Abstract
Indoor fungi are a major cause of cosmetic and structural damage of buildings worldwide and prolonged exposure of these fungi poses a health risk. Aspergillus, Penicillium and Cladosporium species are the most predominant fungi in indoor environments. Cladosporium species predominate under ambient conditions. A total of 123 Cladosporium isolates originating from indoor air and indoor surfaces of archives, industrial factories, laboratories, and other buildings from four continents were identified by sequencing the internal transcribed spacer (ITS), and a part of the translation elongation factor 1α gene (TEF) and actin gene (ACT). Species from the Cladosporium sphaerospermum species complex were most predominant representing 44.7% of all isolates, while the Cladosporium cladosporioides and Cladosporium herbarum species complexes represented 33.3% and 22.0%, respectively. The contribution of the C. sphaerospermum species complex was 23.1% and 58.2% in the indoor air and isolates from indoor surfaces, respectively. Isolates from this species complex showed growth at lower water activity (≥ 0.82) when compared to species from the C. cladosporioides and C. herbarum species complexes (≥ 0.85). Together, these data indicate that xerotolerance provide the C. sphaerospermum species complex advantage in colonizing indoor surfaces. As a consequence, C. sphaerospermum are proposed to be the most predominant fungus at these locations under ambient conditions. Findings are discussed in relation to the specificity of allergy test, as the current species of Cladosporium used to develop these tests are not the predominant indoor species.
Introduction
Indoor fungal growth represents a global problem. For instance, about 25% of dwellings of social housing in the European Union show fungal growth [1, 2]. This causes disfigurement of the building materials and poses a health threat for the occupants and particularly for asthmatic and allergic patients [3, 4]. Indoor fungal growth is strongly increased after incidents of water damage caused for instance by flooding or leakage [5–8].
In outdoor air samples, species belonging to the genera Aspergillus, Penicillium and Cladosporium are commonly occurring. These genera are also predominant in indoor environments, indicating a strong correlation in fungal presence between indoor and outdoor air [9–12]. Penicillium chrysogenum and Aspergillus versicolor are particularly abundant in the indoor environment after water damage, while Cladosporium species dominate under ambient conditions [7, 13–15]. Cladosporium are widespread fungi. In nature, they are often found on dead plant material and on plant surfaces and are therefore considered as phylloplane fungi [16–18]. They have also been isolated from hypersaline environments, hence Cladosporium halotolerans [19, 20], and from soil and rocks such as artic rock [21–24]. These fungi can also be isolated from man-made products such as paint, food, textiles, books, glass windows and wall paper [25].
The genus Cladosporium comprises three species complexes, namely C. cladosporioides, C. herbarum and C. sphaerospermum, representing 169species [25]. The Cladosporium species complexes can be distinguished based on morphology and DNA sequences of the internal transcribed spacer (ITS), translation elongation factor 1α (TEF) and actin (ACT) loci [19, 20, 25, 26]. It has been stated that C. cladosporioides is the most abundant fungus in outdoor air [13]. As the composition of indoor species reflects the composition of the outdoor species we would expect to find C. cladosporioides. However, pilot studies of indoor samples at our institute suggested that members of the C. sphaerospermum species complex were predominant in the indoor environment. This prompted us to study a larger number of isolates collected over 8 years from locations with fungal problems. We identified these isolates using the sequences of 69 species from Bensch et. al. [25]. These 69 species comprise 2 species from outside a species complex and 7, 21 and 39 species from the C, sphaerospermum, C. herbarum and C. cladosporioides species complexes, respectively. We here show that C. sphaerospermum is the most predominant Cladosporium species complex in the indoor environment in general and on indoor surfaces in particular. The latter may be explained by the higher xerotolerance of this species complex when compared to C. cladosporioides and C. herbarum. This is of interest as most allergy tests are focused on Cladosporium herbarum, which has not been identified during this study.
Materials and Methods
Isolation of organisms
Isolates of indoor Cladosporium species were taken from the research collection of the Applied and Industrial Mycology (DTO) group of the CBS Fungal Biodiversity Centre. The isolates were collected during a period of time from 2006 to 2013 on request from residents or owners who noticed indoor fungal problems. The isolates originated from indoor environments including archives, industrial factories, laboratories, and other buildings with or without water damage (Table 1). In these situations usually both swab and air samples were collected. Air samples were taken by using a MAS-100 (Merck) air sampler. This samples an amount of air depending on the size of the room ranging from 15 to 1000 liter. The air is sampled on 2% malt extract agar supplemented with penicillin and streptomycin (MEA p/s) (Oxoid) or on dichloran 18% glycerol agar (DG18) (Oxoid). Samples of indoor surfaces were taken by using a sterile cotton swab or sellotape [27] and were also grown on MEA p/s or DG18. Strains were stored at -80°C in 30% glycerol, 0.025% Tween 80 and 0.025% agar or 30% glycerol, 0.01% Tween 80, 5 mM N-(2-acetamido)-2-aminoethanesulfonic acid (ACES), pH 6.8.
Table 1. Isolates of Cladosporium species identified and used in this study.
| Species complex | Identified species | DTO nr. | Isolation | CBS nr. | Geographical origin | ITS | TEF | ACT | |
|---|---|---|---|---|---|---|---|---|---|
| Cladosporioides | C. acalyphae | 086-C5 | December-08 | S | s Hertogenbosch, The Netherlands | KP701887 | KP701764 | KP702010 | |
| Cladosporioides | C. angustisporum | 127-E6 | April-09 | A | USA | KP701935 | KP701812 | KP702057 | |
| Cladosporioides | C. australiense | 072-C8 | July-08 | 139572 | A | Amsterdam, The Netherlands | KP701873 | KP701750 | KP701996 |
| Cladosporioides | C. australiense | 082-E3 | November-08 | A | Amsterdam, The Netherlands | KP701878 | KP701755 | KP702001 | |
| Cladosporioides | C. australiense | 090-D2 | January-09 | S | Rijswijk, The Netherlands | KP701899 | KP701776 | KP702022 | |
| Cladosporioides | C. australiense | 109-E8 | October-09 | S | Denmark | KP701914 | KP701791 | KP702037 | |
| Cladosporioides | C. australiense | 255-F3 | March-13 | S | Amersfoort, The Netherlands | KP701978 | KP701855 | KP702100 | |
| Cladosporioides | C. cladosporioides | 039-G6 | April-07 | 112388 | A | Germany | KP701868 | KP701745 | KP701991 |
| Cladosporioides | C. cladosporioides | 071-G1 | July-08 | 139571 | Indoor | Greece | KP701872 | KP701749 | KP701995 |
| Cladosporioides | C. cladosporioides | 082-F1 | November-08 | A | Weert, The Netherlands | KP701879 | KP701756 | KP702002 | |
| Cladosporioides | C. cladosporioides | 090-C6 | January-09 | S | Rijswijk, The Netherlands | KP701898 | KP701775 | KP702021 | |
| Cladosporioides | C. cladosporioides | 102-A4 | May-09 | S | Hungary | KP701905 | KP701782 | KP702028 | |
| Cladosporioides | C. cladosporioides | 109-I4 | October-09 | S | Denmark | KP701920 | KP701797 | KP702043 | |
| Cladosporioides | C. cladosporioides | 109-I6 | October-09 | S | Denmark | KP701922 | KP701799 | KP702045 | |
| Cladosporioides | C. cladosporioides | 127-D8 | April-09 | A | The Netherlands | KP701933 | KP701810 | KP702055 | |
| Cladosporioides | C. cladosporioides | 147-A9 | Winter 2009 | Indoor | Hungary | KP701941 | KP701818 | KP702063 | |
| Cladosporioides | C. delicatulum | 082-F3 | November-08 | 139574 | A | Weert, The Netherlands | KP701880 | KP701757 | KP702003 |
| Cladosporioides | C. delicatulum | 134-D3 | June-10 | Indoor | Algeria | KP701939 | KP701816 | KP702061 | |
| Cladosporioides | C. delicatulum | 145-C4 | November-10 | Indoor | Germany | KP701940 | KP701817 | KP702062 | |
| Cladosporioides | C. delicatulum | 167-H5 | May-11 | A | Poland | KP701964 | KP701841 | KP702086 | |
| Cladosporioides | C. globisporum | 220-D4 | August-12 | 139587 | S | Utrecht, The Netherlands | KP701967 | KP701844 | KP702089 |
| Cladosporioides | C. inversicolor | 072-C9 | July-08 | 139573 | A | Amsterdam, The Netherlands | KP701874 | KP701751 | KP701997 |
| Cladosporioides | C. inversicolor | 108-F8 | September-09 | Indoor | France | KP701908 | KP701785 | KP702031 | |
| Cladosporioides | C. perangustum | 220-D5 | August-12 | 139588 | S | Utrecht, The Netherlands | KP701968 | KP701845 | KP702090 |
| Cladosporioides | C. perangustum | 127-E1 | April-09 | A | USA | KP701934 | KP701811 | KP702056 | |
| Cladosporioides | C. pseudocladosporioides | 084-F1 | December-08 | 139575 | Indoor | Germany | KP701881 | KP701758 | KP702004 |
| Cladosporioides | C. pseudocladosporioides | 079-F4 | October-08 | S | s Hertogenbosch, The Netherlands | KP701877 | KP701754 | KP702000 | |
| Cladosporioides | C. pseudocladosporioides | 150-C1 | February-11 | A | Portugal | KP701943 | KP701820 | KP702065 | |
| Cladosporioides | C. pseudocladosporioides | 151-D1 | February-11 | A | Portugal | KP701946 | KP701823 | KP702068 | |
| Cladosporioides | C. pseudocladosporioides | 151-G7 | February-11 | A | Portugal | KP701949 | KP701826 | KP702071 | |
| Cladosporioides | C. subuliforme | 130-H8 | May-10 | A | Thailand | KP701938 | KP701815 | KP702060 | |
| Cladosporioides | C. tenuissimum | 109-A1 | October-09 | A | Thailand | KP701910 | KP701787 | KP702033 | |
| Cladosporioides | C. tenuissimum | 130-F6 | May-10 | A | Thailand | KP701937 | KP701814 | KP702059 | |
| Cladosporioides | Unidentified | 056-H7 | August-07 | S | Utrecht, The Netherlands | KP701871 | KP701748 | KP701994 | |
| Cladosporioides | Unidentified | 072-E4 | July-08 | A | Amsterdam, The Netherlands | KP701875 | KP701752 | KP701998 | |
| Cladosporioides | Unidentified | 084-F2 | December-08 | Indoor | Germany | KP701882 | KP701759 | KP702005 | |
| Cladosporioides | Unidentified | 086-B3 | December-08 | S | s Hertogenbosch, The Netherlands | KP701886 | KP701763 | KP702009 | |
| Cladosporioides | Unidentified | 108-G8 | October-09 | A | Thailand | KP701909 | KP701786 | KP702032 | |
| Cladosporioides | Unidentified | 109-E7 | October-09 | S | Denmark | KP701913 | KP701790 | KP702036 | |
| Cladosporioides | Unidentified | 109-F2 | October-09 | S | Denmark | KP701915 | KP701792 | KP702038 | |
| Herbarum | C. allicinum | 109-I5 | October-09 | 139578 | S | Denmark | KP701921 | KP701798 | KP702044 |
| Herbarum | C. allicinum | 121-H1 | January-10 | 139580 | Indoor | Germany | KP701930 | KP701807 | a |
| Herbarum | C. allicinum | 073-C8 | July-08 | Indoor | Greece | KP701876 | KP701753 | KP701999 | |
| Herbarum | C. allicinum | 084-F3 | December-08 | Indoor | Germany | KP701883 | KP701760 | KP702006 | |
| Herbarum | C. allicinum | 086-D5 | December-08 | S | s Hertogenbosch, The Netherlands | KP701888 | KP701765 | KP702011 | |
| Herbarum | C. allicinum | 089-B9 | January-09 | A | Rijssen, The Netherlands | KP701891 | KP701768 | KP702014 | |
| Herbarum | C. allicinum | 089-G4 | January-09 | A | Eindhoven, The Netherlands | KP701894 | KP701771 | KP702017 | |
| Herbarum | C. allicinum | 089-G6 | January-09 | A | Eindhoven, The Netherlands | KP701895 | KP701772 | KP702018 | |
| Herbarum | C. allicinum | 089-H3 | January-09 | A | Eindhoven, The Netherlands | KP701896 | KP701773 | KP702019 | |
| Herbarum | C. allicinum | 090-D3 | January-09 | S | Rijswijk, The Netherlands | KP701900 | KP701777 | KP702023 | |
| Herbarum | C. allicinum | 101-A1 | May-09 | S | The Netherlands | KP701903 | KP701780 | KP702026 | |
| Herbarum | C. allicinum | 106-C2 | July-09 | A | Amsterdam, The Netherlands | KP701906 | KP701783 | KP702029 | |
| Herbarum | C. allicinum | 109-E6 | October-09 | S | Denmark | KP701912 | KP701789 | KP702035 | |
| Herbarum | C. allicinum | 109-F3 | October-09 | S | Denmark | KP701916 | KP701793 | KP702039 | |
| Herbarum | C. allicinum | 109-F5 | October-09 | S | Denmark | KP701918 | KP701795 | KP702041 | |
| Herbarum | C. allicinum | 110-B7 | October-09 | A | Denmark | KP701923 | KP701800 | KP702046 | |
| Herbarum | C. allicinum | 111-A5 | October-09 | A | Denmark | KP701924 | KP701801 | KP702047 | |
| Herbarum | C. allicinum | 249-G3 | March-13 | S | The Hague, The Netherlands | KP701975 | KP701852 | KP702097 | |
| Herbarum | C. ramotenellum | 089-C1 | January-09 | 139577 | A | Rijssen, The Netherlands | KP701892 | KP701769 | KP702015 |
| Herbarum | C. ramotenellum | 255-G5 | March-13 | 139590 | S | Utrecht, The Netherlands | KP701983 | KP701860 | KP702105 |
| Herbarum | C. ramotenellum | 109-F4 | October-09 | S | Denmark | KP701917 | KP701794 | KP702040 | |
| Herbarum | C. ramotenellum | 151-G3 | February-11 | A | Portugal | KP701947 | KP701824 | KP702069 | |
| Herbarum | C. ramotenellum | 151-G6 | February-11 | A | Portugal | KP701948 | KP701825 | KP702070 | |
| Herbarum | C. ramotenellum | 152-D9 | February-11 | A | Portugal | KP701950 | KP701827 | KP702072 | |
| Herbarum | C. ramotenellum | 249-F5 | March-13 | S | The Hague, The Netherlands | KP701972 | KP701849 | KP702094 | |
| Herbarum | C. sinuosum | 109-I2 | October-09 | S | Denmark | KP701919 | KP701796 | KP702042 | |
| Herbarum | C. tenellum | 127-D7 | January-09 | 139582 | A | USA | KP701932 | KP701809 | KP702054 |
| Herbarum | Unidentified | 090-H8 | January-09 | S | Utrecht, The Netherlands | KP701901 | KP701778 | KP702024 | |
| Sphaerospermum | C. dominicanum | 255-H5 | March-13 | 139591 | S | Utrecht, The Netherlands | KP701987 | KP701864 | KP702109 |
| Sphaerospermum | C. dominicanum | 249-F4 | March-13 | S | The Hague, The Netherlands | KP701971 | KP701848 | KP702093 | |
| Sphaerospermum | C. dominicanum | 255-F7 | March-13 | S | Utrecht, The Netherlands | KP701979 | KP701856 | KP702101 | |
| Sphaerospermum | C. halotolerans | 147-B9 | Winter 2009 | 139583 | Indoor | Hungary | KP701942 | KP701819 | KP702064 |
| Sphaerospermum | C. halotolerans | 161-D3 | May-11 | 139585 | S | Gilze, The Netherlands | KP701955 | KP701832 | KP702077 |
| Sphaerospermum | C. halotolerans | 164-A6 | August-11 | 139586 | S | Veenendaal, The Netherlands | KP701963 | KP701840 | KP702085 |
| Sphaerospermum | C. halotolerans | 220-D7 | August-12 | 139589 | S | Utrecht, The Netherlands | KP701969 | KP701846 | KP702091 |
| Sphaerospermum | C. halotolerans | 049-E7 | September-07 | S | Utrecht, The Netherlands | KP701869 | KP701746 | KP701992 | |
| Sphaerospermum | C. halotolerans | 102-A1 | May-09 | S | Hungary | KP701904 | KP701781 | KP702027 | |
| Sphaerospermum | C. halotolerans | 109-D3 | October-09 | A | Thailand | KP701911 | KP701788 | KP702034 | |
| Sphaerospermum | C. halotolerans | 114-H7 | December-09 | S | Thailand | KP701925 | KP701802 | KP702048 | |
| Sphaerospermum | C. halotolerans | 114-I3 | December-09 | S | Thailand | KP701926 | KP701803 | KP702049 | |
| Sphaerospermum | C. halotolerans | 117-H3 | January-10 | Indoor | The Netherlands | KP701929 | KP701806 | KP702052 | |
| Sphaerospermum | C. halotolerans | 127-E8 | April-09 | A | USA | KP701936 | KP701813 | KP702058 | |
| Sphaerospermum | C. halotolerans | 153-C3 | February-11 | S | Utrecht, The Netherlands | KP701952 | KP701829 | KP702074 | |
| Sphaerospermum | C. halotolerans | 160-I2 | March-11 | S | Utrecht, The Netherlands | KP701953 | KP701830 | KP702075 | |
| Sphaerospermum | C. halotolerans | 161-D5 | May-11 | S | Gilze, The Netherlands | KP701957 | KP701834 | KP702079 | |
| Sphaerospermum | C. halotolerans | 161-D6 | May-11 | S | Gilze, The Netherlands | KP701958 | KP701835 | KP702080 | |
| Sphaerospermum | C. halotolerans | 220-D3 | August-12 | S | Utrecht, The Netherlands | KP701966 | KP701843 | KP702088 | |
| Sphaerospermum | C. halotolerans | 249-F9 | March-13 | S | The Hague, The Netherlands | KP701974 | KP701851 | KP702096 | |
| Sphaerospermum | C. halotolerans | 249-G4 | March-13 | S | The Hague, The Netherlands | KP701976 | KP701853 | KP702098 | |
| Sphaerospermum | C. halotolerans | 255-F8 | March-13 | S | Utrecht, The Netherlands | KP701980 | KP701857 | KP702102 | |
| Sphaerospermum | C. halotolerans | 255-G4 | March-13 | S | Utrecht, The Netherlands | KP701982 | KP701859 | KP702104 | |
| Sphaerospermum | C. halotolerans | 255-G6 | March-13 | S | Utrecht, The Netherlands | KP701984 | KP701861 | KP702106 | |
| Sphaerospermum | C. halotolerans | 255-H3 | March-13 | S | Utrecht, The Netherlands | KP701985 | KP701862 | KP702107 | |
| Sphaerospermum | C. halotolerans | 257-F4 | March-13 | S | Utrecht, The Netherlands | KP701989 | KP701866 | KP702111 | |
| Sphaerospermum | C. langeronii | 124-D5 | February-10 | 139581 | A | Ospel, The Netherlands | KP701931 | KP701808 | KP702053 |
| Sphaerospermum | C. langeronii | 085-H6 | December-08 | A | s Hertogenbosch, The Netherlands | KP701885 | KP701762 | KP702008 | |
| Sphaerospermum | C. sphaerospermum | 084-F4 | December-08 | 139576 | Indoor | Germany | KP701884 | KP701761 | KP702007 |
| Sphaerospermum | C. sphaerospermum | 117-G5 | January-10 | 139579 | Indoor | The Netherlands | KP701927 | KP701804 | KP702050 |
| Sphaerospermum | C. sphaerospermum | 150-H8 | February-11 | 139584 | A | Portugal | KP701944 | KP701821 | KP702066 |
| Sphaerospermum | C. sphaerospermum | 017-C7 | May-06 | S | Eindhoven, The Netherlands | KP701867 | KP701744 | KP701990 | |
| Sphaerospermum | C. sphaerospermum | 049-H5 | September-07 | S | Utrecht, The Netherlands | KP701870 | KP701747 | KP701993 | |
| Sphaerospermum | C. sphaerospermum | 086-E7 | December-08 | Indoor | Utrecht, The Netherlands | KP701889 | KP701766 | KP702012 | |
| Sphaerospermum | C. sphaerospermum | 086-E8 | December-08 | Indoor | Utrecht, The Netherlands | KP701890 | KP701767 | KP702013 | |
| Sphaerospermum | C. sphaerospermum | 089-E9 | January-09 | A | Eindhoven, The Netherlands | KP701893 | KP701770 | KP702016 | |
| Sphaerospermum | C. sphaerospermum | 090-A1 | January-09 | A | Eindhoven, The Netherlands | KP701897 | KP701774 | KP702020 | |
| Sphaerospermum | C. sphaerospermum | 090-I1 | January-09 | S | Utrecht, The Netherlands | KP701902 | KP701779 | KP702025 | |
| Sphaerospermum | C. sphaerospermum | 106-D4 | July-09 | A | Amsterdam, The Netherlands | KP701907 | KP701784 | KP702030 | |
| Sphaerospermum | C. sphaerospermum | 117-H2 | January-10 | Indoor | The Netherlands | KP701928 | KP701805 | KP702051 | |
| Sphaerospermum | C. sphaerospermum | 150-I8 | February-11 | A | Portugal | KP701945 | KP701822 | KP702067 | |
| Sphaerospermum | C. sphaerospermum | 153-B7 | February-11 | S | Utrecht, The Netherlands | KP701951 | KP701828 | KP702073 | |
| Sphaerospermum | C. sphaerospermum | 160-I4 | March-11 | S | Utrecht, The Netherlands | KP701954 | KP701831 | KP702076 | |
| Sphaerospermum | C. sphaerospermum | 161-D4 | May-11 | S | Gilze, The Netherlands | KP701956 | KP701833 | KP702078 | |
| Sphaerospermum | C. sphaerospermum | 161-D7 | May-11 | S | Gilze, The Netherlands | KP701959 | KP701836 | KP702081 | |
| Sphaerospermum | C. sphaerospermum | 161-D8 | May-11 | S | Gilze, The Netherlands | KP701960 | KP701837 | KP702082 | |
| Sphaerospermum | C. sphaerospermum | 161-D9 | May-11 | S | Gilze, The Netherlands | KP701961 | KP701838 | KP702083 | |
| Sphaerospermum | C. sphaerospermum | 194-A4 | April-12 | S | Utrecht, The Netherlands | KP701965 | KP701842 | KP702087 | |
| Sphaerospermum | C. sphaerospermum | 244-C6 | S | Germany | KP701970 | KP701847 | KP702092 | ||
| Sphaerospermum | C. sphaerospermum | 249-F7 | March-13 | S | The Hague, The Netherlands | KP701973 | KP701850 | KP702095 | |
| Sphaerospermum | C. sphaerospermum | 249-G5 | March-13 | S | The Hague, The Netherlands | KP701977 | KP701854 | KP702099 | |
| Sphaerospermum | C. sphaerospermum | 255-G3 | March-13 | S | Utrecht, The Netherlands | KP701981 | KP701858 | KP702103 | |
| Sphaerospermum | C. sphaerospermum | 255-H4 | March-13 | S | Utrecht, The Netherlands | KP701986 | KP701863 | KP702108 | |
| Sphaerospermum | C. sphaerospermum | 255-H7 | March-13 | S | Utrecht, The Netherlands | KP701988 | KP701865 | KP702110 | |
| Sphaerospermum | Unidentified | 162-A4 | June-11 | S | Arnhem, The Netherlands | KP701962 | KP701839 | KP702084 |
Strains with a CBS number are deposited in the CBS strain collection. A and S represent air and swab samples, respectively, and samples taken from unknown indoor sources are indicated with “Indoor”. The GenBank numbers of the ITS, TEF and ACT sequences are shown in the last three columns. Strains used for aw experiments are underlined. aDTO 121-H1 is identified using the sequences of ITS and TEF only.
Growth on media with lowered water activity
Water activity (aw) of the medium was set between 0.99 and 0.75 by substitution of water with 0–50% glycerol (v/v). A volume of 23 ml of medium was poured in Petri dishes with vents (Greiner, Bio-One B. V., Alphen aan de Rijn, The Netherlands) and left to solidify for 24 h on the bench with the lid closed. The aw of control (non-inoculated) plates of the different glycerol-agar mixtures was determined before and after growth experiments using a Novasina labmaster-aw (Novasina, Lachen, Switzerland) and samples with a diameter of 5 cm. The aw changed only marginally during 3 weeks of storage (ranging between a decrease of 0.01 aw unit in 5% glycerol plates and an increase of 0.03 units in 50% glycerol plates).
Agar plates were inoculated with 3 μl spore solution containing 1 x 106 spores ml -1 harvested from 7-days-old MEA-grown cultures. Conidia were collected in 10 mM ACES, 0.02% Tween 80. The spore suspension was filtered over sterile glass wool to remove hyphal fragments. Spores were counted using a Bürker-Türk haemocytometer and the suspension was diluted to 1 x 106 spores ml -1.
DNA sequencing and molecular analysis
Genomic DNA was isolated from 7-days-old cultures using the Ultraclean Microbial DNA isolation kit (MoBio Laboratories, USA) according to the manufacturer’s instructions. ITS, ACT, and TEF loci were amplified, using the primers shown in Table 2 [28–31], as described earlier in Houbraken and Samson (2011) [32]. The fragments were sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The products were analyzed on an ABI Prism 3730 XL DNA Sequencer (Applied Biosystems, USA). Sequences were assembled by using the forward and reverse sequences with the program SeqMan from the LaserGene 9 package (DNAstar, USA). The ITS, ACT, and TEF sequences were concatenated resulting in 1132 nucleotide long sequences. They were aligned to sequences of 70 taxa described by Bench et al. (2012) [25] using the online version of MAFFT [33]. The resulting alignments were manually improved and phylogenetic analyses were conducted using MEGA version 5 software [34]. A phylogenetic tree (S1 Fig) was made using pairwise deletion neighbor joining with 1000 bootstrap repetitions and the nucleotide substitution model, Tamura Nei, with gamma distribution parameter 4.
Table 2. Primers used for PCR and sequencing.
| Primer: | Locus: | Primer sequence: | Reference: |
|---|---|---|---|
| V9G | Internal Transcribed Spacer | TTACGTCCCTGCCCTTTGTA | [29, 31] |
| LS266 | Internal Transcribed Spacer | GCATTCCCAAACAACTCGACTC | [29, 31] |
| ACT-512F | Actin | ATGTGCAAGGCCGGTTTCGC | [28] |
| ACT-783R | Actin | TACGAGTCCTTCTGGCCCAT | [28] |
| EF1-728F | Translation Elongation factor 1 alpha | CATCGAGAAGTTCGAGAAGG | [28] |
| EF2 | Translation Elongation factor 1 alpha | GGARGTACCAGTSATCATGTT | [30] |
Statistics
A Pearson’s Chi-square test was performed using Microsoft Office Professional Plus Excel 2010.
Results
Identification of Cladosporium species of indoor origin
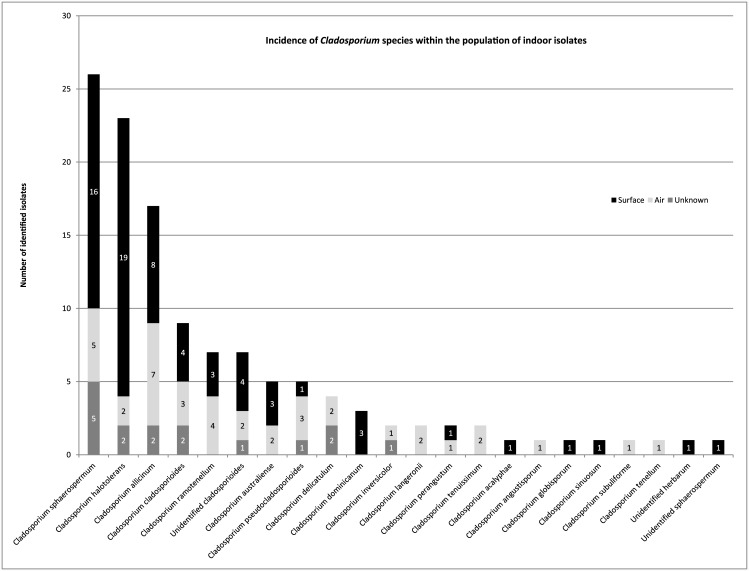
The Cladosporium collection of the Applied and Industrial Mycology group of the CBS Fungal Biodiversity Centre includes 67 strains from indoor surfaces (swab samples), 39 strains from air samples, and 17 isolates from an unknown indoor origin (Table 1). Out of the 123 indoor Cladosporium isolates, 74 originate from the Netherlands, and 49 from Denmark, France, Germany, Greece, Hungary, Poland, Portugal, Thailand, Algeria and The United States of America. The 123 isolates were identified based on the ITS, ACT and TEF sequences and a phylogenetic tree was constructed (S1 Fig). The collection consisted of 55, 41 and 27 isolates belonging to the species complexes of C. sphaerospermum, C. cladosporioides and C. herbarum, respectively (Table 3). Nine isolates could not be identified to species level since the sequences did not align well with any of the type species used for comparison and might represent new species. From these species, seven grouped in the C. cladosporioides species complex, and one in the C. herbarum and one in the C. sphaerospermum species complex. A total number of 19 different species were identified, of which C. sphaerospermum was most common, followed by C. halotolerans and Cladosporium allicinum (Fig 1). To compare the species distribution of swab and air samples, the 17 strains of unknown indoor origin were excluded. The C. sphaerospermum species complex made up more than 44.7% of the total number of 106 isolates and even 58.2% of the swab isolates (Table 3). The C. cladosporioides species complex comprised 33.3% of the indoor isolates and 22.4% of the swab isolates. The C. herbarum species complex comprised 22.0% of the indoor isolates and 19.4% of the swab isolates (Table 3). A Pearson’s Chi-square test shows significant increased numbers (p-value ≤ 0.05) of C. sphaerospermum species complex in swab samples compared to air samples. This was not observed in the case of the C. herbarum and C. cladosporioides species complexes.
Table 3. The number of isolates from indoor air and surface samples from the 3 species complexes.
| Species complex | Surface | % | Air | % | Unknown | % | Total | % |
|---|---|---|---|---|---|---|---|---|
| C. sphaerospermum | 39 | 58.2% | 9 | 23.1% | 7 | 41.2% | 55 | 44.7% |
| C. cladosporioides | 15 | 22.4% | 18 | 46.2% | 8 | 47.0% | 41 | 33.3% |
| C. herbarum | 13 | 19.4% | 12 | 30.8% | 2 | 11.8% | 27 | 22.0% |
| Total | 67 | 100% | 39 | 100% | 17 | 100% | 123 | 100% |
Fig 1. Incidence of Cladosporium species within the population of indoor isolates.
The origin of the samples (air, indoor surface or unknown origin) is also shown.
Growth of indoor Cladosporium species at lower water activity
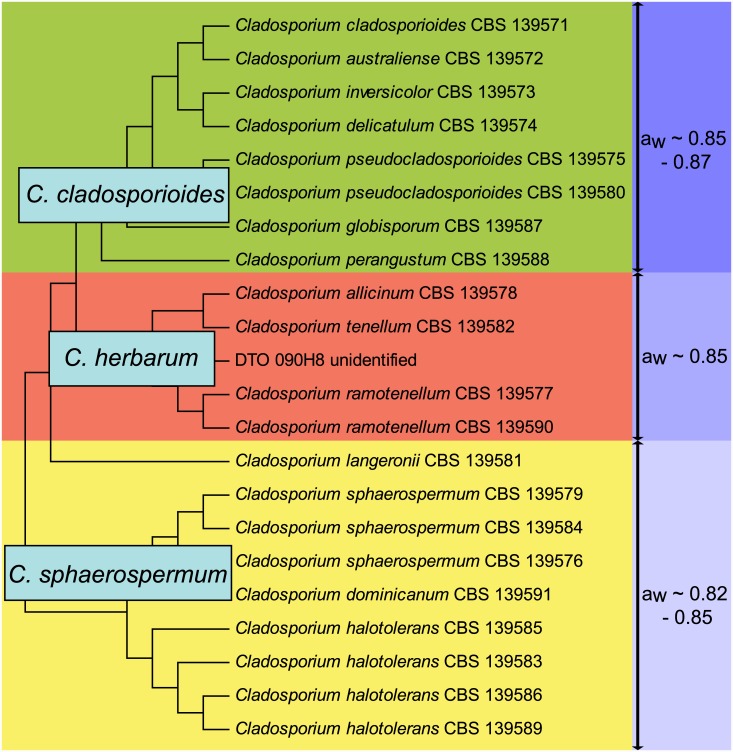
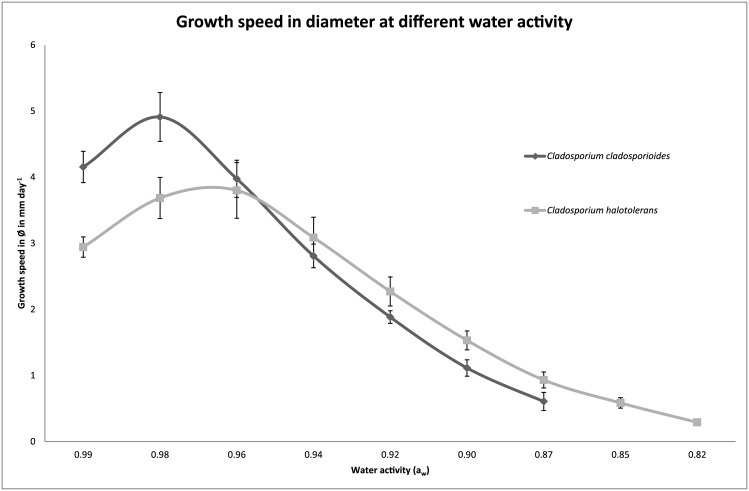
Growth of 22 indoor isolates of Cladosporium was compared on media with water activity (aw) values ranging from 0.75 to 0.99. These isolates include 9, 8 and 5 isolates of the C. sphaerospermum, C. cladosporioides and C. herbarum species complexes, respectively, which represent 14 different species (Fig 2). Moreover, the selection made up 7 air samples, 9 swab samples and 6 indoor samples of which the origin (air or swab) is not known. The minimal aw needed to enable growth during a 3-week-period, ranged from 0.82 to 0.87 for the selected Cladosporium species (Table 4). The air isolates showed a minimal aw of 0.82 to 0.87, while the swab samples showed a minimal aw of 0.82 to 0.85. All isolates of C. sphaerospermum and C. halotolerans grew at aw ≥ 0.82. Interestingly, these are also the two most abundant species in indoor air and swab samples. Cladosporium dominicanum and Cladosporium langeronii that also belong to the C. sphaerospermum species complex grew at aw ≥ 0.85. All selected species from the C. herbarum species complex (i.e. C. allicinum, Cladosporium ramotenellum and Cladosporium tenellum and an unidentified species) also grew at aw ≥ 0.85. Cladosporium globisporum and Cladosporium perangustum of the C. cladosporioides species complex grew at aw ≥ 0.85, while Cladosporium australiense, Cladosporium cladosporioides, Cladosporium delicatulum and Cladosporium inversicolor only grew at aw ≥ 0.87. The ability to grow at a particular water activity even differed within a species. Cladosporium pseudocladosporioides CBS 139575 could grow at aw of ≥ 0.85, while C. pseudocladosporioides CBS 139580 grew at aw of ≥ 0.87 (Table 4). Fig 2 summarizes the data and shows that the isolates from the C. sphaerospermum species complex, that are most abundant on indoor surfaces, grow at the lowest water activity. Notably, growth of C. cladosporioides at water activity of 0.98 is clearly faster compared to that of C. halotolerans (Fig 3 and S2 Fig). This shows that the modest xerophily of the C. sphaerospermum species complex is not the result of its higher growth rates compared to C. cladosporioides and C. herbarum.
Fig 2. Schematic dendrogram showing the minimal water activity needed for growth of 22 selected indoor Cladosporium species.
Green, red and yellow represent the C. cladosporioides, C. herbarum and C. sphaerospermum species complexes, respectively.
Table 4. Growth of 22 isolates of Cladosporium on MEA with different water activity.
| Species | Strain nr. | Growth rate in increase of Ø (mm day -1) at aw opt | aw opt | aw min |
|---|---|---|---|---|
| C. allicinum | 139578 | 4.40 | 0.98 | 0.85 |
| C. australiense | 139572 | 4.84 | 0.98 | 0.87 |
| C. cladosporioides | 139571 | 4.91 | 0.98 | 0.87 |
| C. delicatulum | 139574 | 1.98 | 0.98 | 0.87 |
| C. dominicanum | 139591 | 1.67 | 0.94 | 0.85 |
| C. globisporum | 139587 | 4.24 | 0.98 | 0.85 |
| C. halotolerans | 139583 | 3.98 | 0.98 | 0.82 |
| C. halotolerans | 139585 | 3.13 | 0.96 | 0.82 |
| C. halotolerans | 139586 | 3.80 | 0.96 | 0.82 |
| C. halotolerans | 139589 | 3.91 | 0.96 | 0.82 |
| C. inversicolor | 139573 | 3.51 | 0.98 | 0.87 |
| C. langeronii | 139581 | 1.07 | 0.94 | 0.85 |
| C. perangustum | 139588 | 4.35 | 0.98 | 0.85 |
| C. pseudocladosporioides | 139575 | 5.66 | 0.98 | 0.85 |
| C. pseudocladosporioides | 139580 | 5.53 | 0.98 | 0.87 |
| C. ramotenellum | 139577 | 4.16 | 0.98 | 0.85 |
| C. ramotenellum | 139590 | 4.40 | 0.98 | 0.85 |
| C. sphaerospermum | 139576 | 4.47 | 0.96 | 0.82 |
| C. sphaerospermum | 139579 | 3.72 | 0.96 | 0.82 |
| C. sphaerospermum | 139584 | 3.45 | 0.96 | 0.82 |
| C. tenellum | 139582 | 3.55 | 0.98 | 0.85 |
| Unidentified herbarum | DTO 090-H8 | 2.83 | 0.96 | 0.85 |
Growth rate (increase in diameter, Ø) in mm day-1 is shown for each isolate at the optimal aw at 25°C.
Fig 3. Increase in colony diameter in mm day-1 of Cladosporium cladosporioides CBS 139571 and Cladosporium halotolerans CBS 139586 at varying aw at 25°C.
Discussion
Cladosporium species are among the most abundant fungi in outdoor and indoor air [8, 11, 13]. In fact, C. cladosporioides was reported to be the most predominant fungus in houses in Ontario and Atlanta [11, 13]. Recently, classification of the genus Cladosporium has been revised on the basis of morphology and multi-locus sequencing [19, 20, 25, 26]. This novel classification was used in our study where we sequenced the same genes and compared the data. Our data showed that members of the C. sphaerospermum species complex are the most predominant indoor fungi, which were found in 44.7% of the indoor samples and 58% of indoor swabs. Notably, moderate xerotolerancy of the species (0.82–0.85) correlated with the presence of these species on indoor surfaces.
Generally, indoor levels of fungi are lower than outdoor and are related to the outdoor species composition. The composition of fungi in the indoor environment is highly similar to the outdoor air in well-ventilated houses [6, 8, 9, 13, 35, 36]. Indoor surfaces have been described as passive collectors of airborne fungi of outdoor origin. Yet, different surface types harbor different fungal populations. This indicates that the composition of fungal populations on surfaces can deviate from that of the outdoor and indoor air. This is supported by our finding that C. sphaerospermum species complex isolates are more prominent on indoor surfaces when compared to air samples. These effects might be more pronounced in winter when air replacement with outdoor air in dwellings is markedly lower [8].
Cladosporium species are not reported as producers of mycotoxins. Nonetheless, they may represent a threat to health. Cladosporium species from all three species complexes are reported to cause fungal allergies [4, 37–41], especially in patients with severe asthma [3, 42]. The dominance of the C. sphaerospermum species complex in indoor environments is of interest as C. herbarum is the most studied species in allergy research [43, 44]. Cell extracts of C. herbarum are used for allergy tests, particularly in skin prick tests [37, 44]. Not much is known about the Cladosporium proteins raising allergy. Enolase, aldehyde dehydrogenase and mannitol dehydrogenase (MDH) are known allergens from C. herbarum [43]. The C. herbarum MDH shows an identity of 83.9% with the Cladosporium fulvum MDH [45] and 92.6% identity with the same locus in an isolate of C. sphaerospermum isolated from blood culture and of which the full genome is sequenced [46]. However, we would like to stress that the identification of C. sphaerospermum in the abovementioned study is based on the sequence of the ITS region. By using BLAST we have also identified the sequences of the TEF, ITS and ACT loci. These sequences were aligned to data of Bensch et. al. and group in the phylogenetic tree with C. halotolerans as is shown in S1 Fig. [25]. Therefore, we propose to evaluate cross-reactivity of the allergens of the different Cladosporia. Specifically the common indoor fungi, C. sphaerospermum, C. halotolerans and C. allicinum, should be evaluated to assess whether the screening panels of these fungi have to be adapted.
Supporting Information
Statistical support was calculated by using 1000 bootstrap replicates. Bootstrap values below 70% are not shown. This phylogram was made using pairwise deletion and the nucleotide substitution model, Tamura Nei, with gamma distribution parameter 4.
(EPS)
(EPS)
Acknowledgments
This research is supported by the Dutch Technology Foundation STW, which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs. A part of the isolates were obtained during the experiments of the IMBOL project and we would also like to thank the Alfred Sloan foundation for their support. We received several indoor isolates from the Technical University of Denmark (DTU) and thank Birgitte Andersen for this contribution to the collection. We thank Marina van Houten for technical assistance.
Data Availability
All relevant data and accession numbers are within the paper and its Supporting Information files. All sequence files are available from the GenBank database (accession number(s) TEF: KP701744 - KP701866, ITS: KP701867 - KP701989, Actin: KP701990 - KP702111).
Funding Statement
This research was supported by the Dutch Technology Foundation STW, which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs. Project number 11117 http://www.stw.nl/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bonnefoy X. Inadequate housing and health: an overview. International Journal of Environment and Pollution. 2007;30(3):411–29. [Google Scholar]
- 2. Bonnefoy XR, Braubach M, Moissonnier B, Monollbaev K, Robbel N. Housing and health in Europe: Preliminary results of a pan-European study. Am J Public Health. 2003;93(9):1559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Denning DW, O'Driscoll BR, Hogaboam CM, Bowyer P, Niven RM. The link between fungi and severe asthma: a summary of the evidence. Eur Resp J. 2006;27(3):615–26. [DOI] [PubMed] [Google Scholar]
- 4. Reboux G, Bellanger AP, Roussel S, Grenouillet F, Millon L. Moulds in dwellings: Health risks and involved species. Rev Fr Allergol. 2010;50(8):611–20. [DOI] [PubMed] [Google Scholar]
- 5. Samson RA. Ecology and general characteristics of indoor fungi In: Adan OC, Samson RA, editors. Fundamentals of mold growth in indoor environments and strategies for healthy living: Springer; 2011. p. 101–16. [Google Scholar]
- 6. Amend AS, Seifert KA, Samson R, Bruns TD. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci U S A. 2010;107(31):13748–53. 10.1073/pnas.1000454107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andersen B, Frisvad JC, Sondergaard I, Rasmussen IS, Larsen LS. Associations between fungal species and water-damaged building materials. Appl Environ Microbiol. 2011;77(12):4180–8. 10.1128/AEM.02513-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flannigan B. Microorganisms in indoor air In: Flannigan B, Samson R, Miller D, editors. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control. 2 ed: CRC Press; 2001. p. 17–31. [Google Scholar]
- 9. Adams RI, Miletto M, Taylor JW, Bruns TD. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. Isme J. 2013;7(7):1262–73. 10.1038/ismej.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Ana SG, Torres-Rodriguez JM, Ramirez EA, Garcia SM, Belmonte-Soler J. Seasonal distribution of Alternaria, Aspergillus, Cladosporium and Penicillium species isolated in homes of fungal allergic patients. J Invest Allergol Clin Immunol. 2006;16(6):357–63. [PubMed] [Google Scholar]
- 11. Horner WE, Worthan AG, Morey PR. Air- and dustborne mycoflora in houses free of water damage and fungal growth. Appl Environ Microbiol. 2004;70(11):6394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGinnis MR. Indoor mould development and dispersal. Medical Mycology. 2007;45(1):1–9. [DOI] [PubMed] [Google Scholar]
- 13. Fradkin A, Tarlo SM, Tobin RS, Tucicporretta M, Malloch D. Species identification of airborne molds and its significance for the detection of indoor pollution. Japca-the International Journal of Air Pollution Control and Hazardous Waste Management. 1987;37(1):51–3. [DOI] [PubMed] [Google Scholar]
- 14. Pasanen P, Korpi A, Kalliokoski P, Pasanen AL. Growth and volatile metabolite production of Aspergillus versicolor in house dust. Environ Int. 1997;23(4):425–32. [Google Scholar]
- 15. Polizzi V, Delmulle B, Adams A, Moretti A, Susca A, Picco AM, et al. JEM Spotlight: Fungi, mycotoxins and microbial volatile organic compounds in mouldy interiors from water-damaged buildings. J Environ Monit. 2009;11(10):1849–58. 10.1039/b906856b [DOI] [PubMed] [Google Scholar]
- 16. Park D. Phylloplane fungi: Tolerance of hyphal tips to drying. Transactions of the British Mycological Society. 1982;79(1):174–8. [Google Scholar]
- 17. Moody SA, Newsham KK, Ayres PG, Paul ND. Variation in the responses of litter and phylloplane fungi to UV-B radiation (290–315 nm). Mycol Res. 1999;103:1469–77. [Google Scholar]
- 18. Chauhan D, Navneet Sanjay. Studies on phylloplane microflora of different tree species. Proc Nat Acad Sci India Sect B-Biol Sci. 2010;80:254–8. [Google Scholar]
- 19. Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, et al. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology. 2007;58:105–56. Epub 2008/05/21. 10.3114/sim.2007.58.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zalar P, de Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, Gunde-Cimerman N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Studies in Mycology. 2007;58:157–83. 10.3114/sim.2007.58.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pinar G, Garcia-Valles M, Gimeno-Torrente D, Fernandez-Turiel JL, Ettenauer J, Sterflinger K. Microscopic, chemical, and molecular-biological investigation of the decayed medieval stained window glasses of two Catalonian churches. Int Biodeterior Biodegrad. 2013;84:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sterflinger K. Temperature and NaCl- tolerance of rock-inhabiting meristematic fungi. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 1998;74(4):271–81. [DOI] [PubMed] [Google Scholar]
- 23. Sterflinger K, Pinzari F. The revenge of time: fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ Microbiol. 2012;14(3):559–66. 10.1111/j.1462-2920.2011.02584.x [DOI] [PubMed] [Google Scholar]
- 24. Gunde-Cimerman N, Sonjak S, Zalar P, Frisvad JC, Diderichsen B, Plemenitas A. Extremophilic fungi in arctic ice: a relationship between adaptation to low temperature and water activity. Phys Chem Earth. 2003;28(28–32):1273–8. [Google Scholar]
- 25. Bensch K, Braun U, Groenewald JZ, Crous PW. The genus Cladosporium . Studies in Mycology. 2012;72(1):1–401. 10.3114/sim0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Studies in Mycology. 2010;67:1–94. Epub 2010/09/30. 10.3114/sim.2010.67.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and indoor fungi. Utrecht, The Netherlands: CBS-KNAW Fungal Biodiversity Centre; 2010. [Google Scholar]
- 28. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–6. [Google Scholar]
- 29. de Hoog GS, van den Ende AHGG. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41(5–6):183–9. [DOI] [PubMed] [Google Scholar]
- 30. O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A. 1998;95(5):2044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van den Ende A, de Hoog GS. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana . Studies in Mycology. 1999(43):151–62. [Google Scholar]
- 32. Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70(1):1–51. 10.3114/sim.2011.70.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katoh K, Frith MC. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics. 2012;28(23):3144–6. 10.1093/bioinformatics/bts578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams RI, Miletto M, Taylor JW, Bruns TD. The diversity and distribution of fungi on residential surfaces. PLoS One. 2013;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li D-W, Kendrick B. A year-round comparison of fungal spores in indoor and outdoor air. Mycologia. 1995;87(2):190–5. [Google Scholar]
- 37. Heinzerling L, Mari A, Bergmann K-C, Bresciani M, Burbach G, Darsow U, et al. The skin prick test—European standards. Clinical and translational allergy. 2013;3(1):3 10.1186/2045-7022-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horner WE, Helbling A, Salvaggio JE, Lehrer SB. Fungal allergens. Clin Microbiol Rev. 1995;8(2):161–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mari A, Rasi C, Palazzo P, Scala E. Allergen databases: Current status and perspectives. Curr Allergy Asthma Rep. 2009;9(5):376–83. [DOI] [PubMed] [Google Scholar]
- 40. Sundell J. On the history of indoor air quality and health. Indoor Air. 2004;14:51–8. [DOI] [PubMed] [Google Scholar]
- 41. Simon-Nobbe B, Denk U, Poll V, Rid R, Breitenbach M. The spectrum of fungal allergy. International archives of allergy and immunology. 2008;145(1):58–86. [DOI] [PubMed] [Google Scholar]
- 42. Matheson MC, Abramson MJ, Dharmage SC, Forbes AB, Raven JM, Thien FCK, et al. Changes in indoor allergen and fungal levels predict changes in asthma activity among young adults. Clin Exp Allergy. 2005;35(7):907–13. [DOI] [PubMed] [Google Scholar]
- 43. Breitenbach M. The spectrum of fungal allergy. International archives of allergy and immunology. 2008;145(1):58–86. [DOI] [PubMed] [Google Scholar]
- 44. Poll V, Denk U, Shen HD, Panzani RC, Dissertori O, Lackner P, et al. The vacuolar serine protease, a cross-reactive allergen from Cladosporium herbarum . Mol Immunol. 2009;46(7):1360–73. 10.1016/j.molimm.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 45. Simon-Nobbe B, Denk U, Schneider PB, Radauer C, Teige M, Crameri R, et al. NADP-dependent mannitol dehydrogenase, a major allergen of Cladosporium herbarum . J Biol Chem. 2006;281(24):16354–60. [DOI] [PubMed] [Google Scholar]
- 46. Ng KP, Yew SM, Chan CL, Soo-Hoo TS, Na SL, Hassan H, et al. Sequencing of Cladosporium sphaerospermum, a Dematiaceous Fungus Isolated from Blood Culture. Eukaryot Cell. 2012;11(5):705–6. 10.1128/EC.00081-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical support was calculated by using 1000 bootstrap replicates. Bootstrap values below 70% are not shown. This phylogram was made using pairwise deletion and the nucleotide substitution model, Tamura Nei, with gamma distribution parameter 4.
(EPS)
(EPS)
Data Availability Statement
All relevant data and accession numbers are within the paper and its Supporting Information files. All sequence files are available from the GenBank database (accession number(s) TEF: KP701744 - KP701866, ITS: KP701867 - KP701989, Actin: KP701990 - KP702111).