Abstract
Objective(s):
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression. They have important roles in kidney development, homeostasis and disease, and participate in the onset and progression of tubulointerstitial sclerosis and end-stage glomerular lesions that occur in various forms of chronic kidney disease (CKD). In the present study, we elucidated the role of microRNA 205 (miR-205) in cisplatin-induced renal cell apoptosis and explored the molecular mechanisms.
Materials and Methods:
The chronic interstitial nephropathy rat model was induced, and the miRNA expression profile in the kidney cells from rats with CKD was screened. Cisplatin-induced apoptosis in normal renal HK-2 cells was evaluated using flow cytometry, and regulation of miR-205 on target gene was validated using luciferase assay, western blot and real time PCR assays.
Results:
We found that miR-205 expression was significantly decreased in the cells from kidney of CKD rat (P<0.01). Our data showed that when miR-205 was overexpressed or silenced using the mimic or inhibitor, the percentages of apoptotic cells were suppressed or increased significantly (P<0.05), respectively. Moreover, we have identified CMTM4 gene, which is involved in cell proliferation and apoptosis, as a novel target for miR-205. In addition, miR-205 could inhibit apoptosis by binding to the 3’UTR of CMTM4 mRNA and inhibiting its transcriptional activity.
Conclusion:
This study elucidated that miR-205 plays an important role in the regulation of apoptosis in renal cells, suggesting a potential therapeutic target to hinder CKD development.
Keywords: Apoptosis, Chronic kidney disease, CMTM4, microRNA 205
Introduction
Chronic kidney disease (CKD) is a chronic disease which affects people globally. Based on the latest estimation by the Centers for Disease Control and Prevention, more than 10% of adults in the United States may have various levels of CKD. The patients with CKD have a sustained lower glomerular filtration rate, which is a key measure of kidney function, and they may also exhibit signs of chronic kidney damage, such as the atypical leakage of proteins from plasma into urine (1). The pathological characteristics of CKD include fibrosis of the glomeruli, loss of glomerular capillaries (glomerulosclerosis), and inflammation. Inflammation can also be observed in the rest of the kidney as leukocytes were recruited along with interstitial fibrosis, atrophy (injury with flattening), and loss of either tubule epithelium and/or peritubular capillaries (2). Apoptosis has been associated with the CKD development process, especially in the late stage of renal failure, and much higher apoptosis rate is found in the renal cells. But the detailed molecular mechanism of CKD development is not well understood.
As a family of small, non-coding RNAs, microRNAs (miRNAs) regulate gene expression by binding to the complementary site of their target mRNAs and inhibiting translation of these genes (3). Their primary function is to facilitate the degradation of their target mRNAs, thereby, regulating the expression levels of their target genes. MicroRNAs have also been shown to inhibit protein translation of their target mRNAs (translational suppression) (4). In the kidney, microRNA-205 (miR-205) is highly expressed and very abundant (5); however, the biological function of miR-205 in renal cells is still unknown. It has been reported that miR-205 is one of the most notably down-regulated miRNAs in human breast tumors. In addition, low expression of miR-205 has been associated with a higher chemotherapy relapse in patients who suffered from triple-negative breast cancer (TNBC) with enriched cancer stem cell population (6). It is interesting to note that recent studies have revealed the complex roles of miR-205 as it acts either as an oncogene or as a tumor suppressor gene under different conditions (7). Nonetheless, the role of miR-205 in CKD development is still unclear.
CKLF-like MARVEL transmembrane domain containing member (CMTM) represents a gene family that consists of Chemokine-like factor (CKLF) and CMTM1 to CMTM8. These genes not only play important roles in the immune and male reproductive systems, but also are shown to be involved in tumorigenesis. CMTM4, is the most conserved one among the members of this family. It was first identified in 2009 and its protein is highly expressed in the kidney, brain and heart. It was shown that CMTM4 could exert inhibitory effects on the growth of HeLa cells through the induction of G2/M phase accumulation (8). However, we discovered that CMTM4 is a novel target for miR-205 and could promote renal cell apoptosis.
Here, we examined the microRNAs expression profile in rat CKD model, and found that the expression of miR-205 was significantly up-regulated among all the microRNAs screened in the current study. Furthermore, mechanistic studies indicated that overexpression or knockdown of miR-205 expression in HK-2 renal cells would result in the expression modulation of CMTM4 gene and therefore, affects renal cell apoptosis. These findings will help us to identify the role of miR-205 in the progress and prognosis of CKD.
Materials and Methods
Construct of CKD rats model
The animal experiments were conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Male Sprague-Dawley rats (n=10) with body weight of 200-220 g were fed with a powdered chow containing 0.7% adenine for 2 weeks to induce chronic interstitial nephropathy [9]. Rats were kept in cages (three or four animals per cage) and had free access to food and water (10). They were then anesthetized and euthanized by cardiac exsanguination. Afterwards, the kidneys were removed instantly and processed for the following qRT-PCR and Western blot analysis.
Cell culture
HK-2 cells (ATCC® CRL-2190™) and HEK293 (ATCC® CRL-1573™) were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 U/ml).
Plasmid preparation
Luc-CMTM4-3’UTR was purchased from RiboBio (RiboBio, China), and the mutation of miR-205 binding sites in luc-CMTM4-3’UTR was performed by TransGen (TransGen, China). The pcDNA3.1-CMTM4 (CMTM4 OE) was commercially constructed by GeneChem (GeneChem, China). siRNAs targeting the CMTM4 (si CMTM4) was purchased from Sigma Aldrich (St. Louis, MO, USA), and the non-targeting siRNA control (si Ctrl) from Sigma Aldrich was used as the control.
miR-205 scramble, mimics and inhibitor transfection
The miR-205 scramble, mimics and inhibitor were all purchased from RiboBio (Ribo, China), which were chemically synthesized fragments. miR-205 mimics had the same sequence as the mature miR-205, whereas inhibitor was fragments complementary with mature miR-205, scramble miRNA was 22 nucleotides long and used as the control. Transfection was done according to the manufacturer’s protocol with Lipofectamine 2000 (Life Science, NY, USA).
RNA extraction and qRT-PCR
Total miRNAs were isolated using mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA), followed by reverse transcriptions using Takara RNA PCR kit (Takara Biotechnology, Dalian, China). To quantify the transcriptional levels of the certain genes, real-time PCR was performed using a SYBR Green Premix Ex Taq (Takara, Dalian, China) according to the product specification. Relative expression levels of the miRNA were normalized to that of RNU6, which was routinely used as an internal or endogenous control for miRNA expression. The primers used in qRT-PCR were purchased from RiboBio (RiboBio, China) and the reaction was performed according to the manufacturer’s instructions.
Western blot analysis
Total protein extracts were obtained using 1x lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.2], 5 mM EDTA, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate) containing protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO, USA). The proteins were analyzed by Western blot using antibodies against CMTM4 (Santa Cruz, CA, USA). Antibodies against GAPDH (Santa Cruz, CA, USA) served as the loading control.
Luciferase Assays
HEK293 cells were seeded at a density of 1×105 cells per milliliter in a 24-well plate. When the cells reached about 70% confluence, miR-205 scramble or mimics were co-transfected into cells with luc-CMTM4-3’UTR. Lysates were collected 36 hr after transfection, and luciferase activity was measured using the dual luciferase assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions and normalized against the Renilla luciferase activity.
Flow cytometry analysis of apoptosis
Cultured cells were harvested by trypsinization and washed with 1x PBS. For each sample, 1×106 cells were stained with the Annexin V-FITC/PI apoptosis detection kit (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions, and then analyzed using FACS Calibur (BD Biosciences, CA, USA).
Statistical analysis
Data represent the results of three independent experiments. Differences between groups were analyzed using Student’s t-test analysis and expressed as mean±SD from at least three separate experiments. Data were considered to be statistically significant when P<0.05.
Results
microRNAs were aberrantly expressed in CKD rat models
To study miRNA function in patients with CKD, we used an in vivo model using Sprague-Dawley rats with adenine-induced chronic renal disease. Chronic dietary adenine intake can induce kidney damage in rats, which is a perfect model to mimic CKD in a clinical setting (11). To explore the potential epigenetic mechanism that could be involved in the regulation of this process, we analyzed changes in the global miRNA expression profile in CKD rats using a genome-wide miRNA-PCR array. Among the miRNAs that were down-regulated, miR-205 was identified as the most significant one (P<0.01). Quantitative real time PCR (qRT-PCR) has confirmed our finding. As shown in Figure 1A, the expression level of miR-205 was drastically decreased with maximum inhibition as compared to the other miRNAs, and the miR-205 level retained only about one-fifth of the expression level of the control rats, followed by the expression levels of miR-29, miR-21 and miR-192 (Figure 1A).
Figure 1.
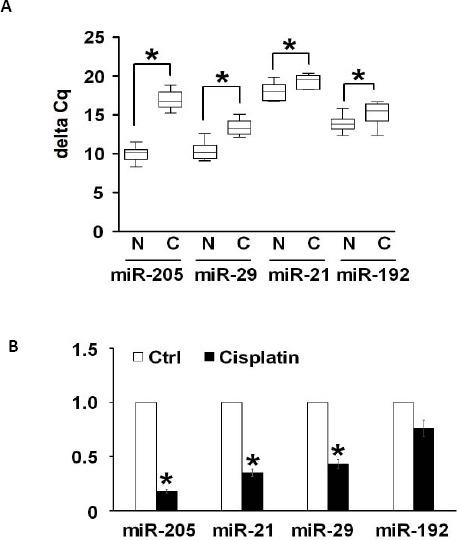
MicroRNAs were aberrantly expressed in CKD rat models
(A) Expression levels of miRNAs were screened by genome-wide miRNA-PCR array in rats CKD models. A set of the most significantly down-regulated miRNAs are selected, and quantitative real-time PCR were used to show their relative expression levels in normal rats (N) and in CKD rats model (C). (B) Expression levels of miR-205, -21, -29 and -192 in renal HK-2 cells with 6 hr of cisplatin treatment. *P<0.05, and **P<0.01 compared with the control
Short exposure of cisplatin can activate apoptosis. We incubated HK-2 cells with 100 μg/ml of cisplatin for 6 hr, and cells were collected to examine the expressions of the above-mentioned miRNAs. Using qRT-PCR analysis, we observed a decreased expression level of miR-205 in cells treated with cisplatin compared with the control cells, and levels of miR-29, miR-21 and miR-192 were also mildly down-regulated following cisplatin treatment (Figure 1B). As it was consistent with our in vivo results, we chose miR-205 with the most significant changes both in vivo and in vitro, as the subject of our study. These results suggested that the changes in the levels of miR-205 expression in response to cisplatin stimulus may have some critical biological functions in the renal cells.
miR-205 could protect renal cells from cisplatin-induced apoptosis
To further gain insight into the biological function of miR-205, the gain-of-function and loss-of-function experiments were designed. We transiently overexpressed miR-205 in HK-2 cells by transfection with miR-205 mimics, and silenced the expression of miR-205 by transfection of miR-205 inhibitor to study its roles in HK-2 cells. The scramble was used as the negative control (Figure 2A). To study the effect of miR-205 in cisplatin-induced apoptosis, flow cytometry analysis was employed with Annexin-V staining. While HK-2 cells treated with mimics were more resistant to cisplatin-induced apoptosis as compared to cells containing the scramble miRNA (P<0.05), knockdown of endogenous miR-205 could induce higher apoptosis rate than cells with scramble (P<0.05) (Figure 2B).
Figure 2.
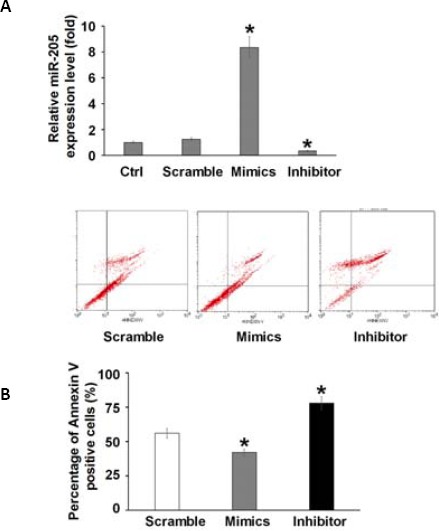
MiR-205 protected renal cells from cisplatin-induced apoptosis
(A) miR-205 was overexpressed by transfection of miR-205 mimics or knockdown by inhibitor against miR-205 as measured by quantitative PCR assay. Scramble served as the control. (B) Representative images of flow cytometry analysis showing Annexin V/PI stained HK-2 cells with scramble, miR-205 mimics or miRNA inhibitor against miR-205, in the presence of cisplatin (100μg/mL) for 6 hr. Results from three independent experiments quantifying the percentage of Annexin V-positive cells are summarized. * P<0.05, and ** P<0.01 compared with the control
CMTM4 is the target formiR-205
To elucidate the detailed molecular mechanisms of the protective effect of miR-205 in renal cell apoptosis, we surveyed its potential targets using TargetScan (Whitehead Institute for Biomedical Research, Cambridge, MA, USA). The CMTM4 was the most possible target with the highest score among all the potential targets. Indeed there are two miR-205 conserved binding sites at the 3’UTR region of CMTM4 gene mRNA (Figure 3A). Western blot analysis showed that overexpression of miR-205 attenuated CMTM4 expression, whereas knockdown of miR-205 elevated the expression of CMTM4 in HK-2 cells (Figure 3B). In addition, luciferase assay showed that addition of 100 nM of miR-205 mimics resulted in decreased luciferase activity of the wild-type CMTM4-3’UTR (luc-CMTM4-3’UTR-WT) in HEK293 cells; However, when the binding site sequence on the 3’UTR of CMTM4 mRNA was mutated from the wild-type “UGAAGG” to “UGAACC”, the inhibitory effect was not observed (Figure 3C). In the presence of miR-205, the cells containing CMTM4 reporter with mutated miR-205-binding sites (luc-CMTM4-3’UTR-Mut) had a similar level of transcriptional activity with that in the absence of miR-205, suggesting the miR-205 could modulate the transcriptional activity of CMTM4 (Figure 3C).
Figure 3.
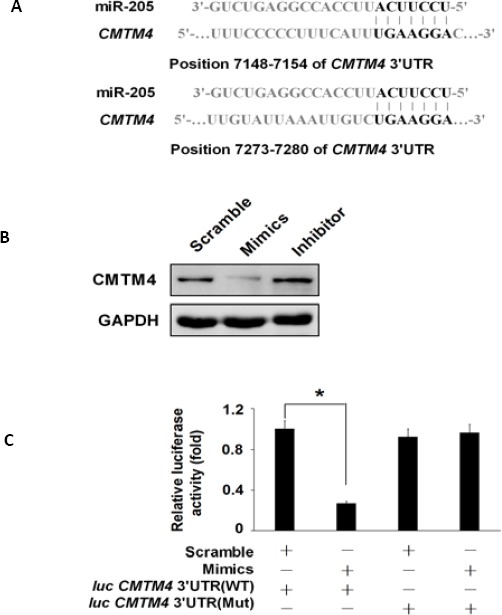
CMTM4 is the target of MiR-205
(A) Putative miR-205 binding sites on the 3’UTR of CMTM4 mRNA with potential complementary residues showing in black. (B) Western blot analysis showing the protein levels of CMTM4 in HK-2 cells treated with scramble, miR-205 mimics and miRNA inhibitor against miR-205. GAPDH was used as a loading control. (C) Luciferase activity was used to measure the transcriptional activity of the luc-CMTM4-3’UTR (WT) or luc-CMTM4-3’UTR (Mut) in HEK293 cells co-transfected with miR-205, and Renilla was used as internal control. * P<0.05, and ** P<0.01 compared with the control. Data represent the results of three independent experiments
CMTM4 participated in renal cell apoptosis
Next, we explored the function of CMTM4 gene in renal cell apoptosis induced by cisplatin. Firstly, we prepared the CMTM4 overexpression plasmid (CMTM4 OE) as well as the siRNA plasmid (si CMTM4) targeting CMTM4, and transfected both constructs into HK-2 cells, respectively. The result showed that CMTM4 was upregulated or silenced in cells with overexpression plasmid or the CMTM4 siRNA, respectively as assessed by Western blot (Figure 4A).
Figure 4.
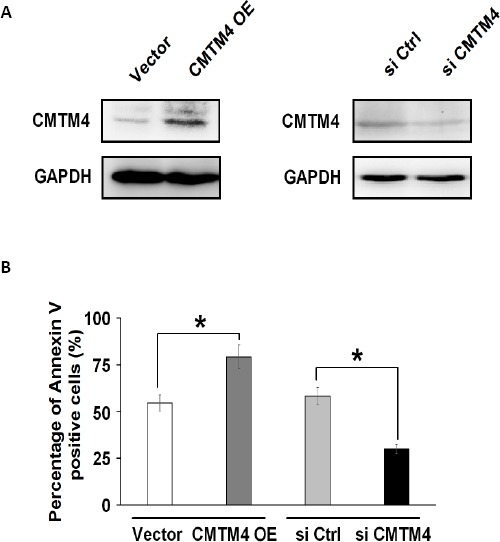
CMTM4 played a role in renal cell apoptosis
(A) Western blot analysis showing the protein levels of CMTM4 in HK-2 cells transfected with CMTM4 overexpression plasmids (OE) (left panel) and siRNA against CMTM4 (right panel). The vector plasmid (Vector) and the scrambled siRNA (NC) served as negative controls, respectively. (B) The result of quantitative analysis from the flow cytometry assay on HK-2 cells transfected with CMTM4 OE plasmid (CMTM4 OE) or siCMTM4 nucleotide fragment (siCMTM4) in the presence of cisplatin (100 μg/ml). * P< 0.05, and ** P<0.01 compared with the control
To understand how CMTM4 exerts its function in cisplatin-induced apoptosis, we conducted flow cytometry analysis. The results showed that in response to cisplatin treatment, overexpression of CMTM4 could enhance apoptosis, the percentage of Annexin V-FITC positive apoptotic cells was increased from 52% to 74%, whereas knockdown of CMTM4 could partially protect cells from apoptosis, the percentage of apoptotic cells was dropped from 56% to 33% (Figure 4B), suggesting that CMTM4 plays a role in renal cell apoptosis.
miR-205 failed to protect renal cell from apoptosis when the binding sites on CMTM4 mRNA were changed
In order to determine whether miR-205 indeed inhibits apoptosis through its target CMTM4, miR-205 mimics and CMTM4 expressing plasmid lacking mir-205 binding sites were co-transfected into HK-2 cells and then, treated with cisplatin. Western blot analysis showed that miR-205 could not decrease the protein level of exogenous CMTM4 because CMTM4 expressing plasmid did not have miR-205 binding sites (Figure 5A). Annexin V-FITC assay was employed to evaluate the effect on renal cell apoptosis, and the results demonstrated that although there were abundant miR-205 in HK-2 renal cells, CMTM4 overexpression could still promote cell apoptosis, and the apoptosis rate remained at normal level (Figure 5B). Without the interaction between miR-205 and CMTM4 3’UTR, the anti-apoptotic effect of miR-205 was diminished, suggesting that miR-205 mediates apoptosis through its transcriptional modulation of CMTM4 gene.
Figure 5.
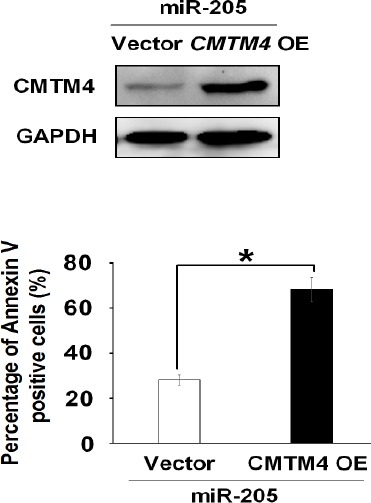
CMTM4 reversed the anti-apoptosis effect of MiR-205.
(A) Western blot analysis showing the protein expression level of CMTM4 in HK-2 cells co-transfected CMTM4 overexpression plasmid with scramble or miR-205 mimics. (B) Quantitative analysis from the results of the flow cytometry assay on HK-2 cells co-transfected with CMTM4 overexpression plasmid (CMTM4 OE) and scramble control or miR-205 mimics in the presence of cisplatin (100 μg/ml)
** P<0.01 compared with the control. Data represent the results of three independent experiments
Discussion
Based on the statistics by the global burden of disease (GBD), diseases of the kidney and urinary tract were ranked the 12th among causes of death which accounts for 1.4 percent of all deaths, and the 17th among causes of disability (12). CKD patients often suffer from cardiovascular or cerebrovascular diseases, and they often die from the complications (13). Patients with different types of CKD usually progressively lose their renal functions, gradually develop glomerular sclerosis and/or renal interstitial fibrosis which are eventually deteriorated with kidney failure (14). In general, the final stage of the renal diseases is called the “end-stage renal disease”, and patients have no choice but undergoing renal replacement therapy to regain renal function.
Emerging evidences showed that miRNAs play important roles in kidney development and kidney diseases. Results from clinical and experimental animal studies demonstrate that miRNAs play essential roles in the pathogenesis of various renal diseases. CKD is characterized by renal fibrosis. Transforming growth factor beta (TGF-β) is recognized as a major mediator of renal fibrosis because it is able to stimulate the accumulation of extracellular matrix (ECM) proteins to impair normal kidney function. In recent years, emerging evidence illustrate the relationship between TGF-β signaling and miRNAs expression during renal diseases development. The expressions of several miRNAs were up-regulated by TGF-β signaling pathway, such as miR-21, miR-29, miR-192, miR-200, and miR-433, in which miR-21, miR-192, and miR-433 are reported to be positively induced by TGF-β signaling, and they play a pathological role in kidney diseases. However, members of both miR-29 and miR-200 families which are inhibited by TGF-β signaling, protect kidneys from renal fibrosis by suppressing the deposition of ECM and preventing epithelial-to-mesenchymal transition, respectively (15). In this paper, we validated the protective effect of miR-205 in the CKD development. It was reported that miR-205 level distinguished squamous cell carcinoma from adenocarcinoma in lung cancer biopsies (16). Also, it was shown that long non-coding RNA MALAT1 could promote aggressive renal cell carcinoma through Ezh2 and interacts with miR-205 (17). Li et al revealed a new potential pathway in which VEGF promoted the invasion of ovarian cancer cells via suppressing the expression of Ezrin and Lamin A/C caused by increased expression of miR-205 (18). In addition, it was found that miR-205 promoted radio-sensitivity and was down-regulated in radio-resistant subpopulations of breast cancer cells, and that loss of miR-205 was highly associated with poor distant relapse-free survival in breast cancer patients (19). However, our study is the first one to discuss the biological function of miR-205 in the chronic kidney disease. Previous studies have reported that miR-205 could regulate cell proliferation and apoptosis through targeting several proteins including Bcl-2, PTEN and ERBB2 (20, 21). In this study, CMTM4 was validated as a novel target of miR-205 in the chronic kidney disease. CMTM4 is the most conserved member of CMTM, which is a novel gene family consisting of CKLF and CMTM1-8. Previous studies demonstrated that the changes in CMTM4 expression may affect cell cycle and division in HeLa cells (8, 22). In our study, we are the first to illustrate another function of CMTM4 in renal cell apoptosis.
MiRNAs represent potential new therapeutic targets for some complex diseases for which effective therapies are lacking while they continue to increase in prevalence worldwide. Several researches aiming at improving common diagnostic markers for CKD, have identified some circulating miRNAs as possible diagnostic and even prognostic biomarkers for kidney disease (23). In this study, we elucidated a novel mechanism via which the anti-apoptosis molecule miR-205, coordinates the expression of its target gene CMTM4 to modulate the renal cells apoptosis, revealing important clinical implications for miR-205 in the prognosis and treatment of CKD. While the animal experiments that investigate the therapeutic effects of miR-205 in CKD are on the way, we hope our findings could potentially help to achieve pain relief in CKD patients using the microRNA molecule drugs.
Conclusion
This study revealed miR-205 as an important inhibitor in the regulation of apoptosis in renal cells. Our results would lead to a better understanding of the pathogenesis of renal tumorigenesis and also, shed light on developing a diagnostic marker or a potential therapeutic target to hinder CKD development.
Acknowledgment
The authors would like to thank Dr Kelsey Moriarty and Dr Zhiqiang Liu at the University of Texas MD Anderson Cancer Center for their kind help in language editing and manuscript preparation.
References
- 1.Duffield JS, Grafals M, Portilla D. MicroRNAs are potential therapeutic targets in fibrosing kidney disease: lessons from animal models. Drug Discov Today Dis Models. 2013;10:e127–e35. doi: 10.1016/j.ddmod.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii Y, Sawada T, Kubota K, Fuchinoue S, Teraoka S, Shimizu A. Injury and progressive loss of peritubular capillaries in the development of chronic allograft nephropathy. Kidney Int. 2005;67:321–332. doi: 10.1111/j.1523-1755.2005.00085.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–92. doi: 10.1016/s0092-8674(04)00035-2. 1 p following S6. [DOI] [PubMed] [Google Scholar]
- 4.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 5.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 7.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, et al. MiR-205 in cancer: an angel or a devil? Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Plate M, Li T, Wang Y, Mo X, Zhang Y, Ma D, et al. Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol cells. 2010;29:355–361. doi: 10.1007/s10059-010-0038-7. [DOI] [PubMed] [Google Scholar]
- 9.Vaziri ND, Liu SM, Lau WL, Khazaeli M, Nazertehrani S, Farzaneh SH, et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One. 2014;9:e114881. doi: 10.1371/journal.pone.0114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inami Y, Hamada C, Seto T, Hotta Y, Aruga S, Inuma J, et al. Effect of AST-120 on Endothelial Dysfunction in Adenine-Induced Uremic Rats. Int J Nephrol. 2014;2014:164125. doi: 10.1155/2014/164125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguy L, Johansson ME, Grimberg E, Lundgren J, Teerlink T, Carlstrom M, et al. Rats with adenine-induced chronic renal failure develop low-renin, salt-sensitive hypertension and increased aortic stiffness. Am J Physiol Regul Integr Comp Physiol. 2013;304:R744–752. doi: 10.1152/ajpregu.00562.2012. [DOI] [PubMed] [Google Scholar]
- 12.Dirks J, Remuzzi G, Horton S, Schieppati A, Rizvi SAH. Diseases of the Kidney and the Urinary System. In: Jamison DT, Breman JG, Measham AR, Alleyne G, Claeson M, Evans DB, et al., editors. Disease Control Priorities in Developing Countries. 2nd ed. Washington (DC): 2006. [Google Scholar]
- 13.Hostetter TH. Chronic kidney disease predicts cardiovascular disease. N Engl J Med. 2004;351:1344–1346. doi: 10.1056/NEJMe048211. [DOI] [PubMed] [Google Scholar]
- 14.National Kidney F. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J kidney Dis. 2002;39:S1–266. [PubMed] [Google Scholar]
- 15.Chung AC, Lan HY. MicroRNAs in renal fibrosis. Front Physiol. 2015;6:50. doi: 10.3389/fphys.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patnaik S, Mallick R, Kannisto E, Sharma R, Bshara W, Yendamuri S, et al. MiR-205 and MiR-375 microRNA assays to distinguish squamous cell carcinoma from adenocarcinoma in lung cancer biopsies. J Thorac Oncol. 2015;10:446–453. doi: 10.1097/JTO.0000000000000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Li L, Li Z, Gong G, Chen P, Liu H, et al. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol. 2015;137:125–133. doi: 10.1016/j.ygyno.2015.01.531. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, et al. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat Commun. 2014;5:5671. doi: 10.1038/ncomms6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30:2897–2902. doi: 10.3892/or.2013.2755. [DOI] [PubMed] [Google Scholar]
- 21.Alla V, Kowtharapu BS, Engelmann D, Emmrich S, Schmitz U, Steder M, et al. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle. 2012;11:3067–3078. doi: 10.4161/cc.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 23.Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]


