Abstract
Background
Osteosarcoma is the most common primary bone tumor in adolescents associated with skeletal development. The molecular pathogenesis of osteosarcoma has not been completely determined, although many molecular alterations have been found in human osteosarcomas and cell lines.
Questions/purposes
We questioned whether (1) we could identify gene expression in osteosarcoma specimens that differs from normal osteoblasts and mesenchymal stem cells and (2) this would provide clues to the molecular pathogenesis of osteosarcoma?
Methods
The whole-genome transcriptional profiles of osteosarcomas, including two primary biopsy specimens, two cell lines, two xenografts derived from patient specimens, and one from normal osteoblasts and from mesenchymal stem cells, respectively, were quantitatively measured using serial analysis of gene expression. A statistical enrichment was performed, which selects the common genes altered in each of the osteosarcomas compared with each of the normal counterparts independently.
Results
Sixty (92%) of 65 total genes that were at least twofold downregulated in osteosarcoma compared with osteoblasts and mesenchymal stem cells, could be classified in four categories: (1) seven genes in the insulin–like growth factor (IGF) signaling axis, including three of the IGF-binding proteins (IGFBP) and three of the IGFBP-related proteins (IGFBPrP); (2) eight genes in the transforming growth factor-β (TGF-β)/bone morphogenetic protein (BMP) signaling cascade; (3) 39 genes encoding cytoskeleton and extracellular matrix proteins that are regulated by TGF-β/BMPs; and (4) six genes involved in cell cycle regulation, including tumor suppressors TP63 and p21.
Conclusions
Based on these transcriptional profiles, a coordinated theme of clustered gene deregulation in osteosarcoma has emerged. Cell proliferation driven by the IGF axes during bone growth is unrestrained owing to downregulation of IGFBPs and cell cycle regulators. Tumor cells may be maintained in an undifferentiated state secondary to impaired TGF-β/BMP signaling. This well-preserved pattern suggests that the alterations in the signaling axes of IGF-1 and TGF-β, in concert with cell cycle regulators, may be an important pathogenic basis of osteosarcoma.
Clinic Relevance
This study provides a possible molecular basis of pathogenesis of osteosarcoma. This may help to develop new therapeutic targets and strategy for this disease. Preclinical and subsequently clinical testing of inhibitors of the IGF-1 and TGF pathways would be warranted.
Introduction
Osteosarcoma is a high-grade primary bone malignancy most commonly seen in children and young adults [18, 19]. The survival of patients with osteosarcoma has not improved during the past three decades since the advent of adjuvant chemotherapy, despite multiple clinical trials of intensified regimens or newer agents. Better understanding of the disease is needed. Although the etiology of osteosarcoma is not well understood [8], several factors suggest a correlation between skeletal development and initiation of osteosarcoma. First, the peak incidence of osteosarcoma coincides with a period of rapid bone growth. In addition, an earlier peak age in girls corresponds to the earlier age of their growth spurt, also suggesting a relationship with skeletal development. Moreover, most osteosarcomas occur near the major growing joints such as the distal femur, proximal tibia, and proximal humerus, which contribute to the majority of longitudinal bone growth. Other aspects of pathogenesis may be inferred from the genetic predisposition syndromes such as Li-Fraumeni syndrome (TP53 mutation; patients with this condition are at increased risk multiple cancers) and hereditary bilateral retinoblastoma (Rb mutation) [10, 11]. It appears that loss of function of tumor suppressor genes played a role in the tumorigenesis in osteosarcoma.
Bone growth is spatially and temporally controlled by systemic endocrine and local growth factors [38]. In response to growth hormone, the multipotent mesenchymal stem cells (MSCs) are recruited to the growth plate and differentiate into proliferating chondroblasts. Chondroblasts undergo hypertrophy and produce cartilage matrix underneath the growth plate. Bone marrow-associated MSCs differentiate into osteoblasts and replace cartilage with osteoid in concert with hematopoietic-derived osteoclasts. Osteoid then is mineralized with calcium and phosphate to form bone [4]. The majority of terminal osteoblasts undergo programmed cell death (apoptosis) with a few becoming osteocytes incorporated in the Haversian system. The transforming growth factor-β (TGF-β) superfamily members, especially the bone morphogenetic proteins (BMPs), are important regulators in this process, as is well documented in animal models and in vitro studies [4, 5]. The biologic action mediated by TGF-β signaling is tightly regulated at multiple levels [29]. However, the molecular basis for the relationship between bone growth and occurrence of osteosarcoma is not well understood.
Bone growth is promoted by insulin-like growth factors (IGF) systematically and locally [33]. IGF-1, the most abundant growth factor in bone, is the mediator of anabolic effects of growth hormone. It mostly is synthesized in the liver and also is secreted by MSCs and osteoblasts in an autocrine fashion. It also stimulates proliferation. In animal models, IGF level has been correlated with size and weight at birth [2, 17] and bone growth [36]. The IGF-signaling axis also includes six high-affinity IGF binding proteins (IGFBPs) and five low-affinity IGFBP-related proteins (IGFBPrPs). IGF signaling is tempered by its binding proteins in a negative feedback loop. It is among the few peptide hormones known to have binding proteins, which is usually characteristic of lipid-soluble hormones. Moreover, IGF-1, along with IGFBPs, reaches a peak level at puberty in response to growth hormone and decreases with age, in parallel with the incidence of osteosarcoma [28]. The mitogenic IGF-1 was reported to be involved in tumorigenesis in several tumor models [16]. In osteosarcoma, IGF-1 levels are associated with metastatic behavior of tumors in animal models [25]. The relationship between IGF signaling and osteosarcoma pathogenesis, however, is not entirely clear.
To better understand the molecular basis of osteosarcoma, we used a comprehensive approach to compare the transcriptional profile of primary tumor cells and cell lines with their normal cellular counterparts. In this study, serial analysis of gene expression (SAGE) was used to directly measure the transcriptome without any prior selection of genes for inclusion [34, 39]. Moreover, the absolute quantity of each transcript obtained for each sample with this approach allows a straightforward comparison to public databases to facilitate the exchange of information regarding rare diseases such as osteosarcoma [13]. We asked the following questions: (1) Can we identify gene expression in osteosarcoma specimens that differs from normal osteoblasts and MSCs; and (2) will this provide clues to the molecular pathogenesis of osteosarcoma?
Materials and Methods
Experimental Design
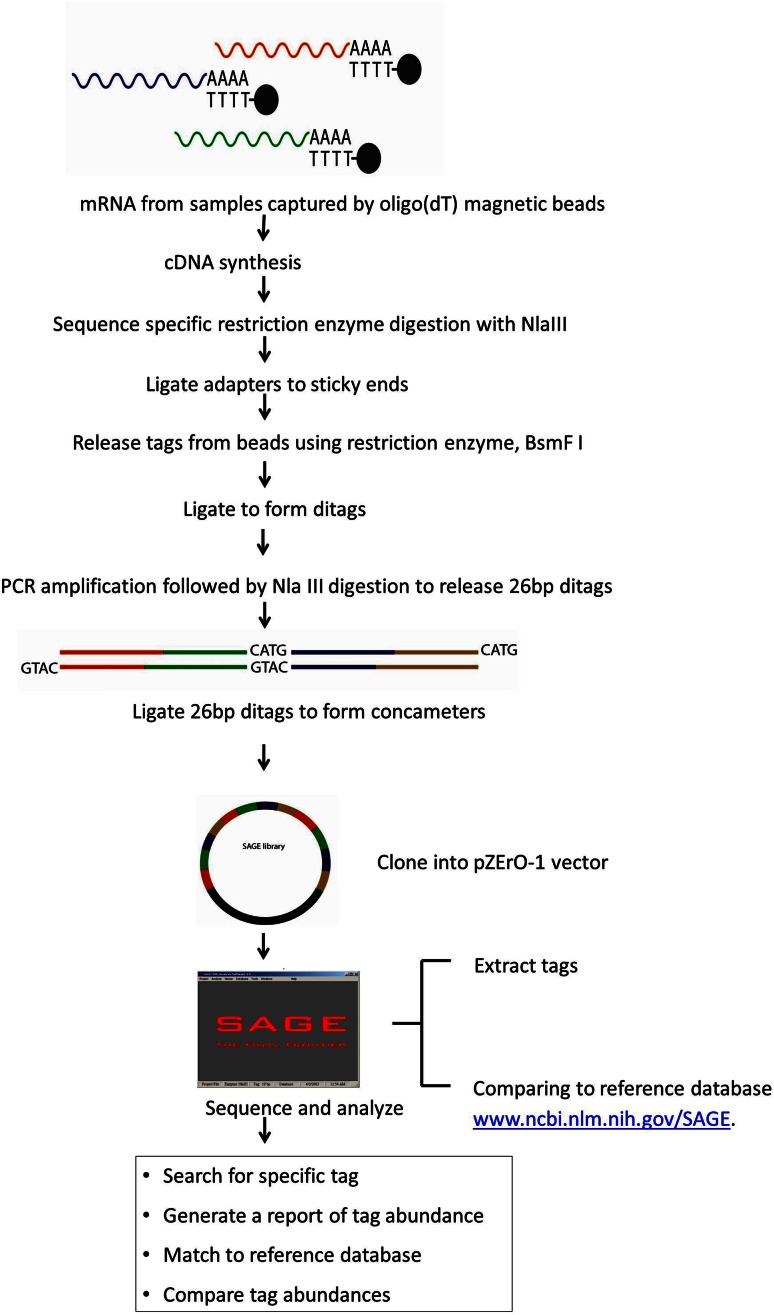
To compare the gene expression profile of osteosarcoma and its normal counterparts, we constructed SAGE libraries from two osteosarcoma biopsy specimens, two cell lines, and two xenografts derived from patient specimens–one from normal osteoblasts and one from MSCs (Fig. 1).
Fig. 1.
A schematic for SAGE library construction is shown.
Osteosarcoma Specimens
The osteosarcoma standard cell line SaOS-2 was purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). In SaOS-2, both alleles of p53 are deleted. All other specimens were derived from patients with osteosarcoma who were treated at Memorial Sloan-Kettering Cancer Center. Specimen OS256 was obtained by biopsy from a 14-year-old female patient with osteosarcoma of the proximal tibia. Specimen OS259 was obtained by biopsy from a 13-year-old male patient with osteosarcoma of the distal femur. The osteosarcoma cell line, OS160CL, was derived from a lung metastatic lesion resected from an 11-year-old female patient with osteosarcoma of the proximal femur using described methods [27]. The OS63CL was derived from the primary surgical specimen resected from a 31-year-old male patient with osteosarcoma of the proximal humerus. Histologic diagnosis was confirmed by a pathologist (HD, AH). Cells were maintained as a monolayer in modified Eagle’s-α media (α –MEM) supplemented with 10% fetal calf serum (Life Technologies, Bethesda, MD, USA), 100 units/mL penicillin, and 3 mg/mL streptomycin at 37°C in a humidified atmosphere with 5% CO2. Human MSCs (provided by Pamela Robey PhD, National Institutes of Health, Bethesda, MD, USA) were further induced toward osteoblastic differentiation with 50 μmol/L ascorbic acid and 10 mmol/L of β-glycerolphosphate for 2 weeks [27] before total RNA was extracted. The osteoblastic differentiation was confirmed by detection of osteoblast-specific markers including alkaline phosphatase, osteocalcin, and collagen type Iα by reverse transcription–polymerase chain reaction (RT-PCR) as described previously [26]. All the cultured cells were checked monthly and shown to be free of mycoplasma contamination. The MSCs, MSC89R, were selected for SAGE analysis based on their high colony-forming efficiency and multilineage differentiation potential [7]. Briefly, the mononuclear cell fraction was recovered from a 10-mL aspirate of the iliac crest obtained from a healthy donor using a Ficoll (Ficoll-PaqueTM; Pharmacia, Piscataway, NJ, USA) gradient, and then cultured in α-MEM supplemented with 100 μg/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L L-glutamine, and 20% fetal calf serum (Atlanta Biologicals, Lawrenceville, GA, USA) lot selected for rapid growth of the cells. After 24 hours the nonadherent cells were removed and the adherent layer cultured until it reached 50% to 70% confluence. Cells subsequently were passed twice and then collected for SAGE analysis or alternatively plated in 100-mm dishes at 10 cell/cm2, cultured for 10 to 14 days, and a single cell-derived colony (clone forming-unit fibroblast 35 [CFUF35]) was isolated using cloning cylinders for analysis by MicroSAGE [32].
Osteosarcoma Xenografts
Tumor cells from patient-derived cell lines OS160CL and OS63CL were used to establish xenografts in mice as previously described according to a protocol approved by the Memorial Hospital Institutional Animal Care and Use Committee [37]. Xenografts were resected when the size was approximately 1.5 cm in diameter. Specimens were histologically confirmed to be osteosarcoma by a pathologist (HD, AH) using standard hematoxylin and eosin staining and were named M160xeno and M63xeno corresponding numerically with their respective parental cell lines.
Construction of SAGE Libraries
Total RNA was extracted with an RNeasy® mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. SAGE libraries of normal human osteoblasts, two primary osteosarcoma tumor specimens, two osteosarcoma xenografts (M160xeno, M63xeno), and two cultured osteosarcoma cell lines (SaOS-2, OS160CL) were constructed with an iSAGETM Kit from InvitrogenTM (Carlsbad, CA, USA) according to the manufacturer’s instructions. Sequencing reactions were performed using a BigDye® Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems®, Foster City, CA, USA), and data were generated with an ABI 3100 sequencer (Applied Biosystems®). To yield tags, the raw data were processed with the SAGE2000 Version 4.5 software (Invitrogen) provided by the kit manufacturer. The SAGE library of mesenchymal stem cells, MSC89R, using the same methodology as described by Tremain et al. [32]. The SAGEmap database, downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SAGE), was used as a reference for tag UniGene identification [13]. Two additional SAGE libraries were included for analysis, including a single cell-derived MSC SAGE library named CFUF35 using micro-SAGE methodology with 16,407 tags in total [32] and a MSC SAGE library using the standard SAGE methodology with 202,962 tags in total [30].
Quantitative Real-time RT-PCR
Real-time fluorescent quantitative RT-PCR was used to validate a fraction of genes identified by SAGE to be differentially expressed using predesigned primers and probes (Applied Biosystems®) including inhibin β A, Assay ID: Hs00170103_m1; Smad4, Hs00232068_m1; CTGF, Hs00170014_m1; IGFBP7 (Mac25), Hs00266026_m1; p63, Hs00186613_m1; IGFBP6, Hs00181853_m1; IGFBP4, Hs00181767_m1; IGFBP3, Hs00181211_m1. The housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control for template loading. Thermal cycling was performed with a 7500-fast real-time PCR system (Applied Biosystems®) using a 2 × TaqMan® Fast Universal PCR Master Mix provided by the manufacturer [37]. The relative quantity was calculated based on standard 2−ΔT method as described previously [37]. In total, eight SAGE libraries were constructed, including six of osteosarcomas, one of MSCs (MSC89R), and one of osteoblasts. At least 30,000 tags were obtained for each library to ensure sufficient coverage for the majority of the transcripts. The osteoblast SAGE library was constructed after 2 weeks of osteoblastic induction of the MSCs. Osteoblastic differentiation was observed by increased mRNA expression of alkaline phosphatase and type Iα collagen by RT-PCR (data not shown). It was further confirmed by a steep increase in the number of tags representing collagen type Iα in the SAGE library of osteoblasts and by lack of cartilage markers such as type II and type X collagen, which were present in the SAGE library of MSC89R. A clustering analysis was performed using BRB-Array Tools Version 3.6 (Biometrics Research Branch, Division of Cancer Treatment & Diagnosis, National Cancer Institute, Bethesda, MD, USA), including eight SAGE libraries established in this study and two additional libraries of MSCs [30, 32] (Fig. 2). Considering the wide range of variation in the six osteosarcomas profiled in this study, a two-step enrichment was adapted in the statistical analyses to identify the common transcriptional alterations in all osteosarcomas and to filter out factors contributed by the individual samples possibly attributable to the growth environment (Fig. 3). A software package, the FosterTool, was designed and developed by one of us (WZ) to facilitate the statistical analyses of the data obtained by SAGE and is available for open access at http://fosterfoundation.com. In the first step, each SAGE library of osteosarcoma was pairwise compared with that of MSC89R and of osteoblast, respectively. In the second step, only gene tags significantly differentially expressed in every osteosarcoma versus osteoblast and MSC89R simultaneously were pooled as true characteristic genes of osteosarcoma. Gene tags were grouped according to their functions as described in the database (http://www.ncbi.nlm.nih.gov).
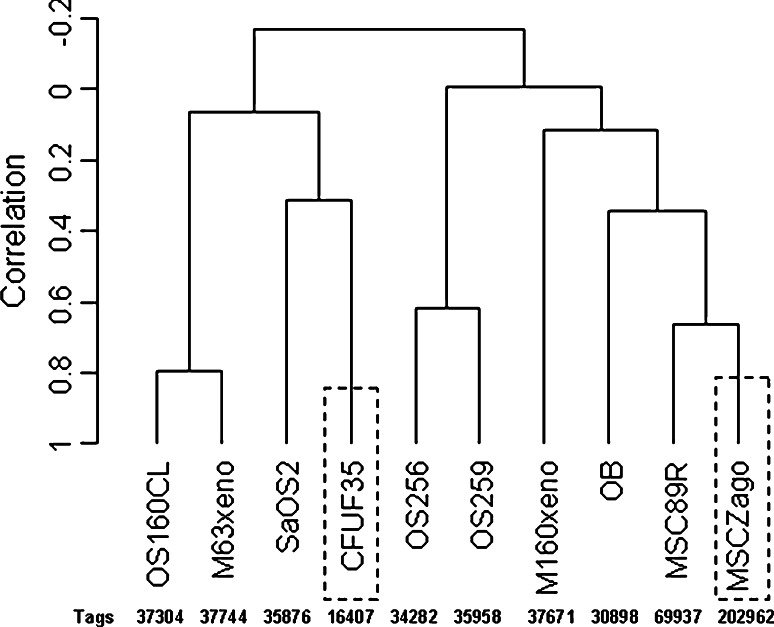
Fig. 2.
Eight SAGE libraries obtained in this study, along with CFUF35, a single cell-derived MSC SAGE library established using MicroSAGE technique [32] (dashed boxes) and another human MSC SAGE library (MSCZago) using standard methodology (dashed boxes) [30] were normalized to tags/50,000 in total and were analyzed using BRB-Array Tools Version 3.6. A dendrogram was drawn to reveal the relationship between the SAGE libraries. The total numbers of tags obtained for each of the libraries before normalization are shown.
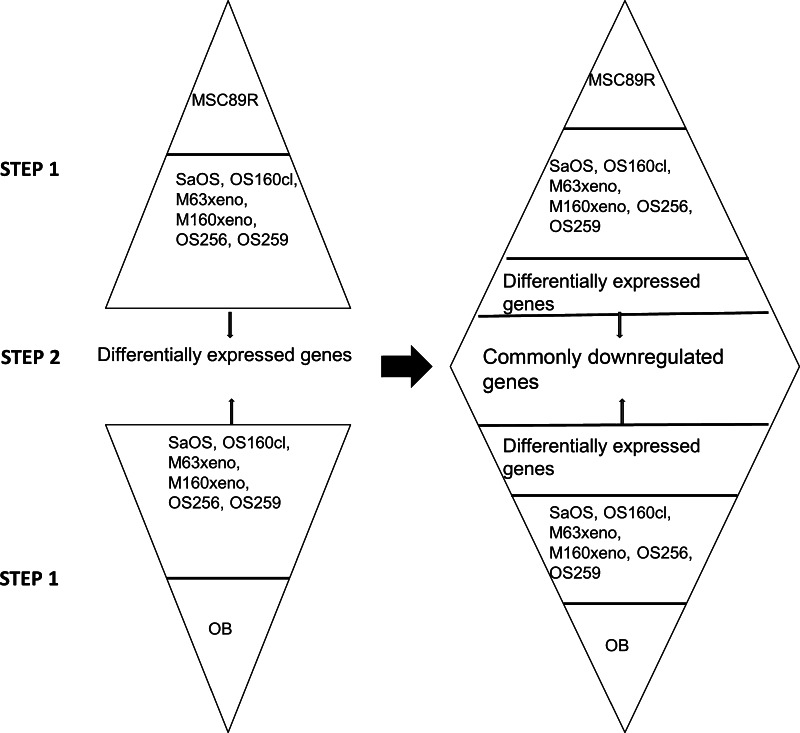
Fig. 3.
A two-step enrichment was adapted in the statistical analyses to identify the common transcriptional alteration in all osteosarcomas and to filter out factors contributed by the individual samples possibly owing to the microenvironment. In Step 1, each osteosarcoma SAGE library was compared with that of osteoblasts and MSC89R independently. Genes (tags), which were differentially present between tumor and normal, are pooled together. A second step identifies genes, which are commonly downregulated in osteosarcoma compared with normal osteoblasts (OB) and MSC89R.
Statistical Analyses
All statistical analyses were performed by statisticians (YZ, WZ) at the Department of Applied Mathematics and Statistics (State University of New York at Stony Brook, Stony Brook, NY, USA). A computer program was developed (NN) at the Department of Computer Science (State University of New York at Stony Brook) to facilitate the parallel comparison between the SAGE libraries. All statistical analyses were two-tailed and a probability less than 0.05 was regarded as statistically significant. The clustering analysis was performed using BRB-Array tools Version 3.6.
Results
Four categories of genes accounted for the dominant majority of the genes that were downregulated in osteosarcoma SAGE libraries (92%; 60 of 65) compared with their normal counterparts. The first group of seven genes identified by SAGE includes those involved in the IGF signaling (Table 1). Three IGFBPs and three IGFBPrPs, seen abundantly in MSCs and osteoblasts, were minimally present in any of the six osteosarcoma SAGE libraries despite IGF-1 levels that were similar in MSC89R (129/50,000 tags), osteoblasts (66/50,000 tags), and osteosarcomas (mean, 67/50,000 tags). The differential expression was validated by TaqMan® real-time RT-PCR in a subset of the genes (Fig. 4), including IGFBP3, IGFBP4, IGFBP6, IGFBPrP1 (IGFBP7), and IGFBPrP3. The fold difference (Fig. 4) measured by SAGE (normalized to tags per 50,000 total tags in each library) and detected by real-time RT-PCR in these eight genes was highly comparable (r2 = 0.8813).The second group of genes found to be downregulated in osteosarcoma includes eight genes involved in TGF- β signaling (Table 1). The identified genes are involved in regulating TGF-β signaling at various levels, including ligand (inhibin β), ligand release (TGF-β latent binding protein), ligand activation (thrombospondin 1), receptor internalization, and signal transduction (caveolin 1). Alterations also are present in extracellular antagonists (follistatin-like 1, Gremlin1, and CSRP1), including members of the IGFBPrPs (CTGF, IGFBP7, and CYR61) in the IGF-1 axis, which are important regulators of TGF-β signaling [1, 5, 14]. The third group tags identified to be downregulated in osteosarcoma encompass a group of genes largely regulated by TGF-β signaling (Table 2). These include genes involved in the extracellular matrix (ECM) formation (Type IV collagens, Type VI collagen, vimentin), ECM remodeling (SERPINs, TIMPs, LOX), and many constituents of the cytoskeleton [15].
Table 1.
The differentially expressed SAGE tags*
| Gene | SAGE tag | MSC | OB | SaOS | M160xeno | OS160cl | M63xeno | OS256 | OS259 | Unigene ID | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IGF | TGGGTGAGCC | 23 | 14 | 2 | 0 | 0 | 0 | 4 | 6 | 520898 | CTSB Cathepsin B |
| CATATCATTA | 309 | 354 | 1 | 1 | 0 | 0 | 13 | 21 | 119206 | IGFBP7 insulin-like growth factor binding protein 7 | |
| TTTGCACCTT | 254 | 274 | 0 | 5 | 25 | 0 | 34 | 9 | 410037 | CTGF Connective tissue growth factor | |
| AGTGTCTGTG | 56 | 20 | 0 | 0 | 0 | 0 | 9 | 1 | 8867 | CYR61 Cysteine-rich, angiogenic inducer, 61 | |
| ACTGAGGAAA | 53 | 9 | 0 | 0 | 0 | 0 | 2 | 3 | 450230 | IGFBP3 Insulin-like growth factor binding protein 3 | |
| ATGTCTTTTC | 90 | 47 | 9 | 3 | 0 | 0 | 10 | 4 | 1516 | IGFBP4 insulin-like growth factor binding protein 4 | |
| GGCCCCTCAC | 42 | 26 | 0 | 1 | 0 | 0 | 5 | 0 | 274313 | IGFBP6 Insulin-like growth factor binding protein 6 | |
| TGF-β | GTGGAATAAA | 53 | 26 | 0 | 0 | 0 | 0 | 5 | 2 | 512776 | LTBP2 Latent transforming growth factor β binding protein 2 |
| CAGTCAATAT | 22 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 28792 | INHBA Inhibin, beta A | |
| AGGTCTTCAA | 34 | 30 | 0 | 0 | 0 | 0 | 2 | 1 | 164226 | THBS1 Thrombospondin 1 | |
| CCGTGACTCT | 60 | 81 | 0 | 4 | 0 | 0 | 11 | 6 | 269512 | FSTL1 Follistatin-like 1 | |
| TCCTGTAAAG | 60 | 18 | 0 | 0 | 0 | 0 | 7 | 0 | 74034 | CAV1 Caveolin 1, caveolae protein, 22 kDa | |
| GGCTGTACCC | 48 | 40 | 2 | 6 | 0 | 0 | 0 | 2 | 108080 | CSRP1 Cysteine and glycine-rich protein 1 | |
| AATCTGTAAC | 25 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 40098 | GREM1 Gremlin 1 homolog, cysteine knot superfamily | |
| GTGTGTTTGT | 563 | 505 | 1 | 34 | 2 | 0 | 30 | 32 | 369397 | TGFBI Transforming growth factor, beta-induced, 68 kDa |
* Original tag number before normalization; SAGE = serial analysis of gene expression; IGF = insulin growth factor; TGF-β = transforming growth factor-β.
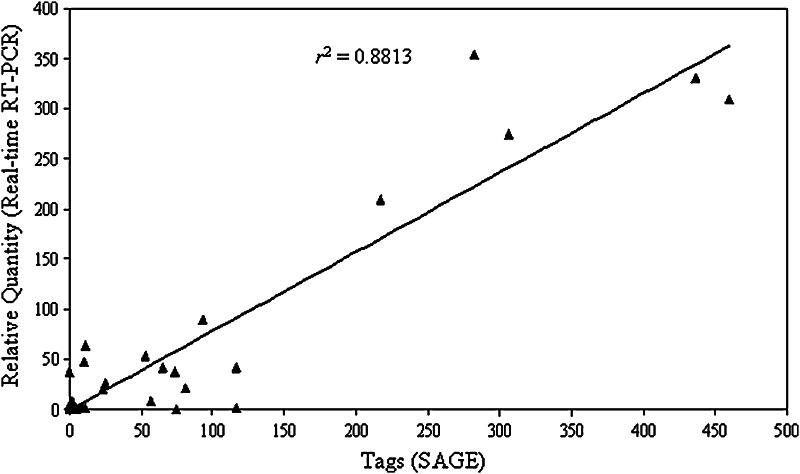
Fig. 4.
The quantities of eight gene tags measured directly by SAGE are shown in the x-axis after normalization (tags/50,000 tags). The fold difference of mRNA expressions as measured by TaqMan® real-time RT-PCR is shown in the y-axis using predesigned primers and probes (Applied Biosystems). The genes were validated included inhibin β A, CTGF, IGFBP7 (IGFBPrP1), p63, IGFBP6, IGFBP4, and IGFBP3. The correlation coefficient between these two methods is calculated as r2 = 0.8813.
Table 2.
Differentially expressed SAGE tags* involved in ECM and cytoskeleton in osteosarcoma as compared with its normal counterparts
| Gene | SAGE tag | MSC | OB | SaOS | M160xeno | OS160cl | M63xeno | OS256 | OS259 | Unigene ID | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ECM | GACCGCAGGA | 60 | 140 | 0 | 0 | 0 | 0 | 1 | 5 | 17441 | COL4A1 Collagen, type IV, alpha 1 |
| GTGCTAAGCG | 27 | 30 | 0 | 1 | 0 | 0 | 1 | 2 | 420269 | COL6A2 Collagen, type VI, alpha 2 | |
| GGAAGCTAAG | 149 | 49 | 0 | 0 | 0 | 0 | 5 | 8 | 136348 | POSTN Periostin, osteoblast specific factor | |
| GCCCCCAATA | 521 | 234 | 36 | 80 | 5 | 0 | 51 | 61 | 445351 | LGALS1 Lectin, galactoside-binding, soluble, 1 (galectin 1) | |
| TCCAAATCGA | 260 | 57 | 0 | 6 | 0 | 0 | 11 | 6 | 533317 | VIM Vimentin | |
| GCCTGTCCCT | 45 | 22 | 0 | 0 | 0 | 0 | 7 | 3 | 821 | BGN Biglycan | |
| GCCATAAAAT | 20 | 21 | 0 | 0 | 0 | 0 | 7 | 1 | 1908 | PRG1 Proteoglycan 1, secretory granule | |
| ECM | TAAAAATGTT | 138 | 116 | 0 | 0 | 0 | 0 | 2 | 1 | 414795 | SERPINE1 (nexin, plasminogen activator inhibitor type 1), member 1 |
| Remodeling | GGTTATTTTG | 29 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 414795 | SERPINE1 (nexin, plasminogen activator inhibitor type 1), member 1 |
| GCTGACGTCA | 42 | 84 | 0 | 0 | 0 | 0 | 2 | 0 | 414795 | SERPINE1 Serine (nexin, plasminogen activator inhibitor type 1), member 1 | |
| TATTCACTAA | 26 | 24 | 0 | 1 | 0 | 0 | 1 | 2 | 38449 | SERPINE2 Serine (nexin, plasminogen activator inhibitor type 1), member 2 | |
| AGCCTTTGTT | 133 | 72 | 5 | 17 | 2 | 0 | 13 | 39 | 241579 | SERPINH1 (heat shock protein 47), member 1, (collagen binding protein 1) | |
| GAGAGTGTCT | 34 | 28 | 1 | 4 | 0 | 0 | 3 | 1 | 522632 | TIMP1Tissue inhibitor of metalloproteinase 1 | |
| TGTCATCACA | 55 | 125 | 0 | 18 | 1 | 1 | 6 | 7 | 116479 | LOXL2 Lysyl oxidase-like 2 | |
| TATGTATTTC | 40 | 24 | 0 | 4 | 0 | 0 | 0 | 0 | 102267 | LOX Lysyl oxidase | |
| TCTTGTGCAT | 72 | 48 | 9 | 15 | 1 | 0 | 3 | 6 | 2795 | LDHA Lactate dehydrogenase A | |
| Cytoskeleton | TGCTAAAAAA | 35 | 16 | 0 | 1 | 0 | 0 | 4 | 3 | 474751 | MYH9 Myosin, heavy polypeptide 9, non-muscle |
| CCCTTAGCTT | 37 | 17 | 5 | 0 | 0 | 0 | 1 | 2 | 190086 | MRCL3 Myosin regulatory light chain MRCL3 | |
| GGAGTGTGCT | 151 | 69 | 0 | 22 | 1 | 0 | 6 | 5 | 504687 | MYL9 Myosin, light polypeptide 9, regulatory | |
| GTGCTGAATG | 203 | 60 | 26 | 26 | 0 | 1 | 11 | 13 | 505705 | Myosin, light polypeptide 6, | |
| TTAAAGATTT | 131 | 41 | 1 | 0 | 0 | 0 | 1 | 1 | 133892 | TPM1 Tropomyosin 1 (alpha) | |
| GACCAGGCCC | 110 | 73 | 0 | 2 | 0 | 0 | 11 | 0 | 300772 | TPM2 Tropomyosin 2 (beta) | |
| AAAATATTTT | 36 | 10 | 0 | 0 | 0 | 0 | 3 | 0 | 509765 | Actinin, alpha 1 | |
| AAGATCAAGA | 34 | 39 | 0 | 1 | 7 | 4 | 0 | 0 | 1288 | ACTA1 Actin, alpha 1, skeletal muscle | |
| CTAGCCTCAC | 271 | 62 | 37 | 6 | 0 | 0 | 14 | 5 | 514581 | ACTG1 Actin, gamma 1 | |
| GCTTTATTTG | 209 | 64 | 5 | 4 | 0 | 0 | 15 | 8 | 456859 | ACTB Actin, beta | |
| TTCTGTGAAT | 64 | 54 | 2 | 3 | 2 | 6 | 5 | 6 | 490203 | CALD1 Caldesmon 1 | |
| ATAGTAGCTT | 28 | 17 | 0 | 2 | 0 | 0 | 8 | 9 | 118400 | FSCN1 Fascin homolog 1, actin-bundling protein | |
| TGCAATATGC | 43 | 38 | 0 | 3 | 0 | 0 | 4 | 1 | 146447 | FBN1 Fibrillin 1 (Marfan syndrome) | |
| CAGTACTGTA | 21 | 27 | 0 | 0 | 0 | 0 | 0 | 6 | 252418 | ELN Elastin | |
| GCCCAAGGAC | 90 | 29 | 8 | 7 | 0 | 0 | 2 | 0 | 195464 | FLNA Filamin A, alpha (actin binding protein 280) | |
| CTGCCAAGTT | 101 | 50 | 1 | 2 | 0 | 0 | 6 | 4 | 413036 | ZYX Zyxin | |
| ACAGGCTACG | 151 | 110 | 1 | 0 | 0 | 0 | 3 | 0 | 4065186 | Transgelin | |
| GTCTGGGGCT | 105 | 71 | 11 | 20 | 1 | 0 | 17 | 15 | 104650 | TAGLN2 Transgelin 2 | |
| TGCCTCTGCG | 84 | 55 | 3 | 32 | 0 | 0 | 1 | 7 | 5098268 | CD151 CD151 antigen | |
| TCTGTGACCT | 20 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 501928 | MICAL2 Microtubule-associated monoxygenase, calponin and LIM domain containing 2 | |
| CTCATCAGCT | 20 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 370581 | CAP1 CAP, adenylate cyclase-associated protein 1 | |
| GCTTGGATCT | 51 | 61 | 0 | 5 | 1 | 0 | 13 | 6 | 250723 | TMAP1 Transmembrane anchor protein 1 | |
| ATCTTGTTAC | 427 | 185 | 0 | 4 | 0 | 0 | 38 | 14 | 203717 | FN1 Fibronectin 1 |
* Original tag number before normalization; ECM = extracellular matrix; SAGE = serial analysis of gene expression.
Six genes involved in cell cycle regulation and apoptosis were downregulated in osteosarcoma (Table 3), including p63 and p21, both of which also potentially are involved in TGF-β signaling [21]. Additional genes (tags) with unknown functions that also were downregulated are shown (Table 3). In contrast to numerous genes downregulated in osteosarcoma, there was no gene identified as being consistently overexpressed in osteosarcomas at the high stringency statistical criteria applied in this analysis.
Table 3.
The differentially expressed SAGE tags* involved in cell cycle regulation and others in osteosarcoma compared with its normal counterparts
| Gene | SAGE tag | MSC | OB | SaOS | M160xeno | OS160cl | M63xeno | OS256 | OS259 | Unigene identification number | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell cycle | GACCAGCAGA | 21 | 20 | 0 | 0 | 0 | 2 | 7 | 6 | 137569 | TP73L Tumor protein p73-like |
| CTGAGAGCTG | 26 | 17 | 1 | 0 | 0 | 0 | 0 | 0 | 369201 | GAS6 Growth arrest-specific 6 | |
| TGTCCTGGTT | 23 | 48 | 9 | 1 | 2 | 4 | 6 | 4 | 370771 | CDKN1A cyclin-dependent kinase inhibitor 1A (p21, Cip1) | |
| GGTTGGCAGG | 39 | 190 | 0 | 3 | 0 | 0 | 5 | 15 | 3745 | MFGE8 Milk fat globule-EGF factor 8 protein | |
| CTTGATTCCC | 28 | 16 | 0 | 2 | 0 | 0 | 1 | 5 | 518374 | QSCN6 Quiescin Q6 | |
| AAAGTCTAGA | 76 | 33 | 6 | 2 | 0 | 0 | 1 | 4 | 523852 | CCND1 Cyclin D1 (PRAD1: parathyroid adenomatosis 1) | |
| Ca++ regulation | CTTCCAGCTA | 52 | 29 | 1 | 3 | 0 | 0 | 8 | 4 | 511605 | ANXA2 annexin A2 |
| CCCCCTGGAT | 56 | 58 | 10 | 4 | 1 | 1 | 4 | 2 | 275243 | S100A6 S100 calcium binding protein A6 (calcyclin) | |
| AGCAGATCAG | 47 | 33 | 14 | 5 | 0 | 0 | 12 | 12 | 143873 | S100A10 S100 calcium binding protein A10 (annexin II ligand, calpactin I) |
|
| Not classified | GCTGGGAGGG | 47 | 23 | 0 | 11 | 1 | 0 | 1 | 4 | 325650 | EHD2 EH-domain containing 2 |
| TTCTTGTTTT | 21 | 13 | 0 | 1 | 0 | 0 | 3 | 3 | 472010 | PRNP Prion protein (p 27–30) |
* Original tag number before normalization is shown; SAGE = serial analysis of gene expression.
Discussion
Osteosarcoma is the most common primary bone tumor in children and young adults. Unfortunately, the prognosis of the patients with this disease has not improved during the past three decades, therefore better understanding of this disease is needed. In theory, osteosarcoma could arise from osteoblasts or the more primitive MSCs [22]. Our study provides a possible molecular basis of the disease by comparing the transcriptional profile of osteosarcoma with that of its potential normal counterparts, osteoblasts and MSCs.
There are limitations to our study. We used libraries from primary materials, cell lines, and xenografts. Although a primary tumor specimen from a biopsy may be the optimal material, these are available only in small amounts because of clinical standards of care and prioritization of tissue for diagnostic purposes. Patient specimens obtained after preoperative chemotherapy usually contain at least partly necrotic tissue. Cultured cell lines and xenograft tissues are more reproducible and can be expanded, but transcriptome differences from the primary material are likely [23]. We therefore used a more stringent criterion for statistical analysis, only genes that were commonly deregulated in all osteosarcoma SAGE libraries were selected. No genes were identified to be consistently overexpressed in osteosarcoma across all the specimens included in this study. This might reflect the variance of the materials we used, including biopsy specimens, cell lines, and xenografts. This also might represent the extreme genetic instability and complexity of the genome in this disease [24].
Our data suggest that dysregulation of the IGF signaling axis might represent an essential step in the tumorigenesis of osteosarcoma. IGF-1 is the mediator of the anabolic effects of growth hormone during rapid bone growth, and IGF-1 levels peak during puberty [28]. IGF-1 level is associated with body weight and bone growth in animal models [2, 17, 36]. Sutter et al. [31] reported that a single IGF-1 allele was a major determinant of the size of dogs. Canine osteosarcoma is most commonly seen in large breeds [20]. The IGFBPs and IGFBPrPs generally are believed to be inhibitory to the mitogenic and antiapoptotic effects of IGF-1, and their expression usually is stimulated by IGF-1 [9]. It appears that this autoregulatory mechanism is disrupted in osteosarcoma. Simultaneous loss of all the IGFBPs and IGFBPrPs in osteosarcoma compared with normal bone cells is striking. This was similarly found in a study using comparable methods [12]. Cells may obtain self-sufficiency by maintaining IGF-1 activity through downregulation of the IGFBPs. This might provide new targets for therapy for this disease [12].
In our study, disruption of TGF-β in osteosarcoma is suggested downregulation of genes of this superfamily at multiple levels (Table 1) and their target genes (Table 2). Even genes such as vimentin, which is widely regarded as a marker for a mesenchymal cellular origin by pathologists, was identified to be downregulated in osteosarcoma in this study. The TGF-β superfamily is involved in virtually every aspect of cellular activity. Osteoblast differentiation is regulated by the TGF-β signaling pathway, including the BMPs [5]. Mesenchymal cells are distinct from epithelial cells in their mechanisms of cell-cell communication. Instead of direct cell-cell contact through various junctions in epithelial cells, mesenchymal cells exist in a large amount of ECM mostly produced by themselves. The ECM and the cell-matrix interactions are important for cell proliferation, survival, and migration [6]. Loss of TGF-β signaling is a hallmark of tumorigenesis in many cancers of epithelial origin [29]. Further study on TGF-β signaling in this disease may shed light on the understanding of the disease.
Some genes consistently downregulated in osteosarcoma, however, were found to be upregulated during the osteoblastic induction from MSCs, including the IGFBPs and genes in the TGF-β/BMP signaling cascade, in agreement with previous studies [1, 2, 5, 9, 14, 28]. This suggests that osteosarcoma perhaps is more primitive than MSCs, the progenitor of osteoblasts, or may suggest a trend of dedifferentiation in osteosarcoma. The question of cell of origin of osteosarcoma is still debated [3, 22, 35]. Our study provides comprehensive evidence that links osteosarcoma with skeletal development. The dysregulated genes identified in osteosarcoma, such as components in IGF and TGF-β/BMP signaling, are major players regulating normal bone growth as suggested in an animal model [35]. These findings suggest that future therapeutic strategies might be directed toward promoting cell differentiation instead of (or in addition to) more-conventional chemotherapeutic approaches. Mutations on p53 and Rb genes were found in osteosarcoma. We did not find the gene expression levels to be different among the osteosarcoma specimens versus normal tissues in this study. This suggests the loss of function of the tumor suppressors is probably the mechanism, instead of expression. Six genes involved in cell cycle regulation and apoptosis were downregulated in osteosarcoma (Table 3), including p63 and p21. Further study is needed to clarify the role of these genes in osteosarcoma.
For the first time, a highly clustered transcriptional profile, which is well preserved in osteosarcomas despite being derived from different materials grown in various conditions, was revealed by SAGE. This well-coordinated expression pattern suggests that profound alterations in the signaling axes of IGF-1 and TGF-β, in concert with cell cycle regulators, may be involved in the pathogenesis of osteosarcoma (Fig. 5). This provides a basis for further investigation to better understand the disease and to identify these pathways as potential new therapeutic targets. This study suggests the therapeutic value of inhibiting the IGF-1 and TGF pathways. Direct inhibitors of the IGF-1 signaling pathway exist in the form of IGF-1R antibodies. Although directly targeting the TGF pathway is not possible, R-spondin is one example of many drugs that target the TGF pathway and could be tested in patients with osteosarcoma. Preclinical testing of inhibitors of the IGF-1 and TGF pathways seems warranted, and with additional promising preclinical data, trials of these inhibitors in patients with osteosarcoma would be warranted.
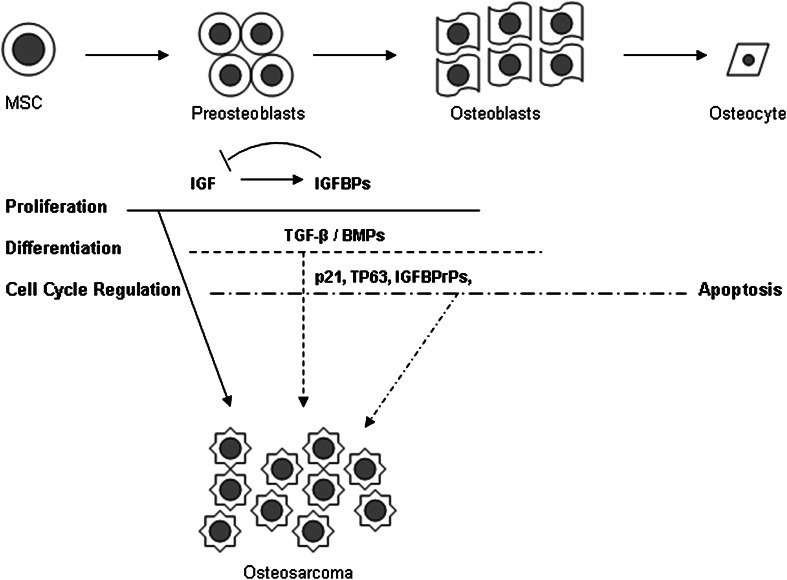
Fig. 5.
During osteogenesis, MSCs are activated by the growth hormone (GH)-IGF axis to enter a proliferation phase and proceed to osteoblastic differentiation regulated by the TGF-β/BMPs. After matrix production, the majority of mature osteoblasts undergo apoptosis with only a few becoming osteocytes. Thus, sequential molecular events may contribute to the pathogenesis of osteosarcoma: (1) downregulation of IGFBPs free IGF-1 to stimulate unopposed cell proliferation; (2) TGF-β/BMPs signaling disrupts differentiation; and (3) alteration of apoptosis and cell cycle regulation.
Acknowledgments
We acknowledge Andrew Huvos, MD (former attending pathologist, Memorial-Sloan Kettering Cancer Center, New York, NY, USA), who performed the histologic examinations.
Footnotes
Funding for this study was received from the Foster Foundation (RG), the Swim Across America Foundation (RG), and the Cure Search Foundation (RG).
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research ® editors and board members are on file with the publication and can be viewed on request.
Clinical Orthopaedics and Related Research ® neither advocates nor endorses the use of any treatment, drug, or device. Readers are encouraged to always seek additional information, including FDA-approval status, of any drug or device prior to clinical use.
This work was performed at The Children’s Hospital of Montefiore, Bronx, NY, USA.
References
- 1.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. doi: 10.1016/S0092-8674(05)80085-6. [DOI] [PubMed] [Google Scholar]
- 3.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci USA. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair HC, Zaidi M, Schlesinger PH. Mechanisms balancing skeletal matrix synthesis and degradation. Biochem J. 2002;364:329–341. doi: 10.1042/bj20020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 6.DeClerck YA, Mercurio AM, Stack MS, Chapman HA, Zutter MM, Muschel RJ, Raz A, Matrisian LM, Sloane BF, Noel A, Hendrix MJ, Coussens L, Padarathsingh M. Proteases, extracellular matrix, and cancer: a workshop of the path B study section. Am J Pathol. 2004;164:1131–1139. doi: 10.1016/S0002-9440(10)63200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digirolamo CM, Stokes D, Colter D, Phinney DG, Class R, Prockop DJ. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res. 2002;397:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Govoni KE, Baylink DJ, Mohan S. The multi-functional role of insulin-like growth factor binding proteins in bone. Pediatr Nephrol. 2005;20:261–268. doi: 10.1007/s00467-004-1658-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks MJ, Roth JR, Kozinetz CA, Wang LL. Clinicopathologic features of osteosarcoma in patients with Rothmund-Thomson syndrome. J Clin Oncol. 2007;25:370–375. doi: 10.1200/JCO.2006.08.4558. [DOI] [PubMed] [Google Scholar]
- 11.Hisada M, Garber JE, Fung CY, Fraumeni JF, Jr, Li FP. Multiple primary cancers in families with Li-Fraumeni syndrome. J Natl Cancer Inst. 1998;90:606–611. doi: 10.1093/jnci/90.8.606. [DOI] [PubMed] [Google Scholar]
- 12.Kuijjer ML, Peterse EF, van den Akker BE, Briaire-de Bruijn IH, Serra M, Meza-Zepeda LA, Hassan AB, Hogendoorn PC, Cleton-Jansen AM. IR/IGF1R signaling as potential target for treatment of high-grade osteosarcoma. BMC Cancer. 2013;13:245. doi: 10.1186/1471-2407-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, Papadopoulos N, Vogelstein B, Kinzler KW, Strausberg RL, Riggins GJ. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 14.Leask A, Abraham DJ. The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochem Cell Biol. 2003;81:355–363. doi: 10.1139/o03-069. [DOI] [PubMed] [Google Scholar]
- 15.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 16.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/S0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 17.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 18.Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44:973–989. doi: 10.1016/S0031-3955(05)70540-X. [DOI] [PubMed] [Google Scholar]
- 19.Meyers PA, Heller G, Healey J, Huvos A, Lane J, Marcove R, Applewhite A, Vlamis V, Rosen G. Chemotherapy for nonmetastatic osteogenic sarcoma: the Memorial Sloan-Kettering experience. J Clin Oncol. 1992;10:5–15. doi: 10.1200/JCO.1992.10.1.5. [DOI] [PubMed] [Google Scholar]
- 20.Misdorp W. Skeletal osteosarcoma: animal model: canine osteosarcoma. Am J Pathol. 1980;98:285–288. [PMC free article] [PubMed] [Google Scholar]
- 21.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 22.Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. doi: 10.1016/j.bone.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB, Reynolds CP, Maris JM, Friedman HS, Dome J, Khoury J, Triche TJ, Seeger RC, Gilbertson R, Khan J, Smith MA, Houghton PJ. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14:4572–4583. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, Ladanyi M, Gorlick R, Levine AJ. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci USA. 2003;100:11547–11552. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollak M, Sem AW, Richard M, Tetenes E, Bell R. Inhibition of metastatic behavior of murine osteosarcoma by hypophysectomy. J Natl Cancer Inst. 1992;84:966–971. doi: 10.1093/jnci/84.12.966. [DOI] [PubMed] [Google Scholar]
- 26.Reseland JE, Syversen U, Bakke I, Qvigstad G, Eide LG, Hjertner O, Gordeladze JO, Drevon CA. Leptin is expressed in and secreted from primary cultures of human osteoblasts and promotes bone mineralization. J Bone Miner Res. 2001;16:1426–1433. doi: 10.1359/jbmr.2001.16.8.1426. [DOI] [PubMed] [Google Scholar]
- 27.Robey PG, Termine JD. Human bone cells in vitro. Calcif Tissue Int. 1985;37:453–460. doi: 10.1007/BF02557826. [DOI] [PubMed] [Google Scholar]
- 28.Rosen CJ. Growth hormone and aging. Endocrine. 2000;12:197–201. doi: 10.1385/ENDO:12:2:197. [DOI] [PubMed] [Google Scholar]
- 29.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 30.Silva WA, Jr, Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JL, Zanette DL, Santos AR, Zago MA. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells. 2003;21:661–669. doi: 10.1634/stemcells.21-6-661. [DOI] [PubMed] [Google Scholar]
- 31.Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremain N, Korkko J, Ibberson D, Kopen GC, DiGirolamo C, Phinney DG. MicroSAGE analysis of 2,353 expressed genes in a single cell-derived colony of undifferentiated human mesenchymal stem cells reveals mRNAs of multiple cell lineages. Stem Cells. 2001;19:408–418. doi: 10.1634/stemcells.19-5-408. [DOI] [PubMed] [Google Scholar]
- 33.Van Wyk JJ, Smith EP. Insulin-like growth factors and skeletal growth: possibilities for therapeutic interventions. J Clin Endocrinol Metab. 1999;84:4349–4354. doi: 10.1210/jcem.84.12.6201. [DOI] [PubMed] [Google Scholar]
- 34.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 35.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, Alt FW, McMahon AP, Orkin SH. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yakar S, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, Ooi GT, Setser J, Frystyk J, Boisclair YR, LeRoith D. Circulating levels of IGF-1 directly regulate bone growth and density. J Clin Invest. 2002;110:771–781. doi: 10.1172/JCI0215463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang R, Hoang BH, Kubo T, Kawano H, Chou A, Sowers R, Huvos AG, Meyers PA, Healey JH, Gorlick R. Over-expression of parathyroid hormone Type 1 receptor confers an aggressive phenotype in osteosarcoma. Int J Cancer. 2007;121:943–954. doi: 10.1002/ijc.22749. [DOI] [PubMed] [Google Scholar]
- 38.Zelzer E, Olsen BR. The genetic basis for skeletal diseases. Nature. 2003;423:343–348. doi: 10.1038/nature01659. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]