Abstract
Viruses are considered key players in phytoplankton population control in oceans. However, mechanisms that control viral gene expression in prominent microalgae such as diatoms remain largely unknown. In this study, potential promoter regions isolated from several marine diatom-infecting viruses (DIVs) were linked to the egfp reporter gene and transformed into the Pennales diatom Phaeodactylum tricornutum. We analysed their activity in cells grown under different conditions. Compared to diatom endogenous promoters, novel DIV promoter (ClP1) mediated a significantly higher degree of reporter transcription and translation. Stable expression levels were observed in transformants grown under both light and dark conditions, and high levels of expression were reported in cells in the stationary phase compared to the exponential phase of growth. Conserved motifs in the sequence of DIV promoters were also found. These results allow the identification of novel regulatory regions that drive DIV gene expression and further examinations of the mechanisms that control virus-mediated bloom control in diatoms. Moreover, the identified ClP1 promoter can serve as a novel tool for metabolic engineering of diatoms. This is the first report describing a promoter of DIVs that may be of use in basic and applied diatom research.
Diatoms are one of the most effective and diverse unicellular photosynthetic eukaryotes, with perhaps as many as 200,000 extant species found in aquatic environments1. There is broad interest in diatoms due to their substantial contributions to global carbon cycling and oxygen production2, their complex evolutionary background as secondary endosymbionts3, and their unique capacities to produce silica-based cell walls4. Moreover, diatoms present great potential as a source of beneficial chemicals for use in human activities because they produce biofuel precursors such as fatty acids and hydrocarbons that may prove useful in solving ecological problems such as the energy crisis5. To understand diatom biology and the means of diatom exploitation in biotechnology, novel genomic resources and molecular tools have been developed in recent years. Databases of genome sequences and the expressed sequence tag (EST) of the model diatoms Pennales Phaeodactylum tricornutum and Centrics Thalassiosira pseudonana have been released for public use, allowing the identification of distinct metabolic features in diatoms6,7,8,9. Recently, the genomic sequences of other diatoms such as Pseudo-nitzschia multiseries (http://genome.jgi-psf.org/Psemu1/Psemu1.home.html) and Fragilariopsis cylindrus (http://genome.jgi-psf.org/Fracy1/Fracy1.home.html) have also been made available on the U.S. Department of Energy Joint Genome Institute’s website (http://jgi.doe.gov/). In addition, methods of genetic transformation via biolistic bombardment and/or electroporation have been developed for several diatom species including Pennales P. tricornutum10,11,12,13,14,15,16, Cylindrotheca fusiformis17,18, Navicula saprophila19, Pseudo-nitzschia arenysensis20, Pseudo-nitzschia multistriata20, Centrics Cyclotella cryptica19, Thalassiosira pseudonana21, Thalassiosira weissflogii22, Chaetoceros sp.23, Cha. gracilis24, and Fistulifera sp.25. Moreover, episomal vector technology based on yeast-derived sequences has recently been developed for P. tricornutum and Thalassiosira pseudonana26.
However, many of the processes that control diatom biology and growth in marine environments remain to be elucidated. Recently, it has been proposed that marine viruses play a key role in controlling the blooms of diatom populations27,28. Viruses may also influence the compositions of marine communities and may act as a major force behind biogeochemical cycles29,30. Thus far, several marine diatom-infecting DNA/RNA viruses that cause the lysis of host diatoms have been isolated from both Centrics and Pennales diatoms28, for example, the Cha. debilis-infecting DNA virus (CdebDNAV)31, the Cha. lorenzianus-infecting DNA virus (ClorDNAV)32, and the Thalassionema nitzschioides-infecting DNA virus (TnitDNAV)33. In silico analysis of these genomes has revealed the presence of putative open reading frames (ORFs) such as the replication-associated protein (VP3) gene, the structural protein (VP2) gene, and genes of unknown function28,32.
To determine the contributions of marine diatom-infecting viruses (DIVs) to phytoplankton biology and ecology, it is important to further examine the DIV genome and to identify mechanisms that control viral gene expression through the characterization of viral promoters. Moreover, DIV promoter characterization may also advance the manipulation and modulation of gene expression in diatoms. Thus far, this strategy is used primarily through endogenous promoters such as the gene-encoding fucoxanthin chlorophyll a/c-binding protein (FCP) gene (fcp, now called Lhcf), which encodes members of the light harvesting complex superfamily10,18,21,24, frustulin17, nitrate reductase (NR)18,21,24, acetyl-CoA acetyltransferase24, acetyl-CoA carboxylase19, β-carbonic anhydrase 1 (CA1)34, long-chain fatty acyl-CoA synthetase24, β-tubulin24, ATP synthase subunit C24, and elongation factor 2 (EF2)16.
Of the endogenous promoters, the fcp promoter has been frequently used in biotechnological applications involving diatoms35,36,37,38,39. However, reports also suggest that fcp promoter activity may not be strong enough to overexpress introduced genes and may promote significant increases in target products35,39. Other studies report that transgene expression follows a circadian pattern due to the presence of light-responsive cis-regulatory elements in the fcp promoter16,40,41. In addition, in some cases, diatom endogenous gene promoters drive species-specific transgene expression and do not function in different diatom species18,20,22. These issues considerably limit the applications of diatom transformation techniques, potentially reducing the utility of diatoms in basic and applied research. To address these problems, the development of promoters that stably induce high-level gene expression is anticipated, which should result in the creation of large accumulations of introduced gene transcripts and proteins. Promoters from viruses that infect plants or mammals have generally been used to transform a wide range of higher plants and mammals, allowing the constitutive and strong expression of introduced genes. For example, the cauliflower mosaic virus 35S (CaMV 35S) and cytomegalovirus (CMV) promoters are known to efficiently facilitate plant and mammalian transformations, respectively42,43. However, only a few reports focus on the transformation of diatoms through the use of CaMV 35S and CMV promoters19,44 in P. tricornutum, and the same promoters do not seem to be effective in other species, such as the Centrics diatom Cyc. cryptica19.
DIV promoters may enable us to solve the above-described limitations in the bioengineering of diatoms as well as to infer the ecological roles of DIVs in marine ecosystems. Thus, in this study, potential promoter regions located upstream of ORFs, such as the replication-associated protein (VP3) gene and the structural protein (VP2) gene in the DIV genome (Fig. 1a), were isolated. DIV promoter-driving gene expression capacities in the Pennales diatom P. tricornutum were evaluated and compared to those of the endogenous fcp promoter (Fig. 1b). We then examined the stability of DIV promoters in cells that were cultured under various growth conditions. The effects of the growth stage, the nutrient concentrations in media, and the photoperiods on viral promoter activity were investigated. The application of viral promoters to a Centrics diatom and to unicellular green alga was also examined. We then discussed the structure of a DIV promoter to show how gene expression mechanisms may play a key ecological role in the control of diatom blooms and how the DIV promoter may be used in applied research on diatoms.
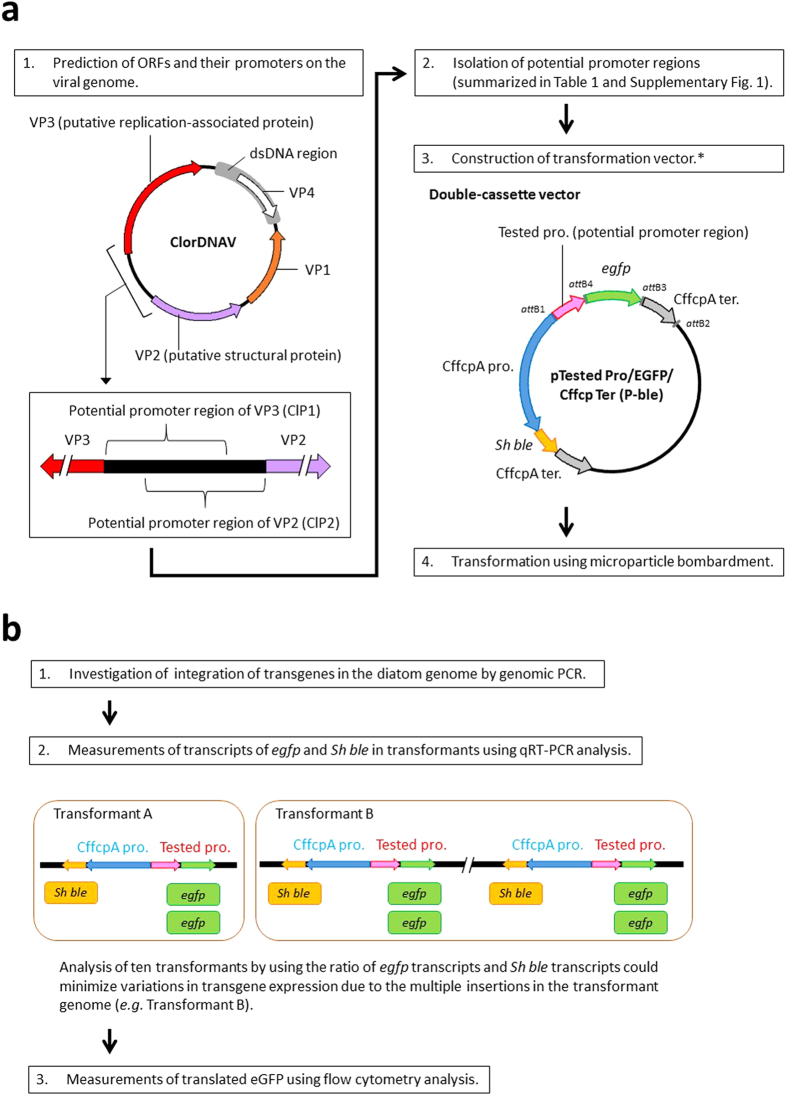
Figure 1. Schematic diagram for the evaluation of promoter activity.
(a) Outline of the construction of transformation vectors and transformations. After predicting putative ORF positions32, upstream regions of the ORFs were determined as potential promoter regions. Potential promoter regions amplified by PCR were used to construct the transformation vectors. The double-cassette vector containing the reporter gene egfp driven by each tested promoter and the antibiotic-resistant gene Sh ble driven by the promoter region of the fucoxanthin chlorophyll a/c-binding protein (FCP) A-1A gene derived from Cyl. fusiformis (termed CffcpA pro.) were constructed. (b) Assessment of promoter activity. Promoter activity was determined by averaging the ratios of egfp mRNA transcript levels to those of Sh ble mRNA transcripts in ten transformants to minimize the effects of copy numbers on the expression of transgenes. These transformants were also used to investigate eGFP protein expression patterns. CffcpA ter.: terminator region of the FCP A-1A gene derived from Cyl. fusiformis. The structure of the ClorDNA genome was modified from Tomaru et al.32. *For the transformation vector of the nitrate reductase gene promoter, we used pNICgfp18 (Supplementary Fig. 5a).
Results
Isolation of potential DIV promoter regions
Potential DIV promoter regions were determined from the genome sequences of CdebDNAV and TnitDNAV via in silico analyses. The ORFs of the two DIVs were identified using an ORF finder (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/gorf/). Three (VP1, VP2, and VP3) and two (VP2 and VP3) ORFs were identified in the CdebDNAV and TnitDNAV genomes, respectively. In regard to ClorDNAV, the four ORFs (VP1, VP2, VP3, and VP4) were previously described by Tomaru et al.32 (Fig. 1a). Regions (approximately 500 bases long) upstream of the translation initiation codon (ATG) of a putative ORF in the DIV genome were predicted to be potential promoter regions (Table 1, Fig. 1a, and Supplementary Fig. 1).
Table 1. Promoters used in this study.
| Promoter | Source organism/virus | Promoter associated gene | Amplifiedsize (bp) | Ref. |
|---|---|---|---|---|
| CdP1 | Chaetoceros debilis-infecting DNA virus (CdebDNAV) | Putative replication-associated protein (VP3) gene | 477 | 31 |
| ClP1 | Chaetoceros lorenzianus-infecting DNA virus (ClorDNAV) | Putative replication-associated protein (VP3) gene | 502 | 32 |
| ClP2 | Chaetoceros lorenzianus-infecting DNA virus (ClorDNAV) | Putative structural protein (VP2) gene | 474 | 32 |
| TnP1 | Thalassionema nitzschioides-infecting DNA virus (TnitDNAV) | Putative replication-associated protein (VP3) gene | 424 | 33 |
| TnP2 | Thalassionema nitzschioides-infecting DNA virus (TnitDNAV) | Putative structural protein (VP2) gene | 424 | 33 |
| PtfcpA pro. | Phaeodactylum tricornutum | Fucoxanthin chlorophyll a/c-binding protein A gene | 444 | 10 |
| Cfnr pro. | Cylindrotheca fusiformis | Nitrate reductase gene | 774 | 18 |
| CaMV 35S pro. | Cauliflower mosaic virus | 35S gene | 454 | 44,81 |
| CMV pro. | Cytomegalovirus | Immediate early promoter regulatory gene | 742 | 44,82 |
| nos pro. | Agrobacterium tumefaciens | Nopaline synthase gene | 338 | 81 |
Construction of transformation vectors
The potential promoter region of each ORF was amplified via polymerase chain reaction (PCR) using DIV genomic DNA and specific primers (Supplementary Table 1). In addition to the potential DIV promoters, the endogenous fcp promoter of P. tricornutum and some extrinsic promoters, such as CaMV 35S, CMV, and the nopaline synthase gene (nos) promoter of Agrobacterium tumefaciens, were amplified from the genomic DNA of P. tricornutum and from commercially available vectors, respectively, via PCR amplification (Supplementary Table 1). To investigate the activity of each tested promoter, we constructed a double-cassette transformation vector (Fig. 1a) that contained the enhanced green fluorescence protein (eGFP) gene (egfp) driven by each tested promoter and the antibiotic-resistant gene Sh ble driven by the Cyl. fusiformis FCP A-1A gene promoter (termed CffcpA pro.). We also constructed a single-cassette transformation vector containing Sh ble driven by each tested promoter (Supplementary Fig. 2) and nat driven by DIV promoters, such as CdP1 and ClP1 (Supplementary Fig. 3), to investigate transformation efficiency.
Confirmation of the introduced gene and promoter in the transformants
Colonies were obtained from a solid f/2 medium containing 500 μg ml−1 antibiotics after its transformation via microparticle bombardment using the double-cassette transformation vectors. To confirm the introduction of the two cassettes, a genomic PCR analysis was performed using different primer sets (Supplementary Tables 1 and 2 and Supplementary Figs 4a and 5a) and using the colony-forming cells as templates. We obtained more than ten transformants with the expected DNA fragment length and containing egfp with the tested promoter, with the exception of transformants that were transfected with pNICgfp18. Six transformants that had been transfected with pNICgfp18 possessed DNA fragments of the expected length, including egfp under the control of the NR gene (nr) promoter from Cyl. fusiformis (termed Cfnr pro.) and the Sh ble gene under the CffcpA pro. A typical electrophoretogram of the amplicons in each transformant is shown in Supplementary Figs 4b and 5b. Ten transformants with each of the tested promoters and six transformants with Cfnr pro. were analysed further.
Quantitative analysis of promoter activity
Variations in the levels of egfp mRNA transcripts normalized to the internal standard gene (ribosomal protein small subunit 30S gene, rps) mRNA transcripts were detected using quantitative reverse transcription-PCR (qRT-PCR) in 6−10 transformants with the egfp driven by each of the tested promoters (Supplementary Fig. 6). Variations in the levels of Sh ble mRNA transcripts driven by the diatom endogenous promoter (CffcpA pro.) that had been normalized to rps were also found amongst the 6−10 transformants (Supplementary Fig. 6). Before determining the activity of each promoter, the egfp mRNA transcript levels in each transformant were divided by those of the Sh ble mRNA transcripts. This method has been used to compare gene expression levels in independent lines and to prevent misinterpretation due to the presence of different transgene copy numbers and possible site integration effects. Figure 2 shows averages and ranges of promoter activity in the 6−10 independent transformants with egfp driven by each tested promoter. We compared the activity of each tested promoter with that of an endogenous promoter PtfcpA pro. because the PtfcpA pro. has generally been used for the transformation of the marine diatom P. tricornutum10,11,13,39. Average promoter activities in the transformants with Cfnr pro. (P = 0.010), TnP1 (P = 0.012), TnP2 (P = 0.016), the CaMV 35S promoter (P = 0.014), the CMV promoter (P = 0.021), and the nos promoter (P = 0.034) (average: 0.0833−0.499) were found to be significantly lower than those in the transformants with PtfcpA pro. (average: 2.08, range: 0.0296−5.97) (Fig. 2). By contrast, the average promoter activity in ClP1 (average: 10.3, range: 2.06−25.1, P = 0.0058) was found to be approximately five times higher than that in PtfcpA pro. (Fig. 2). The average CdP1 (average: 1.99, range: 0.0917−7.34, P = 0.93) and ClP2 activity (average: 3.09, range: 0.352−21.4, P = 0.64) levels showed no significant differences from the average PtfcpA pro. activity level (Fig. 2). The maximum activity levels in the ten transformants of CdP1, ClP1, and ClP2 were higher than that found in the transformants with PtfcpA pro. (Fig. 2).
Figure 2. Relative activities of various promoters including DIV promoters in transformants.
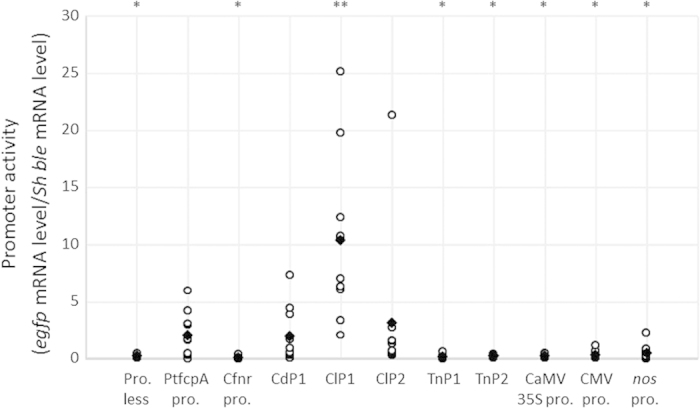
Ten independent transformants for each promoter were analysed. The promoter activity levels were determined by dividing egfp mRNA levels by Sh ble mRNA levels. The circles indicate the mean triplicate measurements of the independent transformants. The diamonds denote the average values of the six transformants from Cfnr pro. or of the ten transformants from the other promoters. Pro.-less denotes cases in which no promoter was linked to the egfp gene in the transformation vector (negative control). PtfcpA pro. shows the positive control in the transformation vector egfp gene driven by PtfcpA pro. Asterisks indicate the presence of a statistically significant difference derived from the PtfcpA pro. transformants (**P < 0.01 and *P < 0.05).
Effect of culture conditions on DIV promoter activity
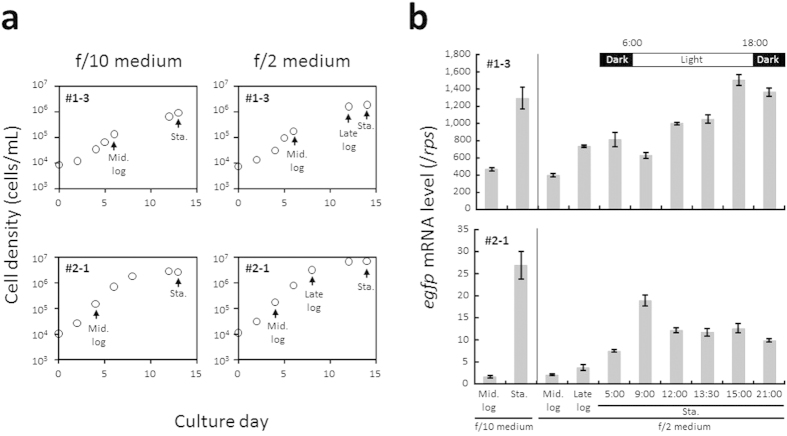
Promoter activities are known to respond to changes in environmental and intracellular conditions. To explore the effects of culture conditions and growth phases on the activity of the DIV promoter (ClP1) that showed the highest levels of activity (Fig. 2), the abundance of egfp mRNA levels in two independent ClP1 transformants was analysed using qRT-PCR under various culture conditions (Fig. 3). Under a low nutrient culture condition (f/10 medium), the abundance of egfp mRNA transcripts driven by ClP1 was almost identical to that reported for the standard nutrient culture condition (f/2 medium) (Fig. 3b). The abundance of egfp mRNA transcripts in the stationary phase seemed to be higher than that found for the log phase under both low and standard nutrient culture conditions (Fig. 3b). ClP1 can effectively drive the expression of egfp mRNA transcripts in both light and dark conditions during the stationary phase (Fig. 3b).
Figure 3. Analyses of viral promoter activities in different culture conditions.
(a) Growth of two transformants with ClP1-driven egfp in f/10 medium and f/2 medium. (b) Relative abundances of egfp mRNA determined by dividing egfp mRNA transcript levels by those of ribosomal protein small subunit 30S gene (rps; internal control gene) mRNA transcripts in the transformant cells incubated in f/10 and f/2 media and harvested at various growth phases and at different times during a light/dark period. The arrows show cell collection points for the qRT-PCR analysis.
Levels of eGFP fluorescence
The fluorescence of the eGFP reporter in the different transgenic lines described above was quantified via flow cytometry. Figure 4 shows normalized eGFP fluorescence levels for cell sizes in the 9−10 independent transformants with egfp driven by each tested promoter. The average eGFP fluorescence levels in the transformants containing TnP1 (P = 0.0071), TnP2 (P = 0.012), the CaMV 35S promoter (P = 0.0063), the CMV promoter (P = 0.021), and the nos promoter (P = 0.012) (average: 2.53−3.24) were found to be significantly lower than those of the transformants containing PtfcpA pro. (average: 6.10, range: 2.21−11.6) (Fig. 4). The statistical analysis results show that the average eGFP fluorescence levels in the transformants containing ClP1 (average: 13.2, range: 2.52−25.0, P = 0.020) were significantly higher than that of the transformants containing PtfcpA pro. The average eGFP fluorescence levels in the transformants with CdP1 (average: 5.27, range: 2.38−16.7, P =0.63) and ClP2 (average: 7.45, range: 2.37−31.8, P = 0.67) showed no significant differences from that in the transformants with PtfcpA pro. (Fig. 4). The maximum levels of eGFP fluorescence in the transformants of CdP1, ClP1, and ClP2 were found to be higher than that in the transformants of PtfcpA pro. (Fig. 4). The eGFP fluorescence levels without normalization were similar to those observed by normalizing fluorescent signals with cell sizes (Supplementary Fig. 7).
Figure 4. Relative abundance of eGFP protein translated from transcripts driven by various promoters in transformants using flow cytometry.
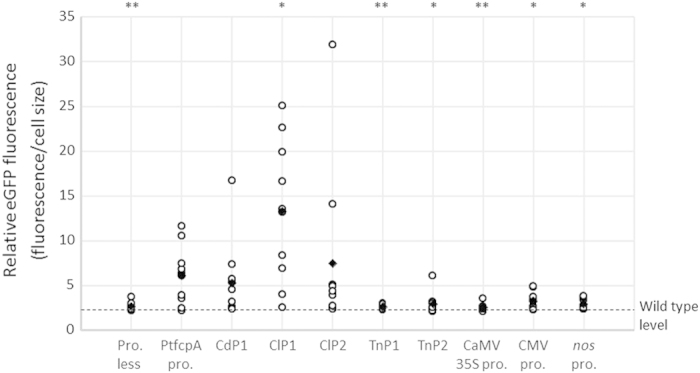
Ten independent transformants derived from various promoters with the exception of the CaMV 35S promoter were analysed. For the CaMV 35S promoter, nine independent transformants were analysed. For normalization purposes, the eGFP fluorescence was divided by the cell size, which was estimated based on values of forward scatter areas (FSC-A) obtained via flow cytometry. The circles denote the mean value for approximately 10,000 cells of independent transformants. The diamonds denote the average values of the transformants. Asterisks indicate statistically significant differences derived from the PtfcpA pro. transformants (**P < 0.01 and *P < 0.05). The broken line denotes the autofluorescence level of wild type cells excited at 488 nm.
Transformation efficiency using DIV promoters
We also assessed whether the different promoters identified in this study may affect transformation efficiency levels in P. tricornutum. To this end, single-cassette vectors containing the selective marker, Sh ble, driven by PtfcpA pro. or ClP1 (Supplementary Fig. 2), were introduced into P. tricornutum via microparticle bombardment. The average transformation efficiencies of PtfcpA pro. and ClP1 reached 13 and 13.5 transformants per 108 cells (n = 4), respectively. Statistical analysis based on the number of colonies formed showed that the transformation efficiency using PtfcpA pro. and ClP1 were not significantly different.
Application of diatom-infecting DIV promoters to other algae
We investigated whether the DIV promoter may also be applied to a species of Centrics diatoms such as Chaetoceros sp. Strain CCK09. To this end, we assayed transformation efficiency levels in this species using single-cassette vectors with the antibiotic-resistant gene nat driven by the DIV promoters CdP1 and ClP1 (Supplementary Fig. 3). The average transformation efficiencies of CdP1 and ClP1 transfected to Chaetoceros sp. were 4 and 2 transformants per 108 cells (n = 3), respectively.
The activity levels of the DIV promoters in the unicellular green alga Chlamydomonas reinhardtii, a species that is phylogenetically different from the diatoms, were also investigated. Single-cassette vectors, in which She ble was driven by a DIV promoter (for example, CdP1 and ClP1) or the Chlamydomonas endogenous promoter rbcS2 (pSP108)45, were used for the assay. A standard electroporation method46 and wall-less Chlamydomonas strain CC503 were used for the transformation. The transformation efficiency of pSP108 was recorded at 168 transformants per 2.5 × 107 cells (n = 14). By contrast, the average transformation efficiency found via CdP1 and ClP1 were recorded at 0 and 0.14 transformants per 2.5 × 107 cells (n = 14), respectively. The transformation efficiency levels obtained using viral promoters were not significantly different from those of the promoter-less vector (average: 0 transformant, n = 14).
In silico analyses of DIV promoter regions
We searched for potential cis-regulatory elements in the DIV promoters using the PLACE47 and PlantCARE48 programs. Both programs revealed the existence of cis-regulatory elements of Myb49, bZIP50, CCAAT-binding51, homeobox52, and E2F-DP53-related transcription factors (TFs) (which have already been found in genomic sequences of the Pennales P. tricornutum and the Centrics Thalassiosira pseudonana54) in the DIV promoters (Supplementary Fig. 1). The PlantCARE program48 also showed that all the DIV promoter regions include a plant-type light-responsive cis-regulatory element (Supplementary Fig. 1). To identify other regulatory sequences that are conserved amongst DIV and endogenous promoters, we examined the frequency of consensus TF binding sites (TFBSs), which are available in the JASPAR database55, through promoterome analyses41 using oPOSSUM version 356. The single site analysis (SSA) algorithm of oPOSSUM version 3 failed to detect any TFBS consensus between the DIV promoters and the potential promoter regions of P. tricornutum. Using the TFBS cluster analysis (TCA) algorithm available in oPOSSUM version 3, sequence motifs of some insect- and vertebrate-type homeobox binding sites were identified in the five DIV promoters and in the 99.9% potential promoter region of P. tricornutum (12,234 of 12,237 sequences, Z-score: 14.406, Fisher score: 0.001) (Supplementary Fig. 1 and Supplementary data). To obtain information on specific mechanisms that drive expression from DIV promoters, we attempted to find conserved motifs among the DIV promoter regions (CdP1, ClP1, and ClP2) using a consensus motif-finding algorithm (CONSENSUS) available through Melina II57 and using default parameters. The “GGCAGGCG” sequence was detected in CdP1, ClP1, and ClP2 through the CONSENSUS algorithm. In the sense strand of ClP1, there are three tandem repeats of the “GGCAGGCG” motif ordered from 5′ to 3′ (Supplementary Fig. 1). In ClP2, there are three tandem repeats, with the direction of the motifs ordered from 3′ to 5′. Only one motif of the 5′ to 3′ direction was found in the sense strand of CdP1 (Supplementary Fig. 1).
Discussion
This is the first report describing the functional characterization of various promoters identified and isolated from DIVs. We tested viral promoter activity levels in diatom cells transformed with specific vectors, in which the viral promoter drove the expression of an eGFP reporter gene. The activity of each tested promoter was estimated from the relative abundance of the eGFP gene transcript relative to that of the antibiotic-resistant gene driven by the endogenous fcp promoter (Fig. 1b). In turn, we identified the strongest promoters and overcame issues of gene expression variability between independent transgenic lines that can be due to differences in the copy numbers of transgenes integrated in a host genome58 and from the positioning of the introduced vector in the genome, i.e., the position effect59. Similar variations have been reported during the characterization of several promoters60. Moreover, variations in fluorescence levels of the eGFP protein were used to assess possible variations in translational efficiency levels in the different transgenic lines. These analyses allowed us to show for the first time that the isolated DIV promoters are functional in P. tricornutum. In particular, we found that the ClP1 promoter of a putative replication-associated (VP3) gene derived from ClorDNAV32 drives the stable expression of the reporter in cells grown in both light and dark conditions, and it shows higher levels of activity than a diatom endogenous promoter.
Interestingly, the activity levels of viral promoters may be heavily modulated in the growth phase. In Schizosaccharomyces pombe, gene expression mediated by the CaMV 35S promoter is induced during the log phase rather than during the stationary phase61. In our study, reporter expression controlled by ClP1 was induced at the stationary phase rather than at the log phase of P. tricornutum growth. In accordance with our data, a study on the relationship between the Cha. tenuissimus-infecting DNA virus (CtenDNAV) type II and the growth phase of Cha. tenuissimus showed that when CtenDNAV type II is inoculated into host cells at the stationary phase, a decrease in host cell quantities occurred one day after inoculation28. However, in host cells at the log phase, a decrease in host cell quantities occurred only after the cells reached the stationary phase28. The infectivity of CtenDNAV type I was also considerable when the host was at a stationary phase62. Moreover, an active replication of the viral genome of CtenDNAV type I was observed after the host cells reached the stationary phase62. Taken together, these results suggest that DIV promoter activation in a host diatom at the stationary phase may facilitate diatom lysis. Although gene expression regulation by DIV promoters in P. tricornutum is not fully understood, these findings suggest that a stronger understanding of the mechanisms that control DIV gene expression may lead to the development of a stronger understanding of diatom population control processes by DIVs in oceans.
Our assessment of viral promoter activity also allowed us to identify ClP1 as a novel DIV promoter that drives strong and stable gene expression in diatoms. ClP1 serves as a useful tool that may be exploited in diatom biotechnology applications for the high-level production of useful materials and for gene functional studies in genetically engineered diatoms63,64. The activity of the “stable” endogenous EF2 gene promoter recently isolated from P. tricornutum16 drove the expression of a transgene 1.2-fold stronger than that driven by the fcp promoter in light conditions16. In comparison, egfp mRNA levels normalized to rps in ClP1 transformants were found to be 3.3 times stronger than those of PtfcpA pro. transformants (Supplementary Fig. 6). This result suggests that ClP1 may induce a stronger expression of transgenes than the EF2 gene promoter throughout a photoperiod. In addition, ClP1 activity remains stable in low nutrient culture conditions. A combination of stable ClP1 activity levels in light/dark cycles under low nutrient conditions and high-level ClP1 activity levels in the stationary phase may have value in biotechnological applications.
In discussing whether DIV promoters may be applied to various diatom species, it is important to consider the phylogeny of diatoms composed of the two orders Pennales and Centrics1,65. In this study, the finding that ClP1 can be applied to both the Centrics diatom Chaetoceros sp. and the Pennales diatom P. tricornutum suggests that the DIV promoter region contains conserved regulatory elements that may be recognized via conserved TFs identified in Pennales and Centrics species54. Considering the phylogenetic relationship, conservation levels in TF families between the Pennales and Centrics diatoms, and our results, DIV promoters including ClP1 may be applied to various diatom species.
Both DIV promoters CdP1 and ClP1 are derived from Chaetoceros spp., although the transformation efficiency of P. tricornutum was found to be higher than that of Chaetoceros sp. It was reported that transformation efficiencies obtained by multi-pulse electroporation using the endogenous fcp promoter in P. tricournutum13 were approximately 10 times higher than those in Chaetoceros gracilis24, suggesting that the transformation efficiency levels in Chaetoceros are lower than those of Phaeodactylum, independent of the promoter used to drive the transgene expression. By contrast, ClP1 cannot function in green algae Chl. reinhardtii, suggesting that regulatory elements in promoter regions and transcriptional regulators differ between the Heterokont and Plantae lineages.
Via an in silico analysis of the DIV promoters, we examined conserved motifs that may be involved in the initiation of diatom virus gene expression. Topically, eukaryotic promoters consist of two regions: a core region and an upstream regulatory region66. The core region consists of an approximately 50−100 base pair (bp) sequence. It includes the transcription start site, known as an initiator (Inr), and flanking sequences such as the TATA box67. The regulatory region includes cis-regulatory elements that control gene expression through interaction with TFs66. Cis-regulatory elements mediate inducible, tissue specific, or development stage-specific gene expression66. In terms of Inr sequences, an Inr consensus has been found for some eukayotes67,68,69. Based on the transcription start site of P. tricornutum, the conserved sequence flanking the transcription start sites (CAY+1A, degenerate bases are described according to the IUPAC nucleotide code) is reported from the upstream regions of some FCP genes70. In addition, −2 to +2 sequences relative to the transcription start site of the CA1 gene (ca1) in P. tricornutum were found to be CACA34. We focused on sequences surrounding CAYA located in the upstream DNA sequence of FCP genes, and other endogenous genes of diatoms were examined via visual observation. We found a “TCAH1W” Inr-like sequence located between bp 26−61 upstream of the translation site in P. tricornutum FCP genes such as the fcpC, fcpD and fcpE, and P. tricornutum CA1 genes (Supplementary Table 3). In the tested DIV promoter regions, we also found this Inr-like sequence between bp 14−65 upstream of the proposed translation site (CdP1: 14 and 35 upstream, ClP1: 36 and 61 upstream, ClP2: 40 upstream, TnP1: 35 upstream, and TnP2: 65 upstream) (Supplementary Table 3 and Supplementary Fig. 1). To determine the universality of “TCAH+1W” in the 5′-flanking sequences (80 bases) of the P. tricornutum genes, we searched for “TCAH+1W” in 12,237 of the 5′-flanking sequences and found “TCAH+1W” in 8,306 of the 12,237 sequences (67.9%) of the potential promoter region of the P. tricornutum genes. From these findings, we present a new consensus motif “TCAH+1W”, which serves as a potential Inr-like sequence in the diatom.
While identifying other regulatory sequences that are conserved amongst DIV and endogenous promoters, we found sequence motifs of some insect- and vertebrate-type homeobox binding sites in the five DIV promoters and in the 99.9% potential promoter region of P. tricornutum via a promoterome analysis41. This finding suggests that these motifs may play a role in P. tricornutum transcription.
In addition to revealing the existence of canonical cis-regulatory elements, several studies have reported on the existence of other cis-regulatory elements in diatom endogenous gene promoters. Three putative cis-regulatory elements from the ca1 promoter of P. tricornutum, which responds to changes in CO2 and cAMP concentrations, have been found34,71. The two iron-responsive conserved cis-regulatory elements were found in upstream DNA sequences of iron starvation-induced protein genes in P. tricornutum72. The expression levels of FCP genes were found to oscillate in a circadian manner in response to a plant-type light-responsive cis-regulatory element in the fcp promoter40,41,73,74,75. Cis-regulatory elements of CO2/cAMP71 or iron72 were not found in any of the DIV promoter regions by visual observation. The PlantCARE program48 showed that all the DIV promoter regions possess a plant-type light-responsive cis-regulatory element. However, ClP1 activity was unaffected by light and dark cycles (Fig. 3), suggesting that the identified elements do not respond to light. EF2 gene promoter activity was also found to be stable in P. tricornutum throughout light and dark cycles. In the sequence of the promoter region of the EF2 gene, a plant-type light-responsive cis-regulatory element was also detected through the PlantCARE program48, supporting the notion that plant-type light-responsive cis-regulatory elements cannot respond to light in P. tricornutum.
The “GGCAGGCG” sequence was detected in CdP1, ClP1, and ClP2 via the CONSENSUS algorithm. The quantity, direction, and proximity of this consensus motif varied among these promoters. ClP1, which showed the highest degree of promoter activity (Fig. 2), possessed three tandem repeats of the “GGCAGGCG” motif in the sense strand in the 5′-to-3′ direction. For ClP2, which showed modest activity levels (Fig. 2), a 3′-to-5′ direction was found, even when three tandem repeats were present in the promoter region (Supplementary Fig. 1). Only one motif was found in the sense strand of CdP1, which also showed modest activity levels (Fig. 2 and Supplementary Fig. 1). However, one and three “GGCAGGCG” sequences were found in the antisense strands of the nos and CMV promoters, respectively, which showed their very weak activity levels (Fig. 2). In the latter case, the three motifs are scattered across the promoter region (Supplementary Fig. 8). These findings suggest that the quantity, direction, and proximity of the novel consensus motif may heavily affect the stability of promoter activity levels though light/dark cycles and of high-level promoter activity levels during the stationary phase in P. tricornutum. In addition to the motifs described above, unknown novel motifs may exist in the promoter regions of DIVs such as ClP1.
In summary, we have isolated promoters from several marine DIVs for the first time. The activity of the DIV promoter of ClP1 in the transformants was higher in the stationary phase than it was in the log phase. Compared to a diatom endogenous promoter, the mediated ClP1 showed a significantly higher level of reporter transcription and translation. Conservative motifs in sequences of the DIV promoters were found. Finally, our findings may help elucidate the mechanism of DIV gene expression, which may play a key role in the formation/decay of diatom blooms in oceans. In addition, the stable and high ClP1 promoter activity levels reported should prove useful for the metabolic engineering of diatoms.
Methods
Algal culture
The Pennales P. tricornutum Bohlin (National Research Institute of Aquaculture, Fisheries Research Agency, Japan, strain NRIA-0065), the Centrics Chaetoceros sp. (Japan Marine Science and Technology Center, Japan, strain CCK09)23, and unicellular green alga Chl. reinhardtii (strain CC-503) were used in this study. P. tricornutum was grown at 20 °C under a light/dark cycle of 12 h light and 12 h dark with ca. 90 μmol photons m−2 s−1 in liquid f/2 medium76. Chaetoceros sp. was grown at 20 °C under continuous illumination at 300 μmol photons m−2 s−1 in SWM3 medium77. The Chl. reinhardtii strain CC-503 was provided by the Chlamydomonas Resource Center (University of Minnesota). The cells were cultivated mixotrophically at 25 °C in Tris-acetate phosphate (TAP) medium78 under constant white fluorescent light conditions (84 μmol photons m−2 s−1) with gentle shaking.
Isolation of DIV promoter regions
DIVs and the sequences of the viral genome were isolated following the method described by Tomaru et al.32. In brief, a 450 ml exponentially growing diatom culture was inoculated with 5 ml of DIV suspension and then lysed. The lysate was filtrated through a 0.4 μm pore size polycarbonate membrane filter (Whatman®; GE Healthcare UK Ltd., Buckinghamshire, England) for the removal of cellular debris. The filtrated lysate was mixed with polyethylene glycol 6000 (final concentration: 10% wt/vol; Wako Pure Chemical Industries, Ltd., Osaka, Japan), and the mixture was stored at 4 °C in the dark overnight. To collect the virus particles, after centrifugation at 57,000 × g at 4 °C for 1.5 h, the pellet was washed with 10 mM phosphate buffer (pH 7.2) and centrifuged at 217,000 × g and 4 °C for 4 h. Viral genomic DNA was extracted from the pellet using a DNeasy Plant Mini Kit (QIAGEN Inc., CA) according to the manufacturer’s instructions. We isolated three viral genomes: the CdebDNAV31 genome, the ClorDNAV32 genome, and the TnitDNAV33 genome. ORFs of ClorDNAV32 have been identified by Tomaru et al.32. We identified ORFs of CdebDNAV31 and TnitDNAV33 using an ORF finder following the method reported by Tomaru et al.32. Fragments of upstream regions of the ORFs, such as the replication-associated protein (VP3) gene and the structural protein (VP2) gene as promoter regions (Fig. 1a), were amplified using PCR. The isolated viral promoters and the source of the promoters are summarized in Table 1. The primers used in this study are shown in Supplementary Table 1. All the DNA fragments, including those in the promoter regions, were amplified using PrimeSTAR® GXL DNA Polymerase (Takara Bio Inc., Otsu, Japan) according to the manufacturer’s instructions.
Amplification of DNA fragments for vector construction
To construct transformation vectors (Fig. 1 and Supplementary Figs 2 and 3), DNA fragments containing various promoter regions, a reporter gene (egfp), antibiotic-resistant genes such as the bleomycin-resistant gene (Sh ble) and the nourseothricin-resistant gene (nat), and a terminator region were cloned into Gateway® Pro Donor vectors using a Gateway® cloning system according to the instructions of MultiSite Gateway® Pro (Life Technologies Corporation, CA) for the preparation of entry vectors containing each fragment. DNA fragments containing PtfcpA pro., a CaMV 35S promoter, a CMV promoter, and a nos promoter were amplified from the genomic DNA of the P. tricornutum UTEX 646 strain (The Culture Collection of Algae (UTEX, The University of Texas at Austin)), pCAMBIA2301 (CAMBIA: Center for Application of Molecular Biology for International Agriculture, Canberra, Australia), phMGFP (Promega, WI), and pBI101.2 (Clontech, CA); these were used as templates. The egfp fragment was amplified from pNICgfp18 as a template. The terminator region of the FCP A-1A gene derived from Cyl. fusiformis (termed CffcpA ter.) was also amplified from pNICgfp18 as a template. The terminator region of the FCP gene derived from Thalassiosira pseudonana (termed Tpfcp ter.) was amplified from pTpfcp/nat21 as a template. Antibiotic genes, Sh ble, and nat were amplified from pNICgfp18 and pTpfcp/nat21 as templates. All the DNA fragments were amplified using PrimeSTAR® GXL DNA Polymerase (Takara Bio Inc., Otsu, Japan), with the primer added to the att sites in the Gateway® system (Supplementary Table 1) according to the manufacturer’s instructions. The amplicons were purified via agarose gel electrophoresis using a QIAquick® Gel Extraction Kit (QIAGEN Inc., CA).
Construction of double-cassette vectors
To evaluate promoter activity levels based on different transgene copy quantities and possible site integration effects, we prepared double-cassette vectors as shown in Fig. 1 using the Gateway® cloning system according to the instructions for MultiSite Gateway® Pro (Life Technologies Corporation, CA). To construct entry vectors, DNA fragments with various promoter regions, a reporter gene (egfp), and CffcpA ter. were cloned into pDONR221P1-P4, pDONR221P4r-P3r, and pDONR221P3-P2, respectively, using the Gateway® BP Clonase® II enzyme mix (Life Technologies Corporation, CA) according to the instructions designated for MultiSite Gateway® Pro (Life Technologies Corporation, CA). In cases involving the construction of the promoter-less vector, the egfp gene and CffcpA ter. were cloned into pDONR221P1-P5r and pDONR221P5-P2, respectively. To construct double-cassette vectors, a destination vector was prepared by cloning the Gateway® cloning cassette and Reading Frame Cassette A (Life Technologies Corporation, CA) into the KpnI site of pCfcp-ble18 that contained the antibiotic gene Sh ble driven by CffcpA pro. The LR recombination reactions, which allow the recombination of three (promoter-reporter gene-terminator) or two fragments (reporter gene-terminator) from the entry vector into the destination vector, were conducted using the Gateway® LR Clonase® II PLUS enzyme mix (Life Technologies Corporation, CA) according to the instructions designated for MultiSite Gateway® Pro (Life Technologies Corporation, CA). Finally, we constructed the transformation vector described as pTested Pro/EGFP/Cffcp Ter (P-ble) (Fig. 1a). To test the activity of the promoter from Cfnr pro., we used pNICgfp18 (Supplementary Fig. 5a).
Construction of single-cassette vectors
To determine transformation efficiency, the single-cassette vectors shown in Supplementary Figs 2 and 3 were constructed following the method for the construction of double-cassette vectors. The antibiotic genes (Sh ble and nat) and terminator regions (CffcpA ter. And Tpfcp ter.) were cloned into pDONR221P4r-P3r and pDONR221P3-P2, respectively, according to the instructions for MultiSite Gateway® Pro (Life Technologies Corporation, CA). To construct the promoter-less vector, the antibiotic gene and terminator were cloned into pDONR221P1-P5r and pDONR221P5-P2, respectively. To construct single-cassette vectors, the destination vector was prepared by cloning the Gateway® cloning cassette, Reading Frame Cassette A (Life Technologies Corporation, CA), into the EcoRI site of the pBluescript SK(-) (Agilent Technologies, CA). The LR recombination reactions, which allow the recombination of three fragments (promoter-antibiotic gene-terminator) or two fragments (antibiotic gene-terminator) from the entry vector into the destination vector, were performed using the Gateway LR Clonase II PLUS enzyme mix (Life Technologies Corporation, CA) according to the instructions for MultiSite Gateway® Pro (Life Technologies Corporation, CA). Finally, we constructed the single-cassette vectors (Supplementary Figs 2 and 3). For the transformation of P. tricornutum, PtfcpA pro. and ClP1 were examined. For the transformation of Chaetoceros sp. and Chl. reinhardtii CC-503, CdP1 and ClP1 were examined.
Transformation of P. tricornutum
Transformation vectors were introduced into P. tricornutum cells using the Biolistic PDS-1000/He particle delivery system (Bio-Rad Laboratories, CA) according to a method previously described10,12,23. Approximately 1 × 108 cells were spread on central areas covering one-third of a plate of solid f/2 medium containing 1% agarose HGS (Nacalai Tesque, Kyoto, Japan) ca. 1 h prior to bombardment. The plate was positioned on the second level (6 cm from the stopping screen) within the Biolistic chamber for bombardment. Tungsten particles M17 (3 mg, 1.1 μm diameter, Bio-Rad Laboratories, CA) were coated with 5 μg of the transformation vector in the presence of CaCl2 and spermidine for five shots, as instructed by the manufacturer. The cells were then bombarded with 600 ng of DNA-coated tungsten particles using a two-ply 650 psi (1300psi) rupture disk. After bombardment, the cells were recovered from the plates and incubated in liquid f/2 medium under standard growth conditions for 24 h. Cultures of P. tricornutum were concentrated by centrifugation and resuspended in 100 μl of f/2 medium. The cell suspensions were spread onto plates of solid f/2 medium containing 0.5% agarose HGS (Nacalai Tesque, Kyoto, Japan) supplemented with 500 μg ml−1 ZeocinTM (InvivoGen, CA). After 5 weeks of selective incubation under standard growth conditions, individual colonies that had formed on the plates were extracted using a platinum loop and suspended into liquid media with 300 μg ml−1 ZeocinTM.
Transformation of Chaetoceros sp
Single-cassette vectors for the transformation of Centrics diatoms (Supplementary Fig. 3) were introduced into Chaetoceros sp. cells using the Biolistic PDS-1000/He particle delivery system (Bio-Rad Laboratories, CA) according to a method previously described23. The cells were bombarded using a two-ply 650 psi (1300psi) rupture disk.
Transformation of Chl. reinhardtii CC-503
Nuclear transformation was performed using the electroporation method46. In brief, the cells were grown to 4 × 106 cells ml−1 in TAP medium. Subsequently, 2.5 × 107 cells were harvested by centrifugation and suspended in 250 μl of TAP medium supplemented with 50 mM sucrose (TAP/sucrose). Electroporation was performed by applying an exponential electric pulse of 0.7 kV at a capacitance level of 50 μF (Electro Cell Manipulator® 600; BTX® Harvard Apparatus, MA) and using 300 ng of non-linearized plasmids according to the manufacturer’s instructions. Transgenic strains were selected directly from TAP/agar plates containing ZeocinTM (10 μg ml−1), and the plates were incubated under continuous fluorescent light conditions (20 μmol m−2 s−1) at 25 °C.
PCR analysis of the introduced genes and of their promoters in the transformed cells
To determine whether the egfp gene with the tested promoters and the Sh ble gene with CffcpA pro. were introduced into colony-forming cells that showed resistance to ZeocinTM, regions containing the promoters and genes were amplified using the cells as a template. Genomic PCR analyses were conducted using Tks GflexTM DNA Polymerase (Takara Bio Inc., Otsu, Japan). The amplicons were analysed using agarose gel electrophoresis. The primers used in the genomic PCR analysis are listed in Supplementary Tables 1 and 2.
RNA isolation
Total RNA was isolated from approximately 5 × 107 transformed cells using an RNeasy Plant Mini Kit (QIAGEN Inc., CA) coupled with a RNase-Free DNase Set (QIAGEN Inc., CA). Reverse transcription (RT) was performed using a PrimeScript® RT reagent Kit (Perfect Real Time) (Takara Bio, Otsu, Japan). We also used a SuperPrepTM Cell Lysis & RT Kit for qPCR (TOYOBO, Osaka, Japan) to isolate total RNA and RT from some of the transformed cells.
Assessment of relative promoter activities
qRT-PCR experiments to quantify egfp and Sh ble transcripts in the transformed cells were performed in triplicate on a Thermal Cycler Dice® Real Time System Single MRQ (Takara Bio Inc., Otsu, Japan) using 2 μl of the cDNA mixture added as a template and SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara Bio Inc., Otsu, Japan). The cycling conditions involved denaturing for 30 s at 95 °C and 40 cycles of melting (5 s at 95 °C) and annealing coupled with an extension (30 s at 60 °C). To determine promoter activities, each qRT-PCR measurement was performed in triplicate using primers to identify the abundance of Sh ble mRNA and egfp mRNA (shown in Supplementary Table 1). The egfp mRNA levels were normalized by dividing by the Sh ble mRNA levels to minimize variations in transgene expression due to multiple insertions (Fig. 1b).
Effect of culture conditions on DIV promoter activity
To explore the effects of nutrient concentrations in media, growth phases of the tested strain, and light cycles on DIV promoter activity, two transformants with ClP1 were cultured in a low nutrient medium (f/10 medium) and standard nutrient medium (f/2 medium). Transformants at the log and stationary growth phases were collected at approximately 13:30. Transformants cultured in f/2 medium at the stationary growth phase were collected during various photoperiods. Total RNA samples from approximately 5 × 107 transformed cells were isolated using the above methods. Using qRT-PCR, we determined the abundance of egfp mRNA and that of rps mRNA. The transcript number of egfp was divided by that of rps, the expression of which was found to be constitutive under various growth conditions74 and which was used as an internal control. The primers for detecting rps mRNA are listed in Supplementary Table 1.
Flow cytometry
To measure the eGFP fluorescence for the transformed cells, approximately 1 × 104 cells were analysed using a BD LSRFortessa™ X-20 flow cytometer system equipped with BD FACS Diva™ software (BD Biosciences, CA). The eGFP fluorescence was analysed using a 488 nm laser and a 530/30 nm bandpass filter. Endogenous chlorophyll fluorescence was analysed using a 488 nm laser and a 610/20 nm bandpass filter. We evaluated cell sizes by measuring the mean forward scatter area (FSC-A) using a 488 nm laser and a 488/10 nm bandpass filter. The data were analysed using FlowJo software (FlowJo, LLC, OR). The cells were gated on endogenous chlorophyll fluorescence so that debris could be eliminated from the analysis conditions. The average values of approximately 1 × 104 cells for each sample were used for the statistical analysis.
Statistical analyses
Statistical analyses were performed on the activity levels of the promoters at the transcriptional and translational levels using Student’s t-test for two group mean values (the PtfcpA pro. transformants and the other promoter transformants), and statistical significance was achieved when P < 0.01 (**) and P < 0.05 (*). In regard to the transformation efficiency of P. tricornutum, statistical analyses were performed using Student’s t-test for two group mean values (the colony number of PtfcpA pro. and that of ClP1). With regard to transformation efficiency in Chl. reinhardtii, statistical analyses were performed using Student’s t-test for two group mean values (the colony number of pSP108 and those of DIV promoters, or that of the promoter-less vector and those of DIV promoters).
In silico analyses
Putative cis-regulatory elements in potential DIV promoter regions and other extrinsic promoters (the CaMV 35 S, CMV, and nos promoters) were identified by searching the PLACE47 and PlantCARE48 databases. The promoterome analysis was performed following the method described by Russo et al.41. In brief, we obtained 12,237 of the 5′-flanking sequences of the P. tricornutum genes from Ensembl Protists BioMart (Dataset: ASM15095v2 (2013-07-EBI-Phatr3))79,80. Using the oPOSSUM version 356 program, SSA and TCA tests were performed using default parameters. The TFBS profile matrix file for vertebrates, insects, nematodes, and fungi was used in the SSA. For the TCA, the TFBS profile matrix file for vertebrates, insects, and nematodes was used. A consensus sequence for the amplified regions of the tested promoters was also analysed using the consensus motif-finding algorithms CONSENSUS from Melina II57 and using default parameters amongst potential DIV and extrinsic promoters (CaMV 35S, CMV, and nos promoters).
Additional Information
How to cite this article: Kadono, T. et al. Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum. Sci. Rep. 5, 18708; doi: 10.1038/srep18708 (2015).
Supplementary Material
Acknowledgments
We thank Nicole Poulsen and Nils Kröger (Technische Universität Dresden) for providing the transformation vectors (pTpfcp/nat, pNICgfp, and pCfcp-ble); Keizo Nagasaki (Fisheries Research Agency, Japan) for critical discussions about DIVs; and Yasufumi Hikichi (Kochi University) for kind advice about the nos promoter and pBI101.2 vector. We also thank Sayo Kataoka and Ken-ichi Yagyu (Science Research Center, Kochi University) for technical supports on the analysis of eGFP fluorescence. We are grateful to Dennis Murphy (Ehime University) for English correction. This study was supported in part by the “Strategic Development of Next Generation Bioenergy Utilization Technology” of New Energy and Industrial Technology Development Organization (NEDO), Japan; in part by a Grant-in-Aid for Scientific Research (Nos. 24580268 and 15K14804) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan to M.A.; and in part by the Kochi University Presidents Discretionary Grant to M.A.; and in part by the Grand Emprunt Project-European Marine Biological Resource Centre-France (EMBRC) and ANR “DiaDomOil” PROGRAMME BIO-MATIERES & ENERGIES to A.F.
Footnotes
Author Contributions Experiments were conceived and designed by T.K., A.M., T. Ohama, A.F., N.K. and M.A. Experiments were performed by T.K., T.Y., A.M., T. Okami, N.K. and L.H. Data were analyzed by T.K., T.Y., T. Ohama and M.A. Reagents, materials, and analysis tools were contributed by T.K., A.M., N.K., Y.T., T. Okami, K.F., M.O., H.Y. and K.O. The final form of the manuscript was written and discussed by T.K., A.M., N.K., T. Ohama, A.F. and M.A.
References
- Mann D. G. & Droop S. J. M. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia 336, 19−32 (1996). [Google Scholar]
- Nelson D. M. et al. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycle 9, 359−372 (1995). [Google Scholar]
- Falkowski P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354−360 (2004). [DOI] [PubMed] [Google Scholar]
- Martin-Jézéquel V., Hildebrand M. & Brzezinski M. A. Silicon metabolism in diatoms: Implications for growth. J. Phycol. 36, 821−840 (2000). [Google Scholar]
- Wijffels R. H. & Barbosa M. J. An outlook on microalgal biofuels. Science 329, 796−799 (2010). [DOI] [PubMed] [Google Scholar]
- Armbrust E. V. et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79−86 (2004). [DOI] [PubMed] [Google Scholar]
- Bowler C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239−244 (2008). [DOI] [PubMed] [Google Scholar]
- Fabris M. et al. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70, 1004−1014 (2012). [DOI] [PubMed] [Google Scholar]
- Maheswari U., Mock T., Armbrust E. V. & Bowler C. Update of the Diatom EST Database: a new tool for digital transcriptomics. Nucleic Acids Res. 37, D1001−1005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt K. E., Kroth-Pancic P. G. & Grossman A. R. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572−579 (1996). [DOI] [PubMed] [Google Scholar]
- Zaslavskaia L. A., Casey Lippmeier J., Kroth P. G., Grossman A. R. & Apt K. E. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J.Phycol. 36, 379−386 (2000). [Google Scholar]
- Miyagawa A. et al. Research note: High efficiency transformation of the diatom Phaeodactylum tricornutum with a promoter from the diatom Cylindrotheca fusiformis. Phycol. Res. 57, 142−146 (2009). [Google Scholar]
- Miyahara M., Aoi M., Inoue-Kashino N., Kashino Y. & Ifuku K. Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Biosci. Biotechnol. Biochem. 77, 874−876 (2013). [DOI] [PubMed] [Google Scholar]
- Weyman P. D. et al. Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol. J. 13, 460−470 (2014). [DOI] [PubMed] [Google Scholar]
- Daboussi F. et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5, 3831 (2014). [DOI] [PubMed] [Google Scholar]
- Seo S., Jeon H., Hwang S., Jin E. & Chang K. S. Development of a new constitutive expression system for the transformation of the diatom Phaeodactylum tricornutum. Algal Res. 11, 50−54 (2015). [Google Scholar]
- Fischer H., Robl I., Sumper M. & Kröger N. Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 35, 113–120 (1999). [Google Scholar]
- Poulsen N. & Kröger N. A new molecular tool for transgenic diatoms: control of mRNA and protein biosynthesis by an inducible promoter-terminator cassette. FEBS J. 272, 3413−3423 (2005). [DOI] [PubMed] [Google Scholar]
- Dunahay T. G., Jarvis E. E. & Roessler P. G. Genetic transformation of the diatoms Cyclotella cryptia and Navicula saprophila. J. Phycol. 31, 1004−1012 (1995). [Google Scholar]
- Sabatino V. et al. Ferrante MI. Establishment of genetic transformation in the sexually reproducing diatoms Pseudo-nitzschia multistriata and Pseudo-nitzschia arenysensis and inheritance of the transgene. Mar. Biotechnol. (NY) 17, 452−462 (2015). [DOI] [PubMed] [Google Scholar]
- Poulsen N., Chesley P. M. & Kröger N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 42, 1059−1065 (2006). [Google Scholar]
- Falciatore A., Casotti R., Leblanc C., Abrescia C. & Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. (NY) 1, 239−251 (1999). [DOI] [PubMed] [Google Scholar]
- Miyagawa-Yamaguchi A. et al. Stable nuclear transformation of the diatom Chaetoceros sp. Phycological Res. 59, 113−119 (2011). [Google Scholar]
- Ifuku K. et al. A stable and efficient nuclear transformation system for the diatom Chaetoceros gracilis. Photosynth. Res. 123, 203−211 (2015). [DOI] [PubMed] [Google Scholar]
- Muto M. et al. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—a high triglyceride producer. Mar. Biotechnol. (NY) 15, 48−55 (2013). [DOI] [PubMed] [Google Scholar]
- Karas B. J. et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6, 6925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehahn Y. et al. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr. Biol. 24, 2041−2046 (2014). [DOI] [PubMed] [Google Scholar]
- Kimura K. & Tomaru Y. Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom. Appl. Environ. Microbiol. 81, 1120−1131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801−812 (2007). [DOI] [PubMed] [Google Scholar]
- de Vargas C. et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). [DOI] [PubMed] [Google Scholar]
- Tomaru Y., Shirai Y., Suzuki H., Nagasaki T. & Nagumo T. Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros debilis. Aquat. Microb. Ecol. 50, 103−112 (2008). [Google Scholar]
- Tomaru Y. et al. Isolation and characterization of a single-stranded DNA virus infecting Chaetoceros lorenzianus Grunow. Appl. Environ. Microbio.l 77, 5285−5293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y. et al. First evidence for the existence of pennate diatom viruses. ISME J. 6, 1445−1448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Nakatsuma D., Ishida M. & Matsuda Y. Regulation of the expression of intracellular beta-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 139, 1041−1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R., Eduafo P. M. & Posewitz M. C. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89−95 (2011). [DOI] [PubMed] [Google Scholar]
- Niu Y. F. et al. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs 11, 4558−4569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentacoste E. M. et al. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. USA 110, 19748−19753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M. L., Haslam R. P., Napier J. A. & Sayanova O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 22, 3−9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadono T. et al. Effect of an introduced phytoene synthase gene expression on carotenoid biosynthesis in the marine diatom Phaeodactylum tricornutum. Mar. Drugs 13, 5334−5357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltjen A., Marquardt J. & Rhiel E. Differential circadian expression of genes fcp2 and fcp6 in Cyclotella cryptica. Int. Microbiol. 7, 127−131 (2004). [PubMed] [Google Scholar]
- Russo M. T., Annunziata R., Sanges R., Ferrante M. I. & Falciatore A. The upstream regulatory sequence of the light harvesting complex Lhcf2 gene of the marine diatom Phaeodactylum tricornutum enhances transcription in an orientation- and distance-independent fashion. Mar. Genomics 24, 69−79 (2015). [DOI] [PubMed] [Google Scholar]
- Schmidt E. V., Christoph G., Zeller R. & Leder P. The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol. Cell Biol. 10, 4406−4411 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P. N. & Chua N. H. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250, 959−966 (1990). [DOI] [PubMed] [Google Scholar]
- Sakaue K., Harada H. & Matsuda Y. Development of gene expression system in a marine diatom using viral promoters of a wide variety of origin. Physiol. Plant. 133, 59−67 (2008). [DOI] [PubMed] [Google Scholar]
- Stevens D. R., Rochaix J. D. & Purton S. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251, 23−30 (1996). [DOI] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A. & Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821−1828 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M. & Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297−300 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325−327 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambawat S., Sharma P., Yadav N. R. & Yadav R. C. MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307−321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M. et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106−111 (2002). [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M. & Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157−166 (2013). [DOI] [PubMed] [Google Scholar]
- Rezsohazy R., Saurin A. J., Maurel-Zaffran C. & Graba Y. Cellular and molecular insights into Hox protein action. Development 142, 1212−1227 (2015). [DOI] [PubMed] [Google Scholar]
- Zheng N., Fraenkel E., Pabo C. O. & Pavletich N. P. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666−674 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayko E., Maumus F., Maheswari U., Jabbari K. & Bowler C. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188, 52−66 (2010). [DOI] [PubMed] [Google Scholar]
- Mathelier A. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142−147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon A. T., Arenillas D. J., Worsley Hunt R. & Wasserman W. W. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2, 987−1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura T., Makiguchi H., Makita Y., Yamashita R. & Nakai K. Melina II: a web tool for comparisons among several predictive algorithms to find potential motifs from promoter regions. Nucleic Acids Res. 35, W227–231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs S. L., Warkentin T. D. & DeLong C. M. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol. Biol. 21, 17−26 (1993). [DOI] [PubMed] [Google Scholar]
- Wakimoto B. T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell 93, 321−324 (1998). [DOI] [PubMed] [Google Scholar]
- Mitra A. & Higgins D. W. The Chlorella virus adenine methyltransferase gene promoter is a strong promoter in plants. Plant Mol. Biol. 26, 85−93 (1994). [DOI] [PubMed] [Google Scholar]
- Smerdon G. R., Aves S. J. & Walton E. F. Production of human gastric lipase in the fission yeast Schizosaccharomyces pombe. Gene 165, 313−318 (1995). [DOI] [PubMed] [Google Scholar]
- Tomaru Y., Kimura K. & Yamaguchi H. Temperature alters algicidal activity of DNA and RNA viruses infecting Chaetoceros tenuissimus. Aquat. Microb. Ecol. 73, 171−183 (2014). [Google Scholar]
- Venayak N., Anesiadis N., Cluett W. R. & Mahadevan R. Engineering metabolism through dynamic control. Curr. Opin. Biotechnol. 34, 142−152 (2015). [DOI] [PubMed] [Google Scholar]
- Pulz O. & Gross W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 65, 635−648 (2004). [DOI] [PubMed] [Google Scholar]
- Theriot E. C., Ashworth M., Ruck E., Nakov T. & Jansen R. K. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol. Evol. 143, 278−296 (2010). [Google Scholar]
- Dutt M., Dhekney S. A., Soriano L., Kandel R. & Grosser J. W. Temporal and spatial control of gene expression in horticultural crops. Horticulture Research 1, 14047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T. & Kadonaga J. T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225−229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod A., Smart C. D. & Fry W. E. Core promoter structure in the oomycete Phytophthora infestans. Eukaryotic Cell 3, 91−99 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston D. R. & Johnson P. J. Gene transcription in Trichomonas vaginalis. Parasitol Today 14, 261−265 (1998). [DOI] [PubMed] [Google Scholar]
- Bhaya D. & Grossman A. R. Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Res. 21, 4458−4466 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno N. et al. CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 158, 499−513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga R., Niwa-Kubota M., Matsui H. & Matsuda Y. Characterization of iron-responsive promoters in the marine diatom Phaeodactylum tricornutum. Mar. Genomics 16, 55−62 (2014). [DOI] [PubMed] [Google Scholar]
- Leblanc C., Falciatore A., Watanabe M. & Bowler C. Semi-quantitative RT-PCR analysis of photoregulated gene expression in marine diatoms. Plant Mol. Biol. 40, 1031−1044 (1999). [DOI] [PubMed] [Google Scholar]
- Siaut M. et al. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23−35 (2007). [DOI] [PubMed] [Google Scholar]
- Brakemann T., Frank B., Peter K. & Erhard R. Structural and functional characterization of putative regulatory DNA sequences of fcp genes in the centric diatom Cyclotella cryptica. Diatom Res. 23, 31−49 (2008). [Google Scholar]
- Guillard R. L. [Culture of phytoplankton for feeding marine invertebrates]. In: Culture of Marine Invertebrate Animals [ Smith W., Chanley M. & Guillard R. L. (eds.)] [26−60] (Springer US, New York, 1975). [Google Scholar]
- Chen L. C.-M., Edelstein T. & McLachian J. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5, 211−220 (1969). [DOI] [PubMed] [Google Scholar]
- Gorman D. S. & Levine R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665−1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J. et al. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42, D546−552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D. et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43, W589−598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. R., Winter J. A., Barnason A. R., Rogers S. G. & Fraley R. T. Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants. Nucleic Acids Res. 15, 1543−1558 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M. et al. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41, 521−530 (1985). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.