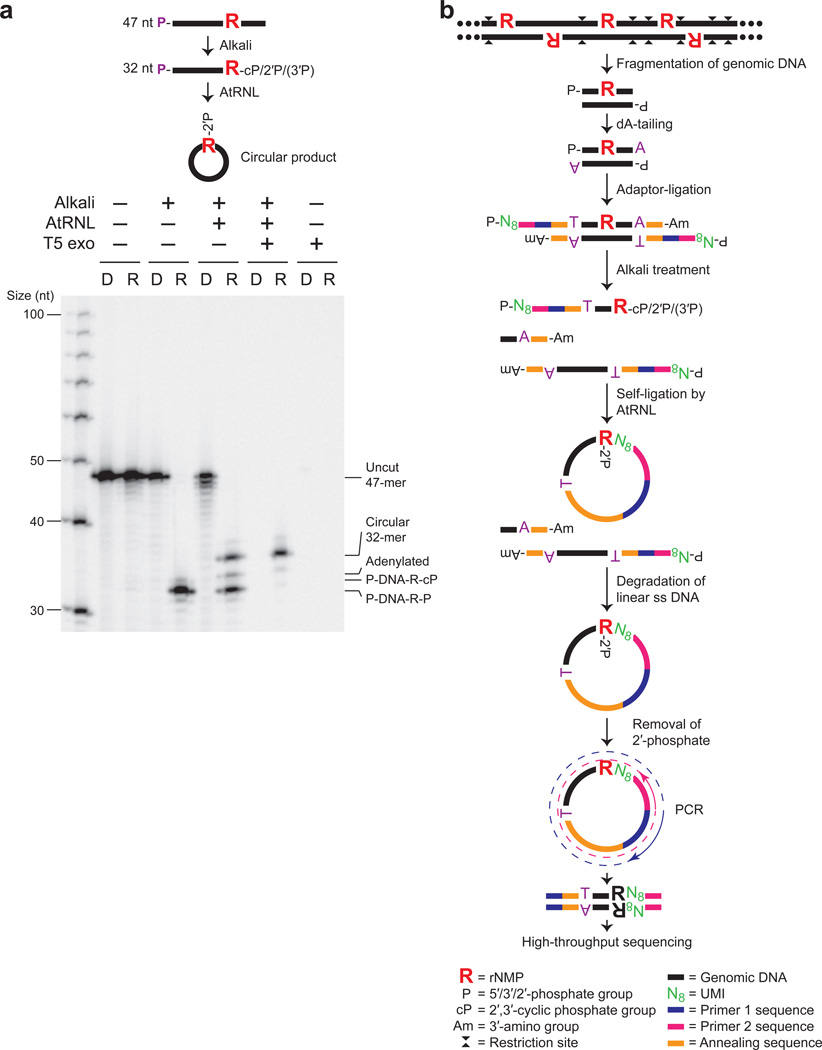
Ribose-seq captures rNMP-terminated single-stranded (ss) DNA fragments generated by alkaline cleavage of rNMPs in DNA (Fig. 1 and Supplementary Fig. 1). We exploited the unique ligation mechanism of Arabidopsis thaliana tRNA ligase (AtRNL), normally involved in tRNA maturation. AtRNL converts 2′,3′-cyclic phosphate ends of RNA to 2′-phosphate and ligates these to 5′-phosphate ends of RNA24,25 or DNA25. We demonstrated that AtRNL captures 2′,3′-cyclic phosphate or 2′-phosphate termini of DNA derived from alkaline cleavage of a DNA oligonucleotide (oligo) at an embedded rNMP, ligating the 2′-phosphate end to the 5′-phosphate terminus of the same DNA molecule and producing a ss DNA circle containing an embedded rNMP. Self-ligation was strongly preferred over dimerization, as linear dimers were not detected (Fig. 1a). Further, these ss DNA circles are resistant to T5 exonuclease enabling their enrichment relative to unligated linear DNA upon exonuclease treatment (Fig. 1a). We did not observe any bias for the 3′ rNMP substrate of AtRNL, which captured an embedded rAMP, rCMP, rGMP, or rUMP with equal efficiency (P values > 0.05 in each case) (Supplementary Fig. 2 and Supplementary Table 1), nor was any bias observed in a previous study26. These data indicate that self-ligation is favored for AtRNL on 2′-phosphate-terminated ss DNA fragments as small as 22 nt (Fig. 1a and Supplementary Fig. 2), thus facilitating library construction and high-throughput DNA sequencing.
Figure 1. Ribose-seq method for mapping rNMPs in genomic DNA.
(a) AtRNL captures 2′,3′-cyclic phosphate or 2′-phosphate DNA termini generated by alkaline cleavage of a single rGMP in a 5′-radiolabeled 47-nt ss DNA oligo (see Supplementary Table 8). R, rGMP or rGMP-bearing oligo. D, DNA-only control oligo. P, 5′-radiolabel. T5 exonuclease treatment confirms the presence of circular ligation product. Left lane, ss DNA ladder. Ligation efficiency was about 50%, as expected due to the mixture of 2′-phosphate and 3′-phosphate ends generated upon alkaline cleavage. (b) Schematic of Ribose-seq protocol. Genomic DNA is fragmented, dA-tailed and ligated to a molecular barcode-containing sequencing adaptor. Alkali treatment denatures the DNA and cleaves at rNMP sites, exposing 2′,3′-cyclic phosphate and 2′-phosphate termini, which are self-ligated to 5′-phosphate ends by AtRNL. Linear, unligated fragments are degraded by T5 exonuclease while the remaining rNMP-captured, circular DNA molecules, upon removal of 2′-phosphate at the ligation junction by 2′-phosphotransferase Tpt1, are PCR-amplified and sequenced.
We applied Ribose-seq to identify rNMPs embedded in nuclear and mitochondrial DNA of RNase H2-deficient budding yeast (strain KK-100, Supplementary Table 2)1. Genomic DNA was extracted from cells grown to stationary phase, and a mixture of three blunt-end restriction enzymes was used to fragment the DNA. Application of our rNMP-capture scheme (Fig. 1b) yields a library of DNA molecules (Supplementary Fig. 3a) with an average size of ~350 bp, each of which maps to a single site of rNMP incorporation and its upstream sequence. In control experiments, we found that exclusion of either AtRNL (Supplementary Fig. 3a) or alkali treatment (Supplementary Fig. 3b) prevented library formation, validating that captured molecules derive from rNMPs embedded in DNA.
Spectrum of rNMPs in S. cerevisiae genome
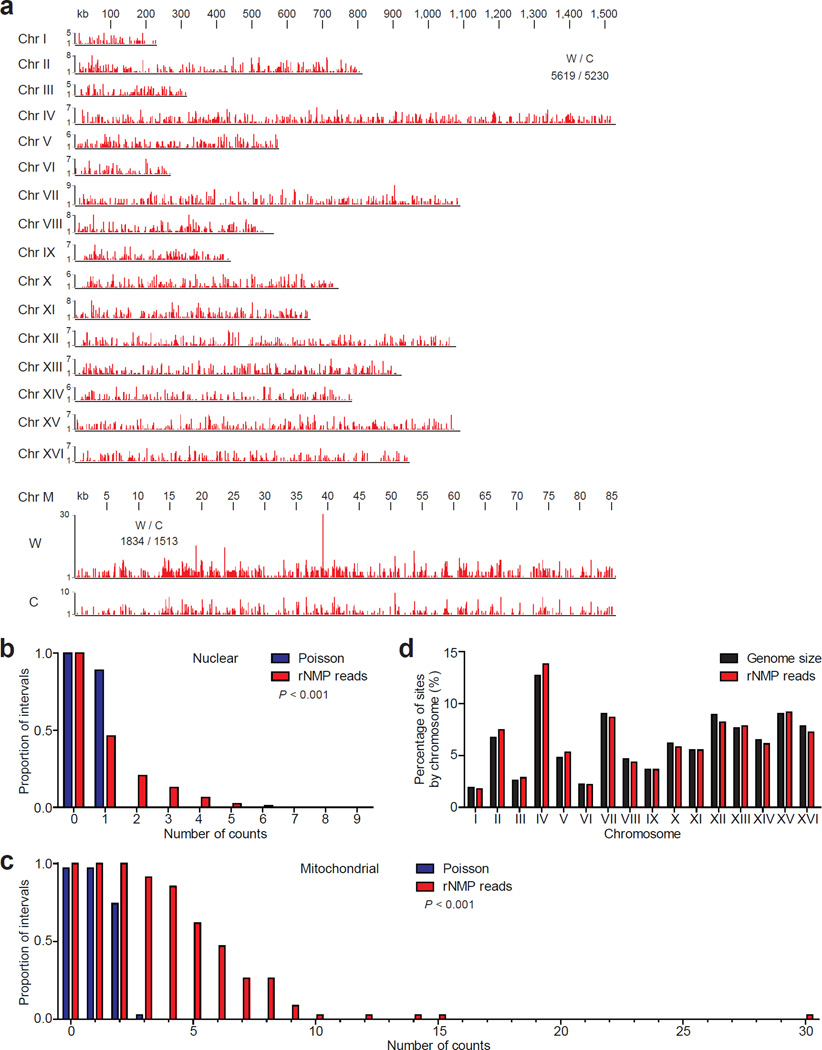
A Ribose-seq library prepared from rnh201Δ cells (KK-100) was sequenced to a depth of ~2 million reads, which were mapped to the yeast S. cerevisiae genome, allowing us to define rNMP locations along yeast nuclear and mitochondrial DNA with single-nucleotide resolution. This analysis uncovered widespread rNMP incorporation with a coverage of 0.449 and 19.5 rNMP reads/kb in the nuclear and mitochondrial genome, respectively (Fig. 2a and Supplementary Table 3). While broadly scattered, the rNMP sites in the nuclear and mitochondrial DNA were not randomly distributed (Fig. 2b,c). We found no major Watson/Crick strand bias in ribonucleotide distribution throughout the genome (Fig. 2a), and the number of rNMPs identified per nuclear chromosome was proportional to chromosome size (Fig. 2d).
Figure 2. Distribution of rNMP incorporation in the S. cerevisiae genome.
(a) Ribose-seq map of rNMPs in genomic DNA from rnh201Δ (KK-100) cells. The data, as peaks of rNMP reads in red, are shown for the individual nuclear chromosomes (Chr I–XVI) and the two strands of mitochondrial DNA (Chr M). The height of each peak corresponds to the number of reads. A comparison of nuclear and mitochondrial rNMP reads for Watson (W) and Crick (C) strands is also displayed. Raw sequencing reads are available at NCBI GEO39 under accession GSE61464. The proportion of 2.5 kb windows containing an observed number of rNMPs was calculated for nuclear (b) and mitochondrial (c) genomes and compared to random expectation based on Poisson frequencies. The P values calculated from chi-square goodness of fit test are shown (n = 10,847 and 3,347 for nuclear and mitochondrial, respectively). (d) Chromosomal distribution of rNMPs is compared to the size of each nuclear chromosome.
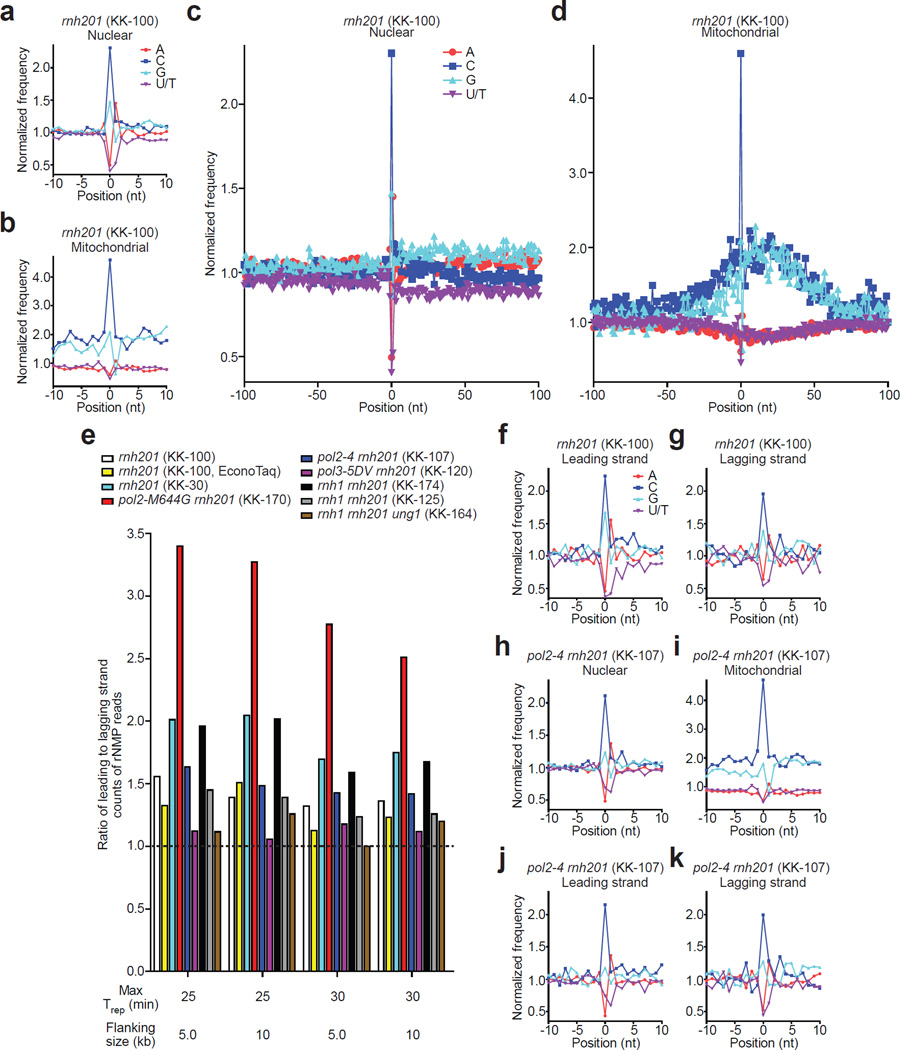
We determined the identity and relative frequencies of incorporated rNMPs, the reverse complement of the 5′ base of each read, as well as flanking bases for the nuclear and mitochondrial genomes in rnh201Δ cells. At the site of rNMP incorporation, we found that rCMP and rGMP were incorporated more frequently than expected from the G+C content, while rAMP and in particular rUMP were incorporated less frequently than expected from the A/T content in both nuclear and mitochondrial genomes, indicating a strong bias in the rNMP spectrum considering the A+T rich content of these genomes in yeast (62% and 83%, respectively) (Fig. 3a,b). Looking at the absolute composition of the genomic rNMPs, we found 44% rC, 28.1% rG, 15.4% rA and 12.5% rU in the nuclear genome, and 36.8% rC, 25.6% rA, 19% rG, and 18.7% rU in the mitochondrial genome (Supplementary Table 4). The difference in the base composition between nuclear and mitochondrial rNMPs is likely due to the higher A+T content of the mitochondrial genome.
Figure 3. Identity and sequence contexts of rNMP incorporation in S. cerevisiae genome.
(a,b) Normalized nucleotide frequencies relative to mapped positions of sequences from Ribose-seq library of rnh201Δ (KK-100) cells. Position 0 is the rNMP. (c, d), Zoom-out of frequencies. (e) Ratios of rNMPs on newly-synthesized leading to lagging strand for all Ribose-seq libraries. Early firing ARSs selected by their replication timing (Trep) were investigated for two different flanking sizes. (f,g) Normalized nucleotide frequencies relative to mapped sequence positions in newly-synthesized leading and lagging strands from the rnh201Δ (KK-100) library. ARSs with Trep of no longer than 25 min were selected with flanking size of 10 kb. (h–k) Normalized nucleotide frequencies relative to mapped sequence positions from a pol24rnh201Δ (KK-107) library. Reads were mapped to (a,c,h) nuclear genome, (b,d,i) mitochondrial genome, (f,j) leading strand or (g,k) lagging strand.
The high level of rCMPs and low level of rUMPs observed both for nuclear and mitochondrial DNA in the rnh201Δ library are not attributable to differential bypass by the Pfu-based DNA polymerase used for PCR (Supplementary Fig. 4 and Supplementary Table 5), as we also observed similar rNMP patterns using a Taq-based DNA polymerase (Supplementary Figs. 5a,b and 6a,b). Similarly, the nucleotide frequency derived from a Ribose-seq library constructed from another rnh201Δ strain (KK-30, Supplementary Table 2) was comparable to that obtained from strain KK-100 both for nuclear and mitochondrial sites (Supplementary Figs. 5c,d and 6c,d). Additional deletion of the gene coding for RNase H1 (rnh1Δ), generating rnh1Δ rnh201Δ strains (KK-174 and KK-125), did not affect the nucleotide frequency of rNMP incorporation (Supplementary Figs. 5e–h and 6e–h). While some variation in the absolute rNMP counts were found among these different libraries in the mitochondrial DNA (Supplementary Table 4), the high level of rCMP and low level of rUMP remained constant, as well as a preferred rNMP incorporation in G+C rich regions of the mitochondrial DNA. These data support a model in which rNMPs in yeast genomic DNA are present as single, di-, or tri-nucleotides, which are not substrates of RNase H114, and indicate that RNase H1 has minor impact on the distribution of genomic rNMPs.
In order to test whether the low frequency of rUMP incorporation was a consequence of removal by the uracil N-glycosylase, Ung1, we deleted the UNG1 gene in the RNases H-defective background (rnh201Δ rnh1Δ ung1Δ, strain KK-164) and mapped rNMP sites in these cells. Ung1 repairs dUMP from nuclear and mitochondrial DNA27. Whereas Ung1 does not act on uracil in RNA (e.g. rRNA)28, it is not known whether Ung1 can act on rUMP embedded in a DNA duplex. We found that the low level of rUMP incorporation in the chromosomal and mitochondrial genome of a rnh1Δ rnh201Δ ung1Δ strain is similar to rUMP incorporation in a rnh1Δ rnh201Δ strain, demonstrating that Ung1 does not target uracil in DNA (Supplementary Figs. 5i,j, 6i,j, and Supplementary Table 4).
Using a yeast assay of chromosomal double-stranded break (DSB) repair by DNA oligos carrying embedded rGMP, rUMP, or deoxyribose nucleotides only (Supplementary Fig. 7a), we demonstrated that Ung1 targets uracil from a dUMP but not an rUMP embedded in DNA, while RNase H2 targets only rNMPs (rGMP and rUMP in this experiment), but not dNMPs (Supplementary Fig. 7b and Supplementary Table 6). We attribute rNMP incorporation frequencies to the levels of corresponding dNTPs. dCTP and dGTP are typically the least abundant dNTPs7,29, and therefore might be depleted faster than dTTP and dATP, increasing the probability of rCMP and rGMP incorporation over rUMP and rAMP. These results are also consistent with the finding that rCMP and rGMP are the most frequently incorporated rNMPs by DNA polymerases in vitro under physiological dNTP/rNTP concentrations5,30. This ability of DNA polymerases to incorporate rNMPs into genomic DNA could potentially serve as a mechanism to continue replication under conditions in which one or more dNTP pools are depleted. In the presence of hydroxyurea, a known ribonucleotide reductase inhibitor, higher level of rNMPs are found incorporated in genomic DNA14. However, extensive rNMP incorporation would also result in increased breaks and genomic instability.
Pattern of sequences flanking rNMPs in S. cerevisiae DNA
Downstream of incorporated rNMPs, we found that the +1 position was most frequently dA and less frequently dG both in the nuclear and in the mitochondrial genomes, with 42 to 52% dA and 6 to 16% dG among all four deoxyribonucleotides (dA, dC, dG, and dT) (Supplementary Table 4). At the +1 position, dT was also frequent (31 – 40%) in the mitochondrial genome. In mitochondrial DNA, high level of dA or dT at the +1 position 3′ from the rNMP could reflect the high A+T content in the mitochondrial genome. It is also possible that the dA in +1 position influences rNMP incorporation by DNA polymerases. Alternatively, we speculate that dA in the +1 position might stabilize incorporated rNMPs, possibly by affecting base stacking and preventing its repair by non-RER mechanisms. It will be interesting to determine the nearest-neighbor thermodynamic parameters for single rNMPs in DNA duplex and, in particular, the stability trend for the base pair 3′ of the rNMP sites. We recently showed that single rGMPs embedded in a short DNA duplex have a marked effect on the elastic properties of DNA by altering the DNA structure at rNMP sites and the nucleotide 3′ to the rNMPs18. Thus, it is reasonable to think that the +1 position 3′ from the rNMP is prone to have altered structure, and it is the most critical site for signaling the presence of an rNMP in DNA because it is the closest nucleotide to the 2′-OH group of the rNMP.
Sites of rNMP incorporation were flanked by sequence contexts that differ between the nuclear and mitochondrial DNA genomes. While nucleotide frequencies up- and downstream of rNMP sites in the nuclear genome were largely similar to background frequencies (Fig. 3c), rNMP sites in mitochondrial DNA were primarily upstream of G+C-rich regions, concentrated in areas in which G+C content was 1.7 to 1.8-fold above the background (Fig. 3d). Interestingly, mitochondrial G+C tracts have been shown to have recombinogenic properties31, and mitochondrial DNA recombination has been suggested to initiate mitochondrial DNA replication in yeast (32 and references therein). Thus, it is possible that the presence of rNMP sites in yeast mitochondrial G+C clusters influences these recombination events in mitochondrial DNA.
rNMP incorporation by replicative DNA polymerases
We next analyzed rNMP incorporation in the newly-synthesized leading and lagging strands of yeast nuclear DNA. We selected 154 to 271 early-firing yeast autonomously replicating sequences (ARSs) (activated in the first 25 or 30 minutes, respectively) based on replication timing (Trep)33. We examined the type and abundance of rNMPs incorporated in regions 5 or 10 kb upstream and downstream from selected ARSs. This analysis was conducted using all our Ribose-seq libraries, including a library derived from yeast RNase H2-deficient cells containing the low fidelity Pol ɛ mutant (rnh201Δ pol2-M644G, Supplementary Table 2). Because yeast Pol ɛ is mainly responsible for leading strand synthesis during DNA replication, yeast cells containing the pol2-M644G mutation, which leads to increased rNMP incorporation, would be predicted to contain more rNMPs on the newly-synthesized leading than on the lagging strand15.
We found higher rNMP incorporation on the newly-synthesized leading strand of DNA replication (Fig. 3e), consistent with previous observations that the leading strand DNA Pol ɛ incorporates more rNMPs than the lagging strand Pol δ7. As expected, analysis of rNMPs from rnh201Δ pol2-M644G cells revealed a stronger bias of rNMP incorporation on the newly-synthesized leading strand compared to all other libraries (Fig. 3e). However, the increase in rNMP incorporation by the low fidelity Pol ɛ mutant did not change the overall rNMP spectrum, having similar patterns of rNMP incorporation to libraries derived from wild-type Pol ɛ strains (Supplementary Figs. 5k,l and 6k,l). Furthermore, we examined whether the spectrum of rNMP incorporation was different between the newly-synthesized leading and the lagging strand of DNA replication. Cells containing either rnh201Δ or rnh1Δ rnh201Δ mutations had a similar spectrum of rNMP incorporation on the leading and the lagging strand (Fig. 3f,g and Supplementary Fig. 8a,b). In contrast, mapping of rNMPs in yeast cells carrying a mutant allele of DNA Pol ɛ that is defective in proofreading activity (pol2–4, Supplementary Table 2), showed a lower frequency of rA versus rU in the nuclear but not in the mitochondrial genome (Fig. 3h,i; Supplementary Fig. 6m,n; and Supplementary Table 4), and showed a bias for lower rA than rU only on the newly-synthesized leading strand (Fig. 3j,k), which was not observed in libraries derived from wild-type nor low fidelity mutant Pol ɛ strains. The rNMP spectra for a strain with proofreading-defective DNA Pol δ, (pol3–5DV, Supplementary Table 2) were not different from those containing the wild-type Pol δ (Supplementary Figs. 5m,n; 6o,p; 8c,d; and Supplementary Table 4). These results suggest that DNA Pol ɛ can proofread rNMPs in DNA, in particular rUMPs, and that this activity is superior to DNA Pol δ, consistent with previous biochemical studies5,30.
Hotspots of rNMP incorporation in the S. cerevisiae genome
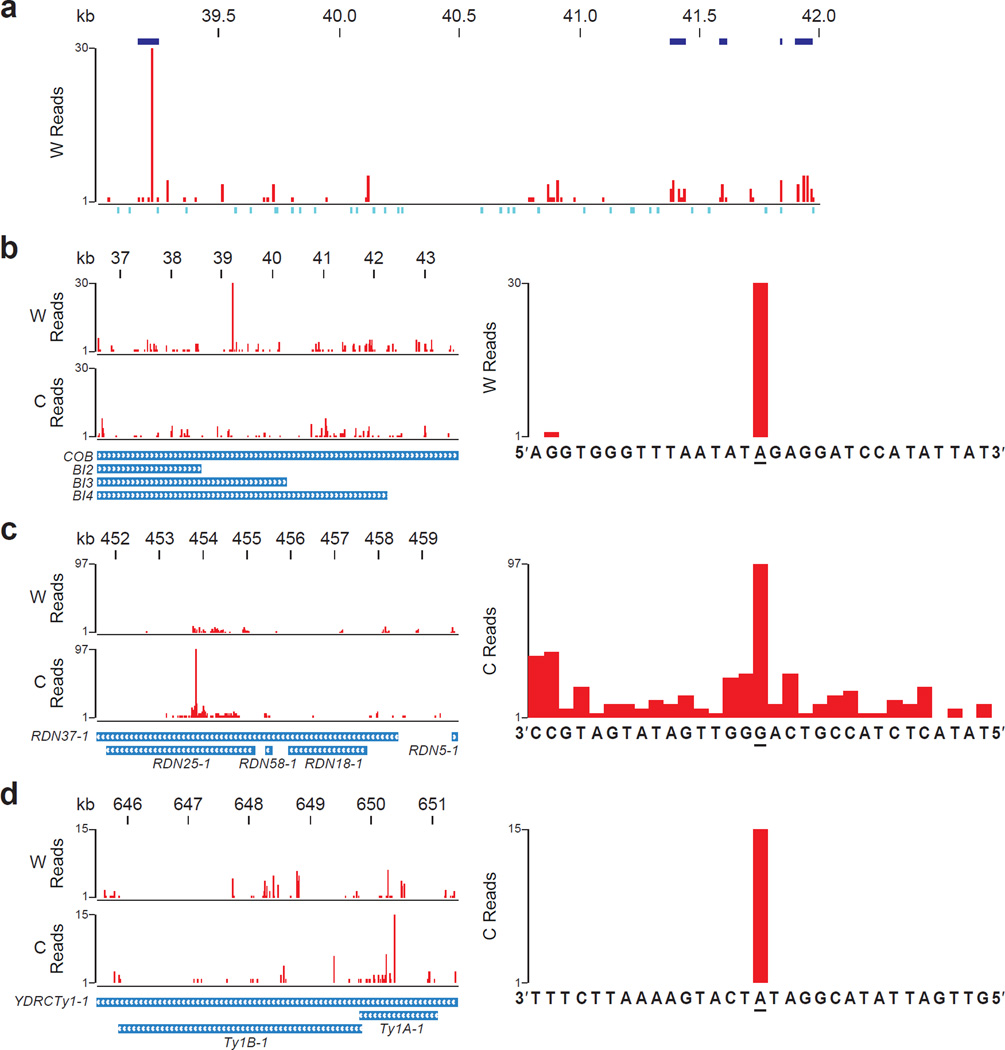
We performed two types of analyses to determine potential hotspots of rNMP incorporation in S. cerevisiae genome. We identify enriched regions of rNMP incorporation in genomic DNA from Ribose-seq data (see Methods). We found several regions of notable rNMP incorporation in mitochondrial DNA for each Ribose-seq library in this study (Supplementary Data; a few regions are displayed in Fig. 4a). Because this analysis excludes all reads aligning to more than one position in the genome, a second analysis was performed with respect to specific loci in order to identify single-nucleotide hotspots that were reproducibly present in multiple Ribose-seq libraries (see Methods). We identified hotspots of rNMP incorporation within sequences present in multiple copies per yeast cell, i.e. the mitochondrial genome (~80 copies)34, the ribosomal DNA (rDNA) repeats (~140) clustered on chromosome XII35, and the yeast retrotransposon (Ty), of which there are ~30 copies encoded on multiple chromosomes36. In mitochondrial DNA, we found a marked hotspot at an rAMP on the Watson strand in the cytochrome oxidase B gene (COB) and the overlapping maturase BI3 and BI4 genes (Fig. 4b and Supplementary Table 7), in addition to several other hotspots (Supplementary Table 7). In the rDNA locus, the strongest hotspot was found in gene RDN37-1 and the overlapping RDN25-1 at an rGMP (Fig. 4c and Supplementary Table 7). In the yeast Ty1 sequence, we found a hotspot at an rAMP in the coding sequence of TY1A-1 (Fig. 4d and Supplementary Table 7).
Figure 4. Hotspots of rNMP incorporation within S. cerevisiae mitochondrial DNA, rDNA repeat, and Ty1.
(a) Ribose-seq map of rNMPs in a 3 kb window (39,001–42,000) of mitochondrial DNA showing enriched regions of rNMP incorporation. W, Watson strand. Enriched regions with q-value < 0.001 are shown in blue above the plot. See Supplementary Data for all enriched regions. Positions of restriction sites used for genomic fragmentation are displayed below the plot in turquoise. (b) Map of rNMPs in COB mitochondrial locus (left).W, Watson, and C, Crick strand. Zoom-in map (right) with sequence at the hotspot site (underlined). (c) Map of rNMPs in the first of two rDNA repeat loci on Chr XII, based on alignment data from both loci of the reference genome (left). Zoom-in map (right) of the rDNA hotspot. (d) Map of rNMPs in the Ty1 locus YDRCTy11 on Chr IV based on multiple-alignment data from several Ty1 loci (left). Zoom-in map (right) of the Ty1 hotspot. Results are shown for rnh201Δ (KK-100) cells.
The occurrence of such hotspots indicates that there are preferred sites for rNMP incorporation in the mitochondrial genome, rDNA, and Ty1 sequences. In addition to the recombinogenic properties of mitochondrial G+C clusters discussed above, yeast rDNA and Ty are also active in recombination35,37. Frequent rNMP incorporation could trigger recombination, similar to rNMPs embedded in the mating type locus of Schizosaccharomyces pombe3. The rNMPs detected in Ty DNA could originate from cDNA rather than genomic DNA. Because rnh201 rnh1 cells have abundant Ty cDNA38, if rNMPs are incorporated in Ty1 during the process of reverse transcription, which forms the cDNA, we would expect a different rNMP pattern in Ty1 DNA in rnh201Δ rnh1Δ vs. rnh201Δ cells. Although we did not observe major differences in the rNMP spectra derived from rnh201Δ single mutant vs. rnh201Δ rnh1Δ cells at the Ty1 locus or in general (Supplementary Table 7, Fig. 3e and Supplementary Figs. 5 and 6), it would be of interest to conduct in vitro tests to determine whether Ty reverse transcriptase incorporates rNMPs frequently opposite to RNA and/or DNA, and whether it has a particular bias for rNMP incorporation.
Discussion
rNMP incorporation has been extensively studied in recent years; however, locating sites of rNMP incorporation in genomic DNA has not yet been possible. Alkaline cleavage of rNMPs and AtRNL ligation exclude Okazaki fragments and DNA abasic sites, allowing the construction of Ribose-seq libraries containing stably incorporated rNMP sites. Ribose-seq enabled us to determine the widespread but nonrandom distribution of rNMPs in budding yeast genomic nuclear and mitochondrial DNA with several hotspots. Our findings both validated the approach and uncovered new aspects of rNMP incorporation in the yeast genome. The observed strand bias incorporation on the newly-synthesized leading strand in wild-type and low fidelity Pol ɛ, and the specific rNMP pattern of proofreading deficient Pol ɛ provide strong support for the in vitro results obtained for these forms of Pol ɛ. rCMPs and rGMPs are more abundant than rAMPs and rUMPs, and frequently downstream of the rNMPs there is a dA. RNase H1 does not contribute substantially to rNMP incorporation, and Ung1 does not remove genomic uracil. It is possible that paucity of rUMP in DNA reflects inherent cleavage bias in other rNMP removal pathways such as Topoisomerase-mediated rNMP cleavage22 . It would be interesting to determine the rNMP spectrum in cells with defects in alternative rNMP removal pathways, either in RNase H2 wild-type or null cells growing under normal and/or stress conditions. Ribose-seq should allow us to better understand the impact of rNMPs on the structure and function of DNA and chromatin, and specific rNMP signatures may represent novel biomarkers for human diseases such as AGS, cancer and other degenerative disorders.
Online Methods
Yeast strain construction
Yeast strains used in this study are presented in Supplementary Table 2. Isogenic yeast haploid strains KK-100, KK-174, KK-107, and KK-120 were derived from E134 (MATα ade5-1 lys2-14A trp1-289 his7-2 leu2-3,112 ura3-52)40. KK-100 was made from E134 by deletion and replacement of RNH201 via transformation with a PCR product containing the hygMX4 cassette flanked by 50 nucleotides of sequence homologous to regions upstream and downstream of the RNH201 ORF. KK-174 was constructed from KK-100 by deletion and replacement of RNH1 via transformation with a PCR product containing the kanMX4. KK-107 was generated by introducing pol2-4 mutation into KK-100 via integration-excision using plasmid YIpJB141. KK-120 was made by introducing pol3-5DV mutation into KK-100 via integration-excision using plasmid p170-5DV42.
Isogenic yeast haploid strains KK-30, KK-125, KK-164, and KK-170 were derived from FRO-767,768 (hoΔ hmlΔ::ADE1 MATa-inc hmrΔ::ADE1 ade1 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL::HO leu2::HOcs mataΔ::hisG)43. KK-30 was made from FRO-768 by reversion of ade3::GAL::HO to intact ADE3 via transformation with a PCR product containing ADE3, followed by replacement of RNH201 with the hygMX4. KK-125 was constructed from KK-30 by replacement of RNH1 with the kanMX4 cassette. KK-164 was generated from KK-125 by replacement of UNG1 with the natMX4 cassette. KK-170 was made by introducing pol2-M644G mutation into KK-30 via integration-excision using plasmid p173-M644G44.
Isogenic yeast haploid strains KK-158,159 were derived from FRO-767,76843. KK-158,159 were constructed from FRO-767–768 by replacement of UNG1 with the hygMX4 cassette.
AtRNL ligation assay
Ribonucleotide (rNMP)-containing DNA oligonucleotide (oligo), Lig.47.R (see Supplementary Table 8), and its DNA-only control, Lig.47.D, were 5′ end-labeled with [γ-32P]ATP (PerkinElmer) by T4 polynucleotide kinase (New England BioLabs). Alkali treatment was carried out in 0.3 M NaOH for 2 hr at 55 °C. The resulting solution was neutralized and diluted. 100 nM of alkali-treated 5’-radiolabeled products were incubated in 50 mM Tris-HCl, pH 7.5, 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, 30 µM ATP (Sigma-Aldrich), and 1 µM AtRNL24 for 1 hr at 30 °C. After dilution, the ligated products and remaining substrates were treated with T5 exonuclease (NEB) for 2 hr at 37 °C. Aliquots were withdrawn after appropriate steps and quenched with 90% formamide. The products were analyzed by 15% (w/v) polyacrylamide, 8 M urea gel electrophoresis (urea-PAGE). 20–100 Oligo Length Standard (Integrated Device Technology) was used as a ladder. Following electrophoresis, gels were exposed to a phosphor screen overnight. Images were taken with Typhoon Trio+ (GE Healthcare) and obtained with ImageQuant (GE Healthcare). Band intensities were quantified by Multi Gauge V3.0 (Fujifilm).
3′ base bias for AtRNL ligation assay
rAMP, rGMP, rUMP, and rCMP-containing DNA oligos (Lig.30.rA, Lig.30.rG, Lig.30.rU, and Lig.30.rC, respectively; see Supplementary Table 8) were 5′ end-labeled with either hot [γ-32P]ATP (PerkinElmer) or cold ATP (Sigma-Aldrich) by T4 polynucleotide kinase (NEB). Each of hot rNMP-containing 5′-radiolabeled DNA oligos was mixed with other three cold equimolar DNA oligos. The mixtures were treated with 0.3M NaOH for 2 hr at 55 °C, neutralized, and diluted. 100 nM of alkali-cleaved products (25 nM of each base) were then incubated in 50 mM Tris-HCl, pH 7.5, 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, 30 µM ATP (Sigma-Aldrich), and either 1 µM or 200 nM AtRNL24 for 1 hr at 30 °C. After dilution, the resulting products were treated with T5 exonuclease for 2 hr at 37 °C. Aliquots were withdrawn after appropriate steps, quenched, and analyzed by urea-PAGE.
rNMP bypass assay
A DNA primer oligo, ByPrim (see Supplementary Table 8), was 5′ end-labeled with [γ-32P]ATP (PerkinElmer) by T4 polynucleotide kinase (NEB). The 5’-radiolabeled primer was annealed to either rCMP- or rUMP-containing template oligo (ByTemp.rC or ByTemp.rU, respectively). 100 nM of annealed substrates were incubated in 1X HF Buffer, 2 mM dNTPs, and 0.2 units of Phusion High-Fidelity DNA Polymerase (NEB) for 30 sec at 72 °C. The reactions were quenched and analyzed by urea-PAGE. Bypass probability was calculated as the band intensity at the +1 position plus all longer products divided by the intensity at the −1 position (preceding the rNMP) plus all longer products, as described in Kokoska et al., 200345.
Double-stranded break repair (DSB) assay with rNMP-containing oligos
Transformation with rNMP-containing DNA oligos, LEU2.rG and LEU2. rU (see Supplementary Table 8) and DNA-only oligos, LEU2.D and LEU2.dU, were done as described in Storici et al., 200743. Cells from each oligo transformation were plated to selective Leu− medium. For each transformation, 20 Leu+ transformants were selected. Colony PCR was performed on those transformants, amplifying with primers, LEU2.3 and LEU2.6, a 900-bp region in LEU2 locus, where a new StuI restriction site is expected. The resulting PCR products were treated with StuI (NEB) and analyzed by agarose gel electrophoresis to check the presence of the StuI restriction site.
Ribose-seq library construction to map rNMPs in DNA
Genomic DNA from S. cerevisiae cells grown in liquid rich medium containing yeast extract, peptone, and 2% (w/v) dextrose (YPD) for 2 days to stationary phase was extracted following the protocol “Preparation of Yeast Samples” in QIAGEN Genomic DNA Handbook. Genomic-tip 500/G (QIAGEN), Genomic DNA Buffer Set (QIAGEN), Proteinase K (QIAGEN), RNase A (QIAGEN), and Lyticase (Sigma-Aldrich) were used to extract genomic DNA from S. cerevisiae cells. Extracted genomic DNA was digested with SspI, DraI, and EcoRV (NEB) overnight at 37 °C to create a population of 500–3,000 bp genomic fragments with an average size of ~1.5 kb. Assuming that rNMPs, if present, could be located in any position of each genomic fragment, an average of 1.5 kb allows a reasonable window of rNMP capture. Following confirmation of digestion by Experion Automated Electrophoresis System (BIO-RAD), the fragments were tailed with dATP (Sigma-Aldrich) by Klenow fragment (exo−) (NEB) for 30 min at 37 °C. The resulting products were purified by spin-column (QIAGEN) and then ligated to preannealed double-stranded (ds) adaptors (Adaptor.L/Adaptor.S, see Supplementary Table 8) that contain single dT overhangs and a randomized 8-base unique molecular identifier (UMI) by T4 DNA ligase (NEB) overnight at 15 °C. The products were purified using AMPure XP beads (Beckman Coulter). All subsequent purifications were done using AMPure XP beads. The adaptor-ligated DNA fragments were incubated in 0.3 M NaOH for 2 hr at 55 °C to expose 2′,3′-cyclic phosphate and 2′-phosphate termini of DNA at rNMP sites, followed by neutralization and purification. The resulting single-stranded (ss) fragments were incubated in 50 mM Tris-HCl, pH 7.5, 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, 30 µM ATP (Sigma-Aldrich), and 1 µM AtRNL24 for 1 hr at 30 °C, followed by purification. The products and remaining fragments of DNA were treated with T5 exonuclease (NEB) for 2 hr at 37 °C to degrade the background of unligated, linear ss DNA, leaving self-ligated ss DNA circles intact. Treatment with the 1 µM Tpt124 in 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.1 mM DTT, 0.4% Triton X-100, and 10 mM NAD+ (Sigma-Aldrich) for 1 hr at 30 °C was used to remove the 2′-phosphate remaining at the ligation junction. After purification and resuspension, the libraries were PCR-amplified with one of the bar-coded primers, PCR.1.Index1–4, and PCR.2 (see Supplementary Table 8) using either Phusion High-Fidelity DNA Polymerase (NEB) or EconoTaq DNA Polymerase (Lucigen), confirmed by 6% polyacrylamide gel electrophoresis (PAGE), purified, and pooled for analysis by Illumina sequencing. 100 bp DNA Ladder (NEB) was used as a ladder. SYBR Gold Nucleic Acid Gel Stain (Life Technologies) was used to stain PAGE gels for visualization under ultraviolet light. Ribose-seq does not capture RNA primers of Okazaki fragments because the 5'-most rNMP is a 5'-triphosphate46, and T4 DNA ligase used to attach the sequencing adaptors absolutely requires a 5’-monophosphate47. Moreover, the rest of the primers are reduced to single nucleotides upon alkali treatment, and they will have no adaptor sequence ligated on. Ribose-seq also does not detect rNMP positions derived from residual RNA molecules or RNA/DNA hybrids not embedded in DNA (like cDNA) nor DNA abasic sites, which could have been ligated to the adaptor sequence by T4 DNA ligase. Following alkali treatment, RNA stretches are reduced to single nucleotides which are removed in subsequent purification steps; even if the 5′-most rNMP is captured, the rNMP-containing ss circle would not have any sequence to be aligned to the reference genome. Abasic sites undergo both β- and δ-eliminations to yield 5′-phosphate and 3′-phosphate ends48,49, which cannot be ligated by AtRNL. Because of the nature of alkaline hydrolysis within a stretch of rNMPs embedded in DNA, our Ribose-seq captures only the 5′-most rNMP of the stretch of two or more rNMPs. Moreover, Ribose-seq does not require rNMPs to be present at the same location from cell to cell, and can identify incorporated rNMPs with single-base precision.
DNA sequencing
Indexed sequencing libraries were mixed at equimolar concentrations and normalized to 10 nM. Libraries were sequenced on an Illumina MiSeq, and 50 cycle single-end reads were collected. Raw sequencing reads are available at NCBI GEO39 under accession GSE61464.
Code availability
Software and pipelines are available in Github (https://github.com/hesselberthlab/modmap) and as Supplementary Software.
Sequence alignment and processing
Reads were aligned to the S. cerevisiae genome (sacCer2) with bowtie using two different settings to report uniquely aligning and multiple aligning reads (`-m 1` and `--all`, respectively). Aligned reads in BAM format were processed to remove PCR duplicates using umitools (https://github.com/brwnj/umitools), which filters reads that contain duplicate UMIs and reports reads with unique UMIs. Reads in this study had an 8-base UMI incorporated during ligation, corresponding to the first 8 cycles of raw FASTQ sequence. Following UMI removal, read depths at each 5′ position were calculated with BEDTools50,51.
Nucleotide frequencies
Nucleotide frequencies for mapped rNMP positions (i.e. the 5′ position of each aligned read) were calculated and normalized to genome frequencies (nuclear and mitochondrial genomes in sacCer2). The identity of the rNMP base is the reverse complement of the 5′ base of each read. Nucleotide frequencies of downstream sequences of incorporated rNMPs, including +1 position, cannot be affected by our approach of capturing rNMPs in DNA because the rNMPs and their upstream sequences are captured, sequenced and aligned to the reference genome.
Replication correlations
The density and identity of rNMPs present on newly-synthesized leading and lagging strands were calculated relative to annotated origins of replication52 and were further categorized by replication timing33. Data were filtered for specified replication timings (e.g. 25 minutes following release into S phase) and distances relative to the middle of each ARS annotation (e.g. 5.0 kb up- and downstream of each ARS).
Determination of hotspots of rNMP incorporation in genomic DNA
Two different analyses were conducted to identify hotspots of rNMP incorporation in genomic DNA. Peak calling was performed with macs2 (version 2.1.0.20140616)53 with specific parameters (--keep-dup all --nomodel -s 25 --extsize 5 --call-summits). Peaks of length greater than 1,000 were filtered from further analysis, and remaining peaks with a q-value less than 0.001 were selected. A second analysis involved finding positions of rNMPs within the locus of interest with Ribose-seq signal greater than the mean plus three standard deviations for each library from rnh201Δ (KK-100), rnh201Δ (KK-100, EconoTaq), rnh201Δ (KK-30), rnh1Δ rnh201Δ (KK-174), and rnh1Δ rnh201Δ (KK-125) cells.
Data presentation and statistics
Graphs were made using GraphPad Prism 5 (GraphPad Software). Nonparametric two-tailed Mann-Whitney U-test54 was implemented for statistical analysis of AtRNL ligation efficiencies, rNMP bypass probabilities, and the percentages of StuI-cut Leu+ transformants in DSB repair assay. Chi-square goodness of fit test54 was used for statistical comparison of the distribution of rNMP reads to the expected Poisson distribution.
Supplementary Material
Acknowledgments
We thank N.V. Hud and L.D. Williams for support with urea-PAGE gels and for advice on this study and the manuscript; M. Goodman, B. Weiss and I.K. Jordan for suggestions on this study and the manuscript and assistance with data analysis; S. Garrey for AtRNL and Tpt1 protein purification; C. Cox for sequencing; Y. Shen for assistance on statistical analysis; A. Gombolay and L. Shetty for technical help; and all members of the Storici laboratory for advice in the course of the study. This research was supported by the National Science Foundation award number MCB-1021763 (to F.S.), the Georgia Research Alliance award number R9028 (to F.S.), an American Cancer Society Research Scholar Grant (to J.R.H.), a Damon Runyon-Rachleff Innovation Award from the Damon Runyon Cancer Research Foundation (to J.R.H.), and the University of Colorado Golfers Against Cancer (to J.R.H.).
Footnotes
Accession Codes
Raw sequencing reads are available at NCBI GEO under accession GSE61464.
Author Contributions
K.D.K. conducted all of the experiments of in vitro biochemical assays and Ribose-seq library construction, and most of the experiments of yeast in vivo DSB repair assay with oligos, with assistance from S.B. J.R.H. conducted the sequencing analysis. F.S., together with K.D.K. and J.R.H., designed experiments, assisted in data analysis, and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potenski CJ, Klein HL. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42:10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem. 2011;286:31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by pol V. PLoS Genet. 2012;8:e1003030. doi: 10.1371/journal.pgen.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3'-OH monoribonucleotide. J. Biol. Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rumbaugh JA, Murante RS, Shi S, Bambara RA. Creation and removal of embedded ribonucleotides in chromosomal DNA during mammalian Okazaki fragment processing. J. Biol. Chem. 1997;272:22591–22599. doi: 10.1074/jbc.272.36.22591. [DOI] [PubMed] [Google Scholar]
- 11.Randerath K, et al. Formation of ribonucleotides in DNA modified by oxidative damage in vitro and in vivo. Characterization by 32P–postlabeling. Mutat. Res. 1992;275:355–366. doi: 10.1016/0921-8734(92)90038-q. [DOI] [PubMed] [Google Scholar]
- 12.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O'Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu HC, et al. RNA intrusions change DNA elastic properties and structure. Nanoscale. 2014;6:10009–10017. doi: 10.1039/c4nr01794c. [DOI] [PubMed] [Google Scholar]
- 19.Caldecott KW. Molecular biology. Ribose--an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 20.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst) 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 24.Schutz K, Hesselberth JR, Fields S. Capture and sequence analysis of RNAs with terminal 2',3'-cyclic phosphates. RNA. 2010;16:621–631. doi: 10.1261/rna.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remus BS, Shuman S. Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases. RNA. 2014;20:462–473. doi: 10.1261/rna.043752.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res. 2014;42:5202–5216. doi: 10.1093/nar/gku118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 29.Fasullo M, Tsaponina O, Sun M, Chabes A. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 2010;38:1195–1203. doi: 10.1093/nar/gkp1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase varepsilon. DNA Repair (Amst) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 32.Gerhold JM, Aun A, Sedman T, Joers P, Sedman J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell. 2010;39:851–861. doi: 10.1016/j.molcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- 34.Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 35.Szostak JW, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 36.Hani J, Feldmann H. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 1998;26:689–696. doi: 10.1093/nar/26.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 38.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Gen. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 40.Clark AB, et al. Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res. 1999;27:736–742. doi: 10.1093/nar/27.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'-5' exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin YH, et al. The 3'-5' exonuclease of DNA polymerase delta can substitute for the 5' flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 46.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CWJ. RNA-protein interactions: a practical approach. 1st edn. New York, U.S.A.: Oxford University Press; 1998. [Google Scholar]
- 48.Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Bailly V, Verly WG. Possible roles of beta-elimination and delta-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 1988;253:553–559. doi: 10.1042/bj2530553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siow CC, Nieduszynska SR, Muller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–D686. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 3rd edn. New York, U.S.A.: W.H. Freeman; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.