Abstract
Telomere crisis occurs during tumorigenesis when depletion of the telomere reserve leads to frequent telomere fusions. The resulting dicentric chromosomes have been proposed to drive genome instability. Here we examine the fate of dicentric human chromosomes in telomere crisis. We observed that dicentric chromosomes invariably persisted through mitosis and developed into 50-200 μm chromatin bridges connecting the daughter cells. Before their resolution at 3-20 h after anaphase, the chromatin bridges induced nuclear envelope rupture in interphase, accumulated the cytoplasmic 3' nuclease TREX1, and developed RPA-coated single stranded (ss) DNA. CRISPR knockouts showed that TREX1 contributed to the generation of the ssDNA and the resolution of the chromatin bridges. Post-crisis clones showed chromothripsis and kataegis, presumably resulting from DNA repair and APOBEC editing of the fragmented chromatin bridge DNA. We propose that chromothripsis in human cancer may arise through TREX1-mediated fragmentation of dicentric chromosomes formed in telomere crisis.
Keywords: telomere crisis, chromothripsis, kataegis, NERDI, TREX1, APOBEC, dicentric chromosome
INTRODUCTION
The view that dicentric chromosomes are broken in mitosis and undergo breakage-fusion-bridge (BFB) cycles originates from McClintock’s cytological observation of corn chromosomes (McClintock, 1938; McClintock, 1941). More recently, the fate of dicentric chromosomes has been studied in yeast as well as plants (reviewed in (Stimpson et al., 2012)). Here, we document the behavior of dicentric chromosomes in human cells.
Dicentric chromosomes can be formed during the early stages of human tumorigenesis when telomere shortening has led to dysfunctional telomeres (reviewed in (Artandi and DePinho, 2010)). Telomere shortening induces senescence or apoptosis when a few telomeres lose the ability to repress DNA damage signaling pathways. Telomere fusions are infrequent in senescence, most likely because of the low frequency of dysfunctional telomeres. Upon by-pass of senescence due to loss of p53 and Rb, further telomere attrition increases the incidence of telomere dysfunction, eventually leading to a telomere crisis where telomeres fuse to form dicentric chromosomes. These dicentrics have been proposed to drive genome instability in cancer. The genomic scars indicative of past telomere crisis have been observed in several types of cancer (Lin et al., 2010; Lin et al., 2014; Roger et al., 2013; Simpson et al., 2015). However, the fate of dicentric chromosomes, including potential BFB cycles, has been elusive.
The genomic footprint of BFB cycles is a ‘fold-back’ inverted rearrangement that demarcates a region of amplification from a terminal chromosomal deletion. Such events have been observed in pancreatic cancer, esophageal cancer, breast cancer and leukemias, among others (Bignell et al., 2007; Campbell et al., 2010; Waddell et al., 2015; Li et al., 2014; Nones et al., 2014). Interestingly, several of these studies have suggested an association between the rearrangements of BFB cycles and chromothripsis (Nones et al., 2014; Li et al., 2014). Chromothripsis is a mysterious mutational process in which one or more localized chromosomal regions undergo catastrophic shattering, triggering a haphazard repair process of stitching chromosomal fragments together in a random order and orientation (Stephens et al., 2011). Chromothripsis has been observed across many tumor types (Forment et al., 2012), especially those with p53 loss (Rausch et al., 2012), as well as occasional occurrence in the germline (Kloosterman and Cuppen, 2013). Chromothripsis breakpoints often show clusters of base substitutions localized nearby (kataegis), exhibiting the C>T and C>G signature at TpC dinucleotides associated with APOBEC-mediated mutagenesis (Nik-Zainal et al., 2012a; Roberts et al., 2012; Roberts et al., 2013; Chan et al., 2015).
The mechanism of chromosome fragmentation that gives rise to chromothripsis in cancer is not known and it is not clear when, where, and how the DNA fragments are rejoined. A proposed explanation of the localized nature of chromothripsis is the sequestration of a chromosome (fragment) in a micronucleus where it is shattered while the rest of the genome remains intact (Zhang et al., 2015). Micronuclei in cancer cell lines show abnormalities in DNA replication, transcription, and nuclear envelope (NE) structure, and display DNA damage (reviewed in (Hatch and Hetzer, 2015). Importantly, micronuclei show frequent nuclear envelope collapse, which could cause the aforementioned abnormalities (Hatch et al., 2013). Chromothripsis was recently shown to arise after rupture of micronuclei containing lagging chromosomes (Zhang et al., 2015). Therefore, a plausible scenario for the origin of chromothripsis involves a lagging chromosome (fragment), formation of a micronucleus that undergoes nuclear envelope collapse, DNA fragmentation due to impaired DNA replication, and random joining of the DNA fragments upon their incorporation into the primary nucleus (Zhang et al., 2015; Hatch and Hetzer, 2015).
Here we present data suggesting a telomere-based mechanism for chromothripsis in cancer. Using inducible telomere crisis in vitro, we document chromothripsis and kataegis in half of the descendant clones sequenced. Dicentric chromosomes formed through telomere fusion persisted through mitosis and cytokinesis to form long chromatin bridges between the daughter cells. The DNA in the chromatin bridges became partially single-stranded due to attack by the major cytoplasmic 3’ nuclease, TREX1 (DNaseIII) (reviewed in (Rice et al., 2015)). TREX1 appeared to gain access to the bridge DNA during transient nuclear envelope rupture during interphase (NERDI) and its nucleolytic activity was required for the timely resolution of the chromatin bridges. After bridge resolution, the partially ssDNA generated by TREX1 rejoined the primary nuclei. We infer from sequence analysis of clones emerging from telomere crisis that the ssDNA is processed by APOBEC3A/B-mediated cytosine deamination (reviewed in (Roberts and Gordenin, 2014)) leading to kataegis and that the DNA fragments are joined randomly to generate the hallmarks of chromothripsis.
RESULTS
An in vitro model for telomere crisis
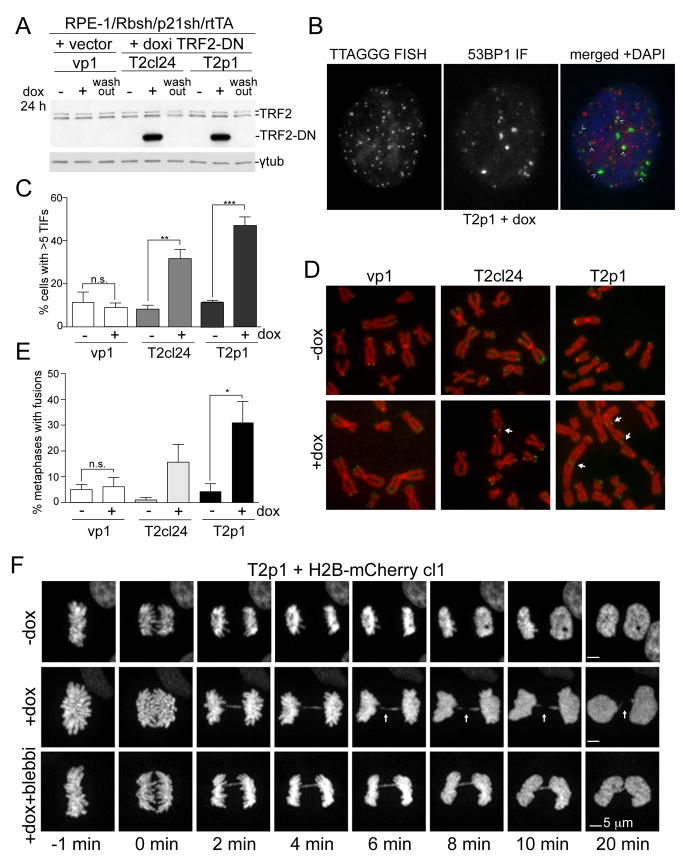
To approximate telomere crisis in vitro, we generated a derivative of the hTERT expressing RPE-1 retinal pigment epithelial cell line in which the Rb and p53 pathways were disabled with shRNAs to Rb and p21 (see Table S1). To induce the telomere fusions typical of telomere crisis, we used a dox-inducible dominant negative allele of TRF2 (TRF2-DN), which deprotects telomeres and induces telomere fusions (van Steensel et al., 1998). As expected, doxycycline induced the 53BP1-containing Telomere dysfunction-Induced Foci (TIFs; (Takai et al., 2003)), which are indicative of TRF2-DN-induced ATM kinase signaling (Karlseder et al., 1999); impaired proliferation; and generated metaphases with telomere fusions (Figure 1A-E; Figure S1A,B). Whereas the dox-inducible clone T2cl24 showed infrequent fusions, a pool of TRF2-DN expressing cells (T2p1) showed telomere fusions in a large fraction of the metaphases, allowing cell biological experiments (Figure 1A-E; Figure S1B). Despite the frequent telomere fusions, the induced T2p1 cells formed micronuclei infrequently (Figure S1C).
Figure 1. Dicentric chromosomes persist through anaphase.
(A) Immunoblotting for TRF2 and TRF2-DN 48 h after dox in the indicated RPE-1 cell lines. Wash-out: 48 h after removal of dox.
(B) Example of 53BP1 TIFs (arrows) in T2p1 48 h after dox. Red: telomeric FISH; green: IF for 53BP1; blue; DAPI DNA stain.
(C) Quantification of TIFs as shown in (B). Bar graphs present mean values from three independent experiments (>49 cells each) and SDs. **, P ≤ 0.01, ***; P ≤ 0.001 (Student’s t-test).
(D) Metaphases with telomere fusions (arrows) in the indicated cells 48 h after dox. Red: DAPI DNA stain; green: telomeric FISH.
(E) Quantification of telomere fusions as determined in (D). Data are means and SDs from three independent experiments (>5600 telomeres per cell line per experiment). n.s., not significant; *, P ≤ 0.05 (Student’s t-test).
(F) Images of H2B-mCherry marked chromatin at the indicated time points from T2p1 with and without induction (+ and -dox) of telomere fusions with and without blebbistatin. Arrows (+dox images) highlight positions with absent H2B signals.
Dicentric chromosomes persist through mitosis and cytokinesis
The behavior of dicentric chromosomes in mitosis was examined using spinning-disk confocal imaging of H2B-marked chromatin in induced T2p1 cells, which developed anaphase bridges in the majority of mitosis (see below Figure 2B) as expected since ~1% of the 92 telomeres in these cells undergo fusion (Figure S1B). The images obtained with these cells were similar to those described by McClintock (McClintock, 1938), showing apparent cleavage of the chromatin bridges immediately before or during cytokinesis (Figure 1F; Figure S1D,E; MovieS1, panel 1). Imaging with myrPALM-mTurquoise2 to mark the plasma membrane (Zacharias et al., 2002) showed that the H2B signal was diminished only at the site of ingression (Figure S1D,E; MovieS1, panel 2), indicating that the disappearance of the H2B signal was likely due to the cleavage furrow pinching the chromatin. Indeed, when ingression was blocked with the actomyosin inhibitor blebbistatin (Straight et al., 2003), the dicentric chromosomes clearly remained intact (0/24 events) (Figure 1F; MovieS1, panel 1). Thus, as is the case in budding yeast (Haber et al., 1984; Lopez et al., 2015; Hill and Bloom, 1989), mammalian dicentric chromosomes can withstand the forces of the mitotic spindle and do not break before cytokinesis. This result is not unexpected given that the spindle force (0.5-1.5 nN) is insufficient to break a mitotic chromosome, which can withstand at least 100 nN (Houchmandzadeh et al., 1997). Cells with dicentrics did not show a delay in their progression through mitosis (Figure S1F) although tubulin remained associated with the midbody slightly longer (Figure S1G,H; MovieS1, panel 3).
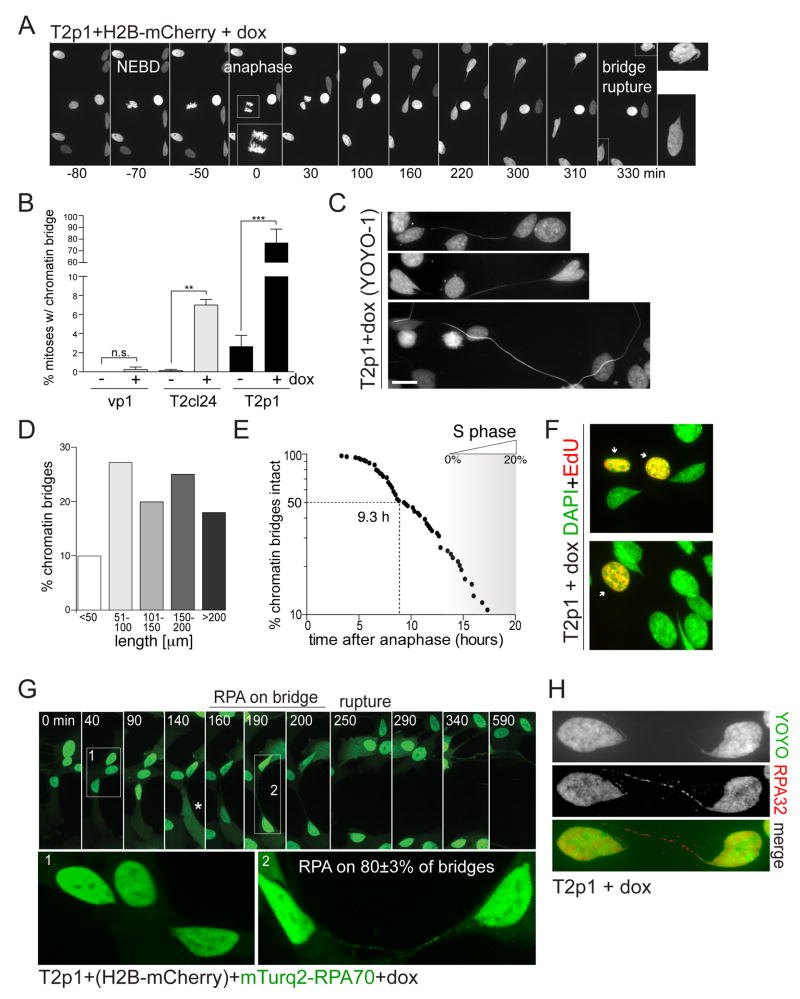
Figure 2. Dicentric chromosomes form RPA-containing chromatin bridges.
(A) Images of chromatin from live-cell imaging of T2p1+H2B-mCherry treated with dox. Time points as indicated. NEBD: Nuclear envelope breakdown. Anaphase is shown in enlarged inset. The bridge resolves ~5.5 h after anaphase. The images of the two daughter nuclei are enlarged on the right.
(B) Quantification of chromatin bridge induction derived from movies as in (A). Bar graphs represent the means and SDs of three independent experiments (>50 cell divisions per experiment). n.s., not significant; **, P ≤ 0.01 (Student’s t-test).
(C) Chromatin bridges with YOYO-1 DNA stain. Scale bar, 10 μm.
(D) Quantification of chromatin bridge length at resolution. Data derived from movies as in (A).
(E) Measurements of the timing of chromatin bridge resolution in h after anaphase. Data obtained from movies as in (A) (n=84 from three independent experiments). Entry into S phase was based on RPA patterns. At 20 h, ~20% of the cells are in S phase.
(F) Two examples of EdU staining (30 min pulse; red) and DAPI stain (green). Note the lack of EdU signal on the chromatin bridges and connected nuclei.
(G) Accumulation of RPA on chromatin bridges before their resolution. T2p1+H2B-mCherry+mTurquoise2-RPA70 cells were examined by live-cell imaging. Stills showing the mTurquoise-RPA70 signal on one chromatin bridge are shown. Enlargements: bridge without and later with RPA70. 80±3% of bridges (mean±SD; 102 bridges from 3 independent experiments) contained RPA. Asterisk: apparent NERDI.
(H) IF for RPA32 (red) on fixed cells with a chromatin bridge. DNA stained with YOYO-1 (green).
Dicentric chromosomes develop into long chromatin bridges
To monitor the fate of the dicentric chromosomes after cytokinesis, we captured 108 adjacent fields by spinning-disk confocal imaging at 10 min intervals for 24-48 h (Figure S2A,B). Computational joining of the fields (Preibisch et al., 2009) allowed ~1,000 cells to be followed for two days (MovieS2). This ‘stitching’ microscopy of H2B-mCherry labeled cells showed that daughter cells migrated away from each other despite their connecting chromatin bridge (Figure 2A-D; MovieS3). The chromatin bridges, which were detectable with YOYO-1 in fixed samples (Figure 2C), developed with high frequency and measured 50-200 μm before breaking (Figure 2B,D). The H2B-mCherry intensity on the bridges appeared diminished and the IF signals for histones H2A, H2B, and H4 were low (Figure S2C), suggesting the loss of nucleosomes, perhaps due to the stretching of the chromatin bridge (Bennink et al., 2001).
The chromatin bridges remained intact for 3-20 h with a median persistence time of ~9.3 h (Figure 2E). Bridge resolution was apparent from a sudden change in morphology, the rapid movement of bridge remnants towards the primary nuclei, and the rapid movement of the daughter cells away from each other (MovieS3). The primary nuclei were often heart-shaped with invaginations opposite from the chromatin bridge, suggesting that the fused chromosome(s) were pulling at the nuclear envelope (Figure 2A,C; Figure S2D; MovieS3). After bridge resolution, a small tail of chromatin was observed that shortened and eventually disappeared, most likely because the nuclear envelope regained its rounded state (Figure S2E). In some cases the bridge remnant persisted until the next mitosis (see below). The chromatin bridges did not appear to give rise to micronuclei (0 out of >100 events scored).
To determine the timing of bridge resolution relative to the cell cycle stage of the connected cells, we examined mTurquoise2-RPA70 patterns in the primary nuclei. At the time of bridge rupture, most primary nuclei had diffusely distributed RPA, indicating that they were not yet in S phase. When >90% of the bridges were resolved at 20 h, fewer than 20% of the primary nuclei showed the punctate S phase RPA pattern indicative of S phase (Figure 2E; Figure S3A-C). Therefore, most chromatin bridges were resolved before DNA replication in the primary nuclei. EdU labeling showed no signal on the bridge DNA, suggesting that the chromatin bridges did not undergo aberrant premature DNA replication (Figure 2F).
Chromatin bridges accumulate RPA-coated ssDNA
Although the chromatin bridges resolved before the primary nuclei entered S phase, ~80% of the bridges contained RPA before and/or at the time of their resolution (Figure 2G,H; Figure S3A-F). IF for endogenous RPA32 and imaging of mTurquoise2-RPA70 showed a punctate pattern on the bridges that developed into bright domains just before resolution (Figure 2G,H; Figure S3A,B; MovieS3, panel 2).
After bridge resolution, the RPA-coated domains became embedded in the primary nucleus or persisted as short connected tails before joining a daughter nucleus after the next mitosis (Figure S3C-F). Once resolved, the bridge remnants showed the presence of γH2AX, 53BP1, and Mre11 (Figure S3G-I), indicating chromatinization of the DNA and activation of the DNA damage response.
RPA-coated chromatin bridges were also observed after deletion of the shelterin protein TIN2 from mouse cells (Takai et al., 2011). Furthermore, RPA accumulated on chromatin bridges induced by TRF2-DN in the HTC75-T4 cell line, which is derived from the human HT1080 fibrosarcoma cell line (Figure S4A-C) (van Steensel et al., 1998). Finally, RPA was present on chromatin bridges resulting from lagging chromosomes induced by either inhibition of the Mps1 kinase (Santaguida et al., 2010) or treatment with nocodazole (Figure S4D-G), suggesting that the formation of ssDNA is not a peculiarity of chromatin bridges formed by dicentrics resulting from telomere fusions.
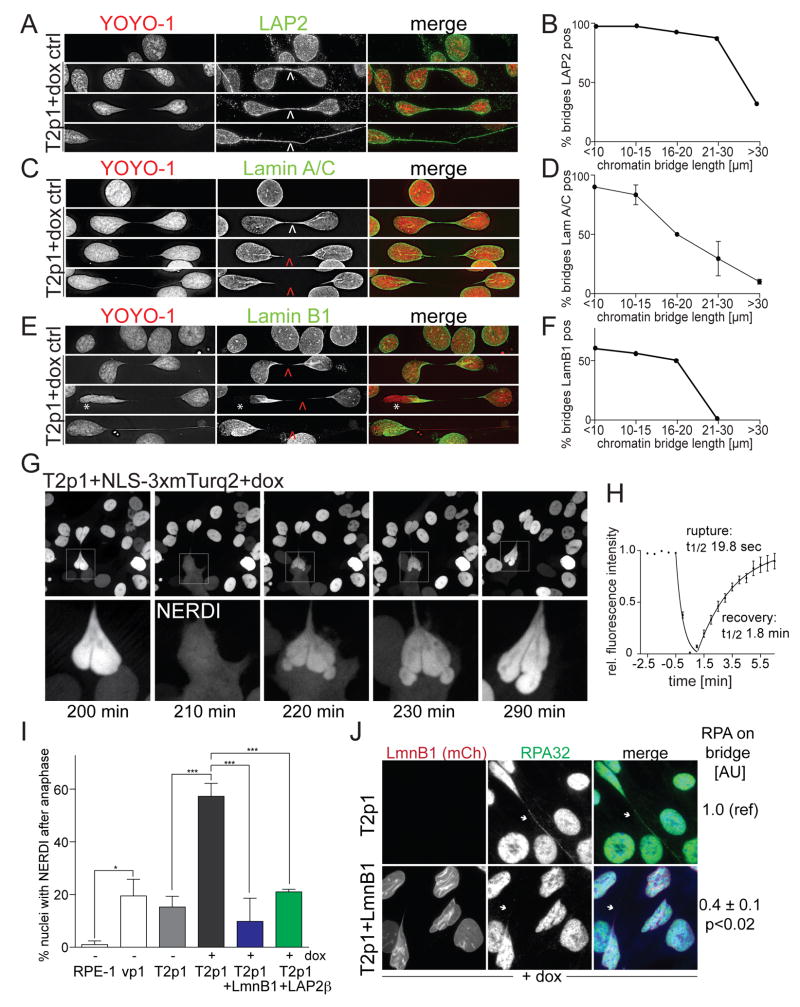
Chromatin bridges have an altered nuclear envelope
IF for the transmembrane nuclear envelope protein LAP2 showed that the chromatin bridges were surrounded by NE (Figure 3A and B). Similarly, BAF1, which binds chromatin and helps assemble the nuclear lamina was detectable on the chromatin bridges (Figure S4F). In contrast, IF for Lamins A/C and B1, Nuclear Pore Complexes (NPCs) (detected by mAb414 and α-TPR), and the NE proteins SUN1, SUN2, and MAD1 suggested that while the chromatin bridges contained an NE, its composition was altered (Figure 3C-F; Figure S4F). Specifically, the intensity of Lamin A/C and Lamin B1 staining gradually diminished as the bridges extended and several NE components (e.g. NPCs, SUN1/2, and MAD1) were not detectable (Figure 3C-F; Figure S4F). Interestingly, Lamin B1 was also depleted from the NE of the primary nuclei (Figure 3E).
Figure 3. Transient NERDI is frequently associated with chromatin bridges.
(A,C,E) IF for LAP2, Lamin A/C, or Lamin B1 IF (green) in T2p1 before and 48 h after induction with dox. DNA stained with YOYO-1 (red). Arrows: white, signals present; red, signals undetectable. Asterisk: loss of Lamin B1 from NE of primary nucleus.
(B,D,F) Quantification of LAP2, Lamin A/C, and Lamin B1 signals on chromatin bridges of the indicated length classes. Chromatin bridges were classified as positive if the IF signal was contiguous across the entire length of the bridge. Data from >100 chromatin bridges in two independent experiments. Error bars: SEMs.
(G) Example of transient NERDI in cells with a chromatin bridge. NLS-3xmTurq2 images at the indicated time points from Movie S4. Bottom: enlargements of the transient NERDI.
(H) Duration of NERDI. Data obtained from movies generated with 30 sec interval imaging on 10 cells as in (G).
(I) Quantification of the frequency of NERDI events occurring in at least one of the two daughter cells within 6 h of anaphase before and after induction with dox. NERDI was assessed as in (G) but at 5 min intervals over 8 h after anaphase. For the +dox samples, only cells with chromatin bridges were scored. Data from at least two experiments with > 40 anaphases each. *, P ≤ 0.05; ***, P ≤ 0.001 (one-way ANOVA with Tukey’s correction for multiple comparisons).
(J) IF of RPA32 (green) with mCherry-Lamin B1 (red) and YOYO-1 stained DNA (blue) in cells with and without Lamin B1 overexpression. Arrows mark chromatin bridges. Note absence of RPA32 on the chromatin bridge in mCherry-Lamin B1 expressing cells. Numbers to the right show quantification (means±SEMs) from >40 chromatin bridges from two independent experiments. P value from Student’s t-test.
Cells with chromatin bridges undergo NERDI
Some of the cells with chromatin bridges appeared to transiently lose RPA from one of the primary nuclei with a concomitant increase of the RPA signal in the cytoplasm (Figure 2G, asterisk). This mis-localization of RPA70 could be explained if the cells experienced NERDI. In micronuclei, NE collapse drives an irreversible loss of compartmentalization (Hatch et al., 2013) whereas in several cancer cell lines, NERDI of the main nucleus is transient (Vargas et al., 2012). Relevant to the anomalous Lamin B1 staining observed in nuclei connected by chromatin bridges (Figure 3E), Lamin B1 depletion exacerbates NERDI (Vargas et al., 2012).
To assay for NERDI, we used a fusion of three tandem copies of mTurquoise2 and the nuclear localization signal (NLS) of SV40 large T, which is confined to the nucleus when the NE is intact. Time-lapse imaging showed that cells with chromatin bridges had frequent and short-lived (~5 min) NERDI as evidenced by the appearance of cytoplasmic NLS-3xmTurq2 and its diminished intensity in the nucleus (Figure 3G,H; MovieS4). After NERDI, the nuclear envelope regained its integrity and retained the NLS-3xmTurq2 marker. Cells that did not contain TRF2-DN (vp1; Table S1) and non-induced T2p1 cells exhibited at least one NERDI event in 20% of daughter cell pairs within 6 h of anaphase (Figure 3I). This high baseline rate of NERDI is likely due to p21 and/or Rb inactivation since NERDI was infrequent in the parental RPE1-hTERT cells (Figure 3I). Importantly, NERDI frequency increased to nearly 60% in dox-induced T2p1 daughter cells with chromatin bridges (Figure 3I). NERDI usually occurred in one of the two connected nuclei. Because NERDI can be induced by Lamin B1 depletion (Vargas et al., 2012), we tested whether Lamin B1 or LAP2β overexpression could repress the nuclear envelope rupture in induced Tp21 cells (Figure 3I). Overexpression of both proteins diminished the frequency of NERDI (Figure 3I). Lamin B1 overexpression strongly diminished RPA accumulation on chromatin bridges suggesting that the formation of ssDNA on chromatin bridges depended on NERDI (Figure 3J). We conclude that telomere dysfunction in this cell system induces a significant increase in NERDI. As a result, at least one of the two cells connected by a chromatin bridge is likely to experience a NERDI event within the time period preceding bridge resolution.
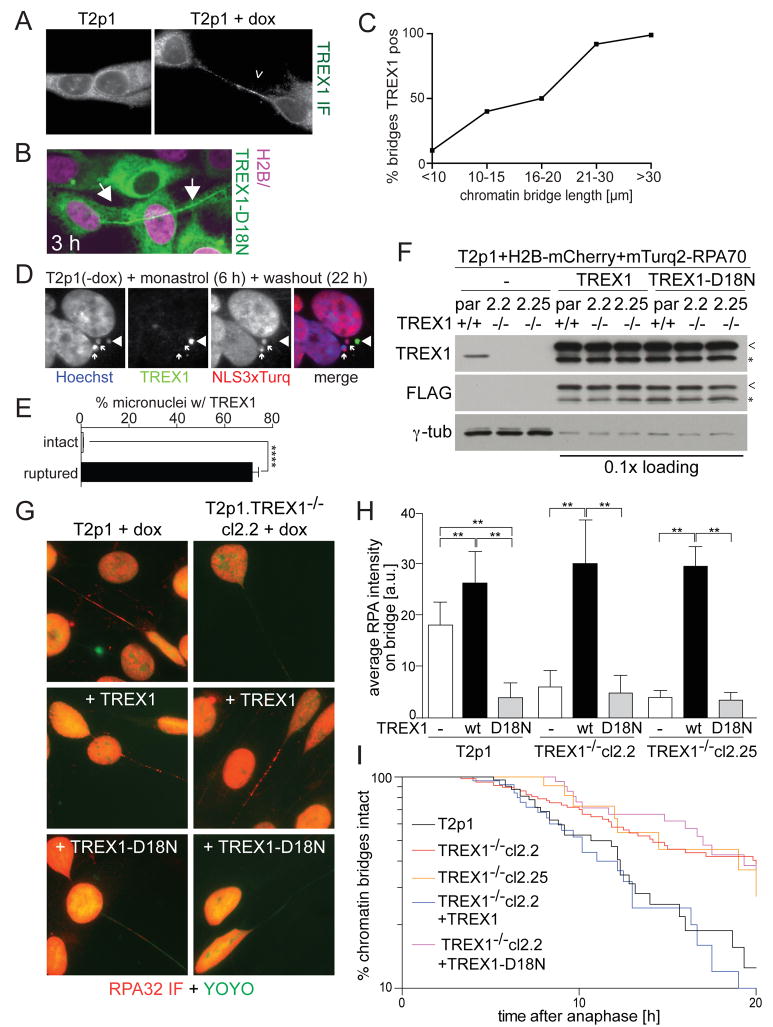
Chromatin bridge DNA is processed by the cytoplasmic 3’ exonuclease TREX1
Although we initially queried nuclear nucleases, the frequent NERDI suggested that cytoplasmic nuclease might attack the chromatin bridges to generate ssDNA. IF showed that the major cytoplasmic 3’ exonuclease TREX1 was present on the chromatin bridges in cells undergoing telomere crisis whereas TREX1 was only observed in the cytoplasm of control cells (Figure 4A). TREX1 also localized to chromatin bridges induced by telomere dysfunction in the HTC75-T4 cell line and to chromatin bridges formed by lagging chromosomes induced by nocodazole (Figure S4A and G). IF analysis and imaging of the inactive mTurq2-tagged TREX1-D18N (Lehtinen et al., 2008) indicated that this nuclease often appears on the chromatin bridges before their rupture (Figure 4B,C; MovieS5). IF for TREX1 in micronuclei induced by monastrol also showed TREX1 accumulation specifically in micronuclei that had undergone NE rupture (Figure 4D,E).
Figure 4. TREX1 generates ssDNA in chromatin bridges and promotes resolution.
(A) IF for TREX1 (white) on Tp21 cells with and without dox.
(B) Images from live-cell imaging of mTurq2-TREX1-D18N on a chromatin bridge (Movie S5).
(C) Quantification of TREX1 positive chromatin bridges of the indicated length classes. Positively scored chromatin bridges had at least 5 TREX1 foci. Data from three independent experiments with 100 chromatin bridges each.
(D) IF for TREX1 (green) in T2p1 cells with intact (NLS-3xTurq+; arrows) and disrupted (NLS-3xTurq-; arrowhead) micronuclei induced with monastrol.
(E) Quantification of TREX1 positive micronuclei as in (D). Over 300 micronuclei were analyzed from three independent experiments. ****, P ≤ 0.0001 (Student’s t-test).
(F) Immunoblotting for endogenous TREX1 and exogenous wild type and mutant TREX1 (FLAG) in the indicated cell lines. Par: parental T2p1+H2B-mCherry+mTurq2-RPA70 cells. Cl.2.2 and cl.2.25: TREX1 CRISPR KO clones. Arrowheads: full length FLAG-TREX1. Asterisks: degradation products.
(G) Examples of the RPA32 IF in cells as in (F).
(H) Quantification of the RPA32 IF intensity on chromatin bridges in cells as in (F). Data was obtained from 55 chromatin bridges from three independent experiments. Bars indicate SDs. **, P ≤ 0.01 (Student’s t-test).
(I) Timing of chromatin bridge resolution after anaphase in the indicated cell lines. See legend to Figure 2E.
To test the role of TREX1 in the generation of ssDNA, we used CRISPR/Cas9 to derive TREX1 KO subclones from the T2p1 telomere crisis cell line (Figure S5A,B). Loss of TREX1 was demonstrated by immunoblotting and IF (Figure 4F; Figure S5C,D) and sequence analysis revealed bi-allelic CRISPR gene editing (Figure S5E). The TREX1 KO cell lines showed normal proliferation and, after induction of TRF2-DN, displayed the expected reduced proliferation and chromatin bridges (Figure S5F,G). Importantly, IF showed a nearly complete abrogation of the accumulation of RPA on the chromatin bridges in the TREX1 KO cells (Figure 4G,H). This absence of RPA accumulation could be reversed by reintroduction of wild type TREX1, whereas the inactive TREX1-D18N did not have this effect (Figure 4F-H). Consistent with a previous report (Lehtinen et al., 2008), TREX1-D18N had a dominant-negative effect in the TREX1-proficient parental cell line, significantly reducing the appearance of RPA on the chromatin bridges (Figure 4F-H). In contrast, overexpression of the wild type allele in the parental Tp21 cell line slightly increased the accumulation of RPA on the chromatin bridges. As a control, we confirmed that TREX1 deficiency did not diminish RPA foci formed during replication stress (Figure S5H,I).
TREX1 deletion from the HTC75-T4 cell line also strongly diminished RPA accumulation on chromatin bridges (Figure S4B,C). Furthermore, chromatin bridges resulting from lagging chromosomes induced by Mps1 kinase inhibition failed to accumulate RPA when TREX1 was absent (Figure S4D-F). Thus, TREX1 deficiency generally affects the formation of RPA-containing ssDNA in chromatin bridges.
Cells lacking TREX1 showed a significant delay in the resolution of chromatin bridges (Figure 4I). At 20 h post-anaphase, only 15% of the chromatin bridges in the TREX1-proficient cells remained whereas more than 40% of the bridges of the TREX1 KO cell line were still intact. The timing of bridge resolution was restored by wild type TREX1 but not by TREX1-D18N. TREX1-independent mechanisms also contribute to the resolution of the chromatin bridges, since resolution was not abrogated by TREX1 deficiency.
Frequent chromothripsis in post-crisis clones
To determine the genetic alterations induced by telomere crisis, we isolated post-crisis subclones of T2p1 and T2cl24. Clones were karyotyped to exclude those that had escaped the telomere dysfunction (Figure S6 and Table S2). Clones with aneuploidy and/or marker chromosomes were analyzed by telomere fusion PCR to confirm the presence of telomere fusion (Figure S6C). Ten such post-crisis clones were selected for sequence analysis, as were the parental T2p1, one post-crisis subclone with the parental karyotype (24.2), and seven control subclones derived from uninduced T2p1 cells. We performed whole genome sequencing on all 19 lines, comparing post-crisis whole genome sequencing data with that from the parental lines, in order to identify mutations acquired during the telomere crisis. None of the subclones derived from the uninduced T2p1 cells showed genomic alterations.
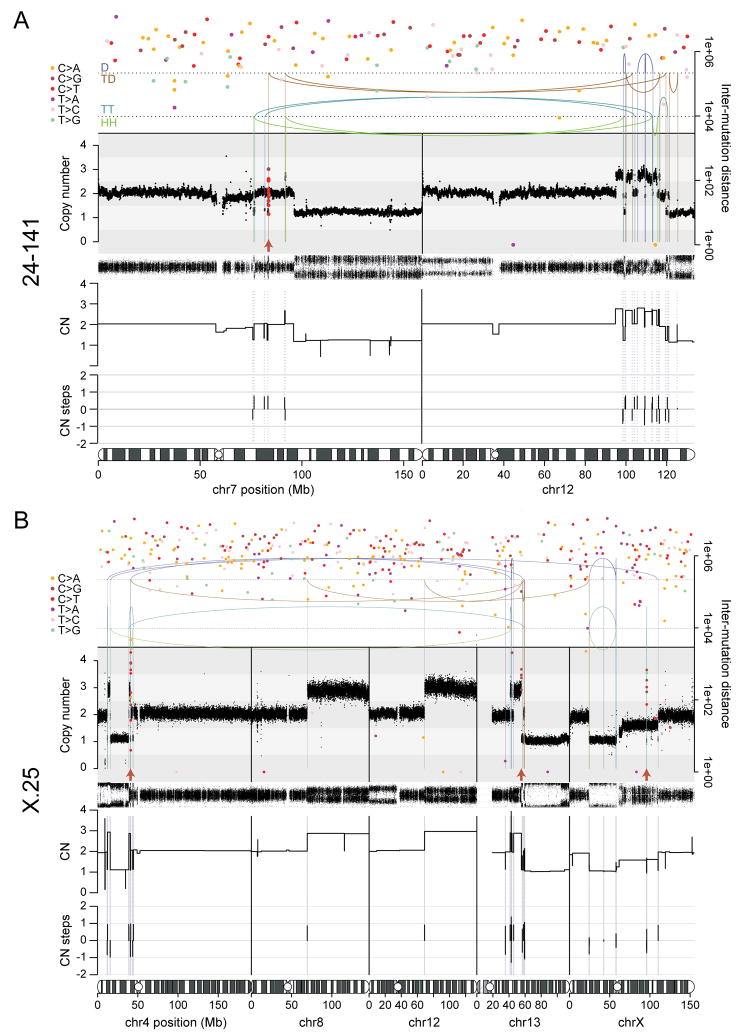
Strikingly, 5 of 10 post-telomere crisis lines showed clusters of genomic rearrangements affecting one or more chromosomes (Figure 5; Figure S7A). These rearrangements exhibited the hallmarks of chromothripsis, including spatial clustering, randomness of fragment orientation and oscillating copy number states (Figure 5A,B; Figure S7A) (Korbel and Campbell, 2013). In several examples, the rearrangements were near telomeres and associated with terminal deletions of the chromosome, consistent with products of telomere fusion. Chromothripsis events that are more internal in the chromosomes can also result from telomere fusions since the genomic region in the bridge could be far from the telomere depending on the structure of the dicentric. Sometimes the clustered rearrangements affected one chromosome, sometimes two or three, as has been observed in cancers (Stephens et al., 2011). Importantly, chromothripsis never involved whole chromosomes, but rather was localized to the specific regions that presumably resided in the chromatin bridge. Consistent with these results, chromothripsis was recently reported in cells that were subjected to TRF2 inhibition and an Mps1 kinase inhibitor (Mardin et al., 2015).
Figure 5. Chromothripsis and kataegis in post-crisis clones 24-141 and X-25.
(A) Chromothripsis and rainfall plot of sample 24-141 involving chromosomes 7 and 12.
(B) Chromothripsis and rainfall plot of sample X-25 involving chromosomes 4, 13 and X. The unbalanced rearrangements involving chromosomes 8 and 12 may have taken place together with the chromothripsis event. In (A) and (B), top: the arcs represent the two ends of rearrangements. Arcs are grouped from top to bottom by the type of rearrangement orientation as follows: deletion (D; +-); tandem duplication (TD; -+); tail-tail (TT; ++); head-head (HH; --). Middle: estimated copy number over genomic windows. The variant allele frequency (VAF) track is shown below the copy number track. Inferred copy number segments are shown below the VAF track. Bottom: amount of copy number change between copy number segments. Chromothripsis after a duplication will yield three copy number states with copy number steps of +1 or -1. Duplication after chromothripsis will yield some copy number steps of +2 or -2. Filled circles: positions of point mutations colored by mutation type. The Y-axis shows the distance of each mutation to the next on the same chromosome, with the respective axis on the right-hand side of the graph. Red arrows: kataegis clusters.
The oscillations of copy number in regions of apparent chromothripsis often sampled three copy number states. Such a scenario can arise either from a chromothripsis event simultaneously affecting two copies of the same genomic region or from a subsequent duplication of part of a chromothripsis chromosome. These two possibilities can be distinguished by the patterns of copy number changes across breakpoints in the region (Li et al., 2014). In all examples observed here, the patterns of copy number and rearrangements implied that two copies of the affected genomic regions were simultaneously subjected to the catastrophic shattering and repair of chromothripsis (Figure 5; Figure S7). Two copies could derive from the end-to-end fusion of sister chromatids that form the chromatin bridge.
Kataegis accompanies chromothripsis
The recruitment of RPA suggested that chromatin bridges contain extensive ssDNA. As ssDNA represents one of the target substrates for APOBEC enzymes, we hypothesized that the regions caught up in the chromatin bridge would show clusters of point mutations, known as kataegis, analogous to those seen in cancers (Nik-Zainal et al., 2012a; Nik-Zainal et al., 2012b; Roberts et al., 2012).
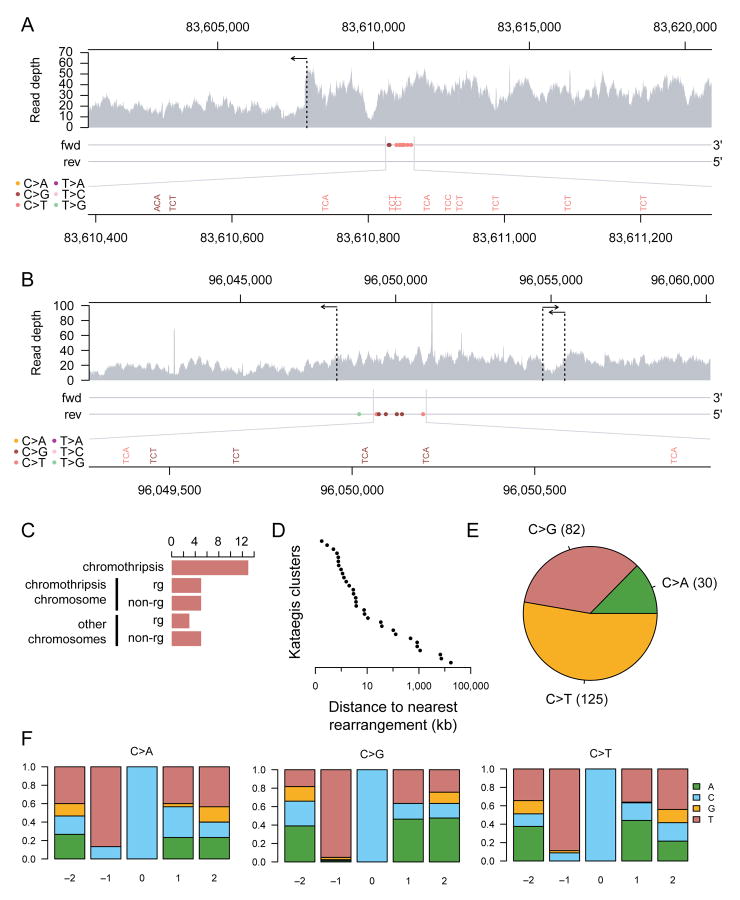
We observed 29 clusters of point mutations from 7 of the 10 post-crisis samples sequenced here that were absent from the parental cell lines. These clusters exhibited the cardinal features of kataegis observed in human cancers. First, clusters were often found within a kilobase or two of genomic breakpoints (Figure 6A-D). This was predominantly in association with chromothripsis rearrangements, although occasional clusters were also found near simpler structural variants (Figure 6C). Secondly, the clusters exhibited a pronounced preference for C>T and C>G mutations occurring in a TpC context (Figure 6E,F), the classic signature of APOBEC3A/B activity ((Roberts et al., 2013; Burns et al., 2013a; Burns et al., 2013b); reviewed in (Roberts and Gordenin, 2014)). Thirdly, the mutation clusters were processive, indicative of the damage occurring on a single strand of DNA (Figure 6A,B).
Figure 6. Mutational patterns of kataegis in post-crisis clones.
(A) A chromothripsis-associated kataegis in sample 24-141 on chromosome 7.
(B) A kataegis event in sample X-25 on chromosome X. This kataegis event took place on a chromosome with evidence for chromothripsis, but the rearrangements associated with the kataegis event do not appear to be part of the chromothripsis (Figure 5B). For both (A) and (B), the top panel shows raw read coverage of the region. The horizontal arrows indicate the positions of rearrangements. The two horizontal lines in the middle panel represent the forward and reverse strands. The pyrimidine strands of the mutations called are indicated by their placement on one of the two strands. Mutations are colored by mutation type. The bottom panel magnifies the mutation cluster regions and shows mutation contexts.
(C) The number of kataegis events grouped by their association with rearrangements as follows. From top to bottom: kataegis events within 10 kb of a chromothripsis rearrangement; kataegis events on a chromothripsis chromosome within 10 kb of a non-chromothripsis rearrangement; kataegis events on a chromothripsis chromosome with no rearrangements within 10 kb; kataegis events on a non-chromothripsis chromosome within 10 kb of a rearrangement; and kataegis events on non-chromothripsis chromosome with no rearrangements within 10 kb.
(D) The distance of each of the 31 detected kataegis events to their nearest respective rearrangement breakpoint.
(E) The frequency distribution of mutation types in the detected kataegis clusters.
(F) The nucleotide context around the mutated cytosine grouped by cytosine mutation type. The relative positions shown are on the pyrimidine (cytosine) strand. The Y-axes show the fraction of each nucleotide on the pyrimidine strand.
See also related Figure S7.
DISCUSSION
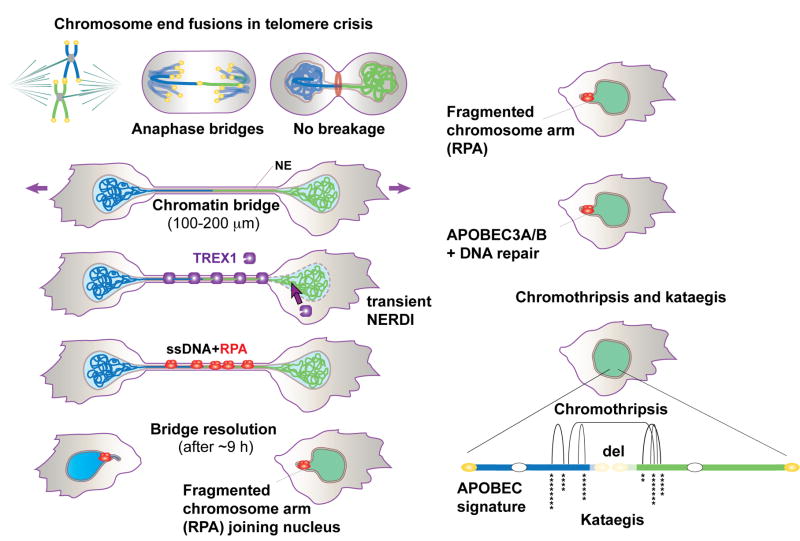
The findings reported here suggest that chromothripsis and kataegis can arise as a consequence of telomere crisis in the early stages of human tumorigenesis (Figure 7). The dicentric chromosomes formed in telomere crisis developed into long chromatin bridges that connect the two primary nuclei until the bridge breaks. Three important events in cells with chromatin bridges can explain the observed chromothripsis and kataegis. First, one of the connected primary nuclei undergoes NERDI that allows entry of the cytoplasmic 3’ exonuclease TREX1. Second, TREX1 generates extensive ssDNA in the chromatin bridges. Third, TREX1-mediated processing contributes to the resolution of the bridge, leaving two bridge DNA remnants that each join their primary nucleus. Subsequent repair of the bridge remnant DNA results in random joining of DNA segments typical of chromothripsis. In addition, APOBEC-derived hypermutation is prominent at the boundaries of the rearranged sequences. Since APOBEC enzymes act on ssDNA, the observed kataegis is consistent with the extensive single-stranded nature of the bridge remnants.
Figure 7. The fate of dicentric chromosomes formed in telomere crisis.
Telomere fusions in telomere crisis give rise to anaphase bridges that persist and develop into chromatin bridges. Cells with chromatin bridges undergo frequent NERDI and TREX1 accumulates on the chromatin bridge. TREX1 generates RPA-marked ssDNA in the chromatin bridge before their resolution. The RPA marked bridge remnants eventually join the primary nucleus where DNA repair and APOBEC3A/B editing are inferred to take place. Clonal descendants derived from telomere crisis cells show chromothripsis and kataegis.
Complex clusters of structural variants have been observed in many cancer genomes. Chromothripsis is an extreme example of such clusters, with tens to hundreds of genomic rearrangements affecting one or a few chromosomes or chromosome regions. Especially high rates have been observed in sarcomas, esophageal cancers and neuroblastomas (Nones et al., 2014; Stephens et al., 2011; Garsed et al., 2014; Mehine et al., 2013), and there appears to be an association with telomere crisis, especially BFB cycles (Li et al., 2014). Our study strengthens this association, suggesting that one of the routes to the chromosome damage that precipitates chromothripsis could be chromatin bridges formed by dicentric chromosomes.
While this study suggests that telomere crisis can precipitate chromothripsis events in cancer, many mechanistic questions remain. These issues are briefly discussed below.
Formation of extended chromatin bridges
Chromatin bridges have been documented and observed numerous times upon induction of telomere fusions in a wide variety of cell lines, but their significance, duration, and eventual fate had not been determined. In all cases, the chromatin bridges were observed in adherent tissue culture cells. We do not know whether chromatin bridges also develop if cells are grown in soft agar or indeed if such bridges would occur when incipient cancer cells undergo telomere crisis in vivo. Presumably, cell migration is not limited to tissue culture settings and takes place in the mass of cells that eventually gives rise to overt cancer. Cell motility is well documented in wound healing and is promoted by the epithelial-mesenchymal transition (EMT) in cancers of epithelial origin (Scheel and Weinberg, 2012). Furthermore, modeling suggests that cell migration is an important contributor to cancer development (Waclaw et al., 2015).
NERDI in cells with chromatin bridges
In cells with chromatin bridges, NERDI is more frequent and occurs sooner after anaphase. What is the mechanism by which chromatin bridges induce NERDI? One possibility is that the long bridges simply deplete critical NE components (e.g. Lamin B1, Lamin A/C) from the primary nuclei, thus inducing the transient failure in compartmentalization. A second possibility is that the stretching of the bridge exerts mechanical forces on the primary nuclei that overwhelm the resilience of the NE. Indeed, the shape of many of the primary nuclei suggests that considerable pulling forces are exerted by the chromosome(s) in the bridge but further work is required to understand the exact mechanism by which NERDI occurs. The attachment of human chromosomes to the nuclear lamins (Guelen et al., 2008) could play a role in generating the observed distortions and contribute to NERDI induction.
Preferential TREX1 action on bridge DNA
Why does TREX1 accumulate on the bridge and attack the bridge DNA rather than affecting the chromatin of the primary nucleus undergoing NERDI? One explanation could be that the bridge DNA is in a non-canonical chromatin state. It is possible that the pulling force of the migrating daughter cells results in loss of nucleosomes from the chromatin in the bridge (reviewed in (Chien and van Noort, 2009)). This mechanical nucleosome removal is consistent with the diminished staining for histones on the chromatin bridges. If TREX1 preferentially binds naked dsDNA, it would be expected to accumulate more on the non-nucleosomal bridge DNA than on the chromatin in the primary nucleus. Indeed, TREX1 degrades naked DNA much faster than nucleosomal DNA (Chowdhury et al., 2006) but whether this effect is due a higher affinity for naked DNA is not known.
Since TREX1 is a 3’ exonuclease, it will require nicked DNA substrates for the generation of ssDNA. Indeed, TUNEL staining has previously shown free 3’ ends on a chromatin bridge (Gisselsson et al., 2001). The nicks in the bridge DNA could originate from RNaseH2-mediated removal of misincorporated ribonucleotides (Reijns et al., 2012). Another possibility is that the TREX1 associated endonuclease NM23-H1 generates the 3’ ends used by the exonuclease (Chowdhury et al., 2006). So far, we have failed to detect NM23-H1 on the chromatin bridges but its abundance there may simply be too low for detection.
Bridge resolution
Bridge resolution is strongly correlated with a sudden increase in RPA staining, suggesting that the formation of ssDNA is a critical step. In the absence of TREX1, no or very low amounts of RPA are detected on the chromatin bridges. The residual RPA staining could be due to another nuclease or could be due to the over-stretching of the DNA, which can lead to RPA-coated denatured DNA in vitro (van Mameren et al., 2009). Although TREX1 is not solely responsible for bridge resolution, it significantly enhances the rupture of the bridges. We imagine that rupture occurs when two TREX1 3’ exonucleases acting on the Watson and Crick strands meet.
Conclusions
Telomere crisis has previously been shown to give rise to aneuploidy, non-reciprocal translocations, and whole genome reduplication (Artandi et al., 2000; Davoli and de Lange, 2012; Davoli et al., 2010). The data presented here suggest that chromothripsis and kataegis can be added to the list of cancer-relevant genome alterations that could be the consequence of telomere crisis. It will therefore be of interest to develop better tools to detect telomere crisis in the early stages of cancer. Methods to detect telomere crisis in pre-cancerous lesions may have predictive power with regard to disease progression and outcome.
EXPERIMENTAL PROCEDURES
(additional Experimental Procedures available in Supplemental Materials)
Cell Culture Procedures and plasmids
RPE1-hTERT and Phoenix cells from the American Type Culture Collection (ATCC) were cultured as described by the ATCC. Drug treatments, retroviral transduction, and cell cloning were performed using standard conditions (see Supplemental Experimental Procedures). CRISPR/Cas9 knockouts were generated with sgTREX1-2, 5’-GAGCCCCCCCACCTCTC-(PAM)-3’ using the gRNA cloning vector (Addgene) and co-transfection with an hCas9 plasmid (Addgene) by nucleofection (Lonza apparatus). Clones were isolated by limiting dilution, screened for TREX1 deletion by immunoblotting, and sequenced.
Immunoblotting and immunofluorescence
Immunoblotting and IF were performed using standard protocols. Cells were incubated with EdU for 30 min prior to PFA fixation and EdU detection with a Click-iT EdU Alexa Fluor 647 imaging kit (Life Technologies).
Live-cell Imaging
Cells were plated onto 35 mm glass bottom dishes (MatTek) 48 h before imaging. One h before imaging, the media was replaced with phenol red-free DMEM/F12 medium. Live cell imaging was performed at 37°C and 5% CO2 using a CellVoyager CV1000 spinning-disk confocal system (Yokogawa, Olympus) equipped with 445, 488, and 561 nm lasers, a Hamamatsu 512 × 512 EMCCD camera, and pinhole size of 50 μm. Details of image acquisition, processing, and quantification are given in the Supplemental Materials.
Telomere fusion PCR and karyotypic analysis
PCR for telomeric fusions and metaphase telomere fusion assays were performed essentially as described (Letsolo et al., 2010; Capper et al., 2007). Karyotypes were determined using standard protocols.
X-ten sequencing and mapping
Genomic DNA sequencing libraries were synthesized on robots and cluster generation and sequencing were performed using the manufacturer pipelines. Average sequence coverage across the samples was 33.3x (range, 27.4-35.9x). Mapping to genome build hs37d5 was performed using the BWA algorithm (BWA mem 0.7.8 (Li and Durbin, 2010)).
Copy number analysis
The reference genome divided into windows of equivalent read numbers (Campbell et al., 2008; Li et al., 2009) was used to extract reads with a mapping quality of at least 35 and the following flags: Properly paired; Non-secondary; QC-pass; Non-duplicate; Non-supplementary. Reads overlapping with each window were counted using BEDTools (Quinlan, 2014) and copy numbers were inferred from read depth data (Li et al., 2014).
Rearrangement calling and chromothripsis
Clusters of abnormally paired read pairs were identified from the merged sequence data using an in-house algorithm ‘Brass’. Raw rearrangement calls supported by clusters of abnormally mapped read pairs were called if the clusters were formed of at least four read pairs all from the same sample. For X-37, which yielded noisier data, at least six read pairs were required. The raw rearrangements were filtered as described previously (Li et al., 2014).
Mutation calling and kataegis
Point mutations were called using an in-house algorithm ‘Caveman’ as before (Nik-Zainal et al., 2012a) with RPE-1/Rbsh/p21sh/rtTA (Table S1) as reference. Raw mutations filtering is described in the Supplemental Materials. Kataegis mutation clusters were detected using visual inspection based on the criteria of short inter-mutation distance (generally <2kb) between cytosine mutations that were processive and enriched with TpC context.
Supplementary Material
Acknowledgments
We thank Prasad Jallepalli for reagents, Martin Hetzer for discussion, and Zhe Yang for performing PCR validation of chromothriptic breakpoints. We thank S. Yu and the de Lange lab for discussion and help with this manuscript. P. Ariel, T. Tong and Dr. A. North of the RU Bio-imaging Core Facility provided expert assistance with microscopy. The MSKCC Cytogenetics Core acknowledges NIH Cancer Center grant P30 CA008748. JM is a Merck Fellow of The Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by grants from the NCI (5R01CA181090) and the Breast Cancer Research Foundation to TdL. TdL is an American Cancer Society Research Professor.
Footnotes
AUTHOR CONTRIBUTIONS JM and TdL designed the experiments. JM performed all the cell biological experiments with the help of NB. YL and PC performed the genomic analysis on clones derived by JM and NB. TdL and PJC wrote the paper with the help of the other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013a;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013b;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien FT, van Noort J. 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr Pharm Biotechnol. 2009;10:474–485. doi: 10.2174/138920109788922128. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Garsed DW, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Jonson T, Petersén A, Strömbeck B, Dal Cin P, Höglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci U S A. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. Linking Micronuclei to Chromosome Fragmentation. Cell. 2015;161:1502–1504. doi: 10.1016/j.cell.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Hill A, Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Marko JF, Chatenay D, Libchaber A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J Cell Biol. 1997;139:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Cuppen E. Chromothripsis in congenital disorders and cancer: similarities and differences. Curr Opin Cell Biol. 2013;25:341–348. doi: 10.1016/j.ceb.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J Biol Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic Acids Res. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000, G. P. D. P. S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, Fegan C, Pepper C, Baird DM. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- Lin TT, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167:214–223. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- Lopez V, Barinova N, Onishi M, Pobiega S, Pringle JR, Dubrana K, Marcand S. Cytokinesis breaks dicentric chromosomes preferentially at pericentromeric regions and telomere fusions. Genes Dev. 2015;29:322–336. doi: 10.1101/gad.254664.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The Production of Homozygous Deficient Tissues with Mutant Characteristics by Means of the Aberrant Mitotic Behavior of Ring-Shaped Chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehine M, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369:43–53. doi: 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012a;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012b;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nones K, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Rodero MP, Crow YJ. Human disease phenotypes associated with mutations in TREX1. J Clin Immunol. 2015;35:235–243. doi: 10.1007/s10875-015-0147-3. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger L, Jones RE, Heppel NH, Williams GT, Sampson JR, Baird DM. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. J Natl Cancer Inst. 2013;105:1202–1211. doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K, Jones RE, Grimstead JW, Hills R, Pepper C, Baird DM. Telomere fusion threshold identifies a poor prognostic subset of breast cancer patients. Mol Oncol. 2015;9:1186–1193. doi: 10.1016/j.molonc.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Matheny JE, Sullivan BA. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 2012;20:595–605. doi: 10.1007/s10577-012-9302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci U S A. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.