SUMMARY
The cleft is an integral part of synapses, yet its macromolecular organization remains unclear. We here show that the cleft of excitatory synapses exhibits a distinct density profile as measured by cryo-electron tomography (cryo-ET). Aiming for molecular insights, we analyzed the synapse-organizing proteins SynCAM 1 and EphB2. Cryo-ET of SynCAM 1 knock-out and overexpressor synapses showed that this immunoglobulin protein shapes the cleft’s edge. In agreement, SynCAM 1 delineates the postsynaptic perimeter as determined by immuno-electron microscopy (EM) and superresolution imaging. In contrast, the EphB2 receptor tyrosine kinase is enriched deeper within the postsynaptic area. Unexpectedly, SynCAM 1 can form ensembles proximal to postsynaptic densities, and synapses containing these ensembles were larger. Postsynaptic SynCAM 1 surface puncta were not static but became enlarged after a long-term depression paradigm. These results support that the synaptic cleft is organized on a nanoscale into sub-compartments marked by distinct trans-synaptic complexes.
Keywords: synapse, synaptic cleft, adhesion, SynCAM, CADM, Nectin-like protein, EphB2
INTRODUCTION
Neuronal transmission requires precise organization of pre- and post-synaptic specializations (Harris and Weinberg, 2012; Sigrist and Sabatini, 2012). Limited structural insights are available into the synaptic cleft, the third compartment of a synapse. Current results are that the complexes spanning the cleft form net-like structures that can be periodically arranged (Lucic et al., 2005; Zuber et al., 2005; High et al., 2015).
Trans-synaptic interactions modulate synapse development and plasticity (Missler et al., 2012). Ultrastructural localization of N-cadherin shows that it is expressed throughout the cleft of developing synapses and present at the edge of mature synapses (Elste and Benson, 2006; Uchida et al., 1996; Yamagata et al., 1995). N-cadherin does not induce synapses and comparable insights into synaptogenic proteins are lacking, though immuno-EM studies have demonstrated the differential expression of neuroligins at excitatory and inhibitory synapses (Song et al., 1999; Varoqueaux et al., 2004; Mortillo et al., 2012). Synaptogenic proteins may demarcate and function at specialized synaptic zones, yet the limited understanding of cleft topography restricts addressing these questions.
We here delineated macromolecular properties of the excitatory synaptic cleft. To gain molecular insights, we investigated two proteins that form trans-synaptic complexes to promote excitatory synapse number, the immunoglobulin adhesion protein SynCAM 1 (Synaptic Cell Adhesion Molecule 1, also named nectin-like 2 or Cadm1) (Biederer et al., 2002; Fogel et al., 2007; Robbins et al., 2010) and the EphB2 receptor tyrosine kinase (Sheffler-Collins and Dalva, 2012). Analysis of excitatory synapses by cryo-ET, immuno-EM, and STED (Stimulated Emission Depletion) and STORM (Stochastic Optical Reconstruction Microscopy) super-resolution imaging supports that the synaptic cleft is comprised of structurally and molecularly defined and dynamic sub-compartments.
RESULTS
Structural organization of the cleft of excitatory synapses
Cryo-ET enables high-resolution imaging of the entire cleft in a fully hydrated, physiologically relevant state (Lucic et al., 2013). We recorded tomograms of neocortical synaptosomes from adult mice (Figures 1A and S1A). All analyzed synapses were asymmetric with a postsynaptic density (PSD) and likely corresponded to excitatory synapses. The mean cleft width of wild-type (WT) synapses was 22.0±0.5 nm (Figure S1C), as described (Rees et al., 1976). Numerous complexes spanned the cleft and often assumed the shape of a laterally extended, net-like density (Figure 1A and Supplemental Media File 1), as described (Lucic et al., 2005). Trans complexes and net-like structures were seen in all analyzed tomograms.
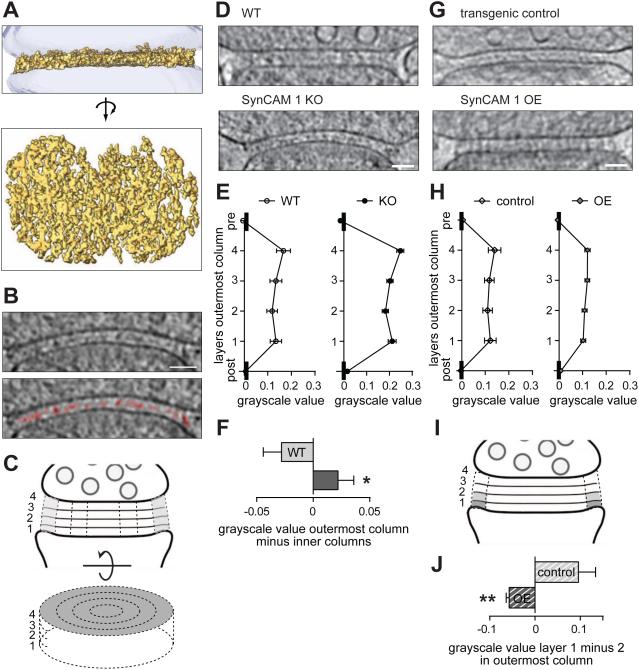
Figure 1. The excitatory synaptic cleft is structurally organized and SynCAM 1 shapes the edge.
(A) Top, side view of a segmented synaptic cleft. Bottom, top view.
(B) Top, tomographic slice from a synaptosome at 4 voxels depth (9.2 nm). Bottom, segmented net-like structures closer to the postsynaptic (lower) side are marked in red. The asterisk marks a gold particle for tomogram alignment. Scale bar, 50 nm.
(C) Cleft separation into four layers and concentric columns. The outermost column in shown in grey.
(D) WT and SynCAM 1 KO cleft tomograms at 4 voxels depth (9.2 nm). Arrowheads mark the less dense central density towards the edge of the KO cleft. Scale bar, 50 nm.
(E) Profiles of the outermost WT and KO cleft columns. Lower grayscale values correspond to higher densities. Mean layer values were calculated in each tomogram and averaged per genotype (N=7 WT, 8 KO synapses).
(F) SynCAM 1 KO synapses have a higher grayscale value differential and hence lower relative protein density in the outer column compared to the inner columns (N=7 WT, 8 KO synapses).
(G) Tomograms of transgenic control and SynCAM 1 overexpressor (OE) clefts at 4 voxel depth (9.2 nm). The central cleft density of the control is barely visible in OE synapses. Scale bar, 50 nm.
(H) lat profile of the SynCAM 1 OE cleft. Greyscale values are shown as in (E) (N=5 synapses each).
(I) Data in (J) was calculated by subtracting grayscale values of volumes depicted in light gray from those in dark gray.
(J) SynCAM 1 OE synapses have higher density in layer 1 relative to layer 2 in the outer cleft column compared to controls (N=5 synapses each).
See also Figure S1 and Supplemental Media Files 1 and 2.
Observation of tomograms indicated an increased central density in the cleft, closer to the postsynaptic side (Figure 1B and Supplemental Media File 2). To measure whether this density is off-set from the middle, we separated the cleft into four layers because this gave the most robust results with low noise (Figure 1C). Each cleft was further divided into four concentric columns with even radii (Figure 1C). Lower cryo-ET greyscale values correspond to higher protein densities, and mean greyscale values exhibited a minimum, i.e. highest density, in the second layer counted from the post-to pre-synaptic membrane when all columns were combined (layer 2 vs. 1/3/4; paired t-test, p=0.0007/0.015/0.0001; N=7 WT synapses) as well as in the outermost column (Figure 1D, E left; layer 2 vs. 1/3/4; paired t-test, p=0.015/0.019/0.0003; N=7 WT synapses). Densities of the four cleft columns were indistinguishable (not shown).
The edge of the synaptic cleft is shaped by SynCAM 1
We next tested whether SynCAM 1 affects the makeup of the synaptic cleft, choosing this immunoglobulin adhesion protein due to its expression across excitatory forebrain synapses, its ability to increase excitatory synapse number in cultured neurons and the brain, and the high synaptic membrane content of SynCAMs (Biederer et al., 2002; Fogel et al., 2007; Robbins et al., 2010). Neocortical synaptosomes from adult SynCAM 1 knock-out (KO) mice had the same cleft width as WT (Figures 1D and S1A,C). Loss of SynCAM 1 did not alter the layer profile when data of all columns were averaged (data not shown) or in the outermost column (Figure 1D, E right). The higher grayscale values in the KO could not be interpreted with certainty as lower total cleft protein amounts because of the inability to determine absolute values with cryo-ET. However, relative changes can be robustly compared. This showed that synapses lacking SynCAM 1 exhibited a loss of relative protein density, i.e. an increased grayscale differential, in the outermost cleft column compared to the inner columns (Figure 1F; t-test, p=0.037; N=7 WT and 8 KO synapses). Loss of SynCAM 1 therefore lowers the density distribution towards the synaptic edge.
Because SynCAM 1 loss preserved the highest density in layer 2, other complexes likely establish this profile. We asked whether those interactions can be imbalanced by elevating SynCAM 1. We recorded cryo-ET images of synaptosomes from transgenic mice overexpressing (OE) SynCAM 1 in excitatory neurons and from littermates lacking the SynCAM 1 transgene (transgenic controls) (Figure 1G). Cleft width was unaffected by elevated SynCAM 1 (Figure S1B,C). Control synapses showed the layer profile expected from WT synapses (Figure 1H left vs. 1E left). In contrast, the profile of OE synapses was flat (Figure 1H right). We measured an inverted difference (higher density in layer 1 than 2) in the outermost column of OE synapses, different from controls (t-test, p=0.0044; N=5 synapses each) (Figure 1I,J). This inversion only occurred in the outermost column (data not shown). Elevated SynCAM 1 therefore disrupts the layer profile in the outer cleft column, possibly through its increased expression at the postsynaptic edge. These structural aberrations after loss and overexpression of SynCAM 1 indicated that this adhesion protein organizes the outer zone of the cleft.
SynCAM 1 localizes to the postsynaptic edge of excitatory synapses
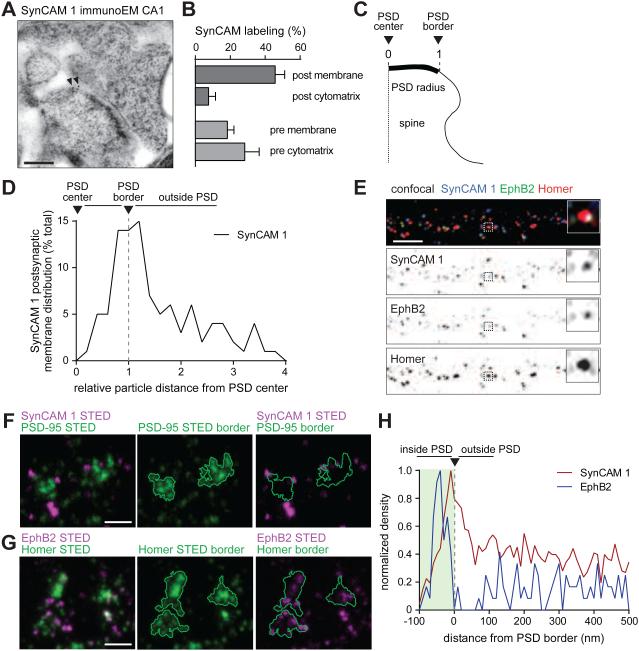
We next localized endogenous SynCAM 1 using immuno-EM of high-pressure frozen hippocampal slices from adult mice (Figure 2A). In CA1 stratum radiatum, anti-SynCAM 1 immuno-gold particles labeled most frequently excitatory synapses. This was expected from an immuno-EM study with antibodies detecting the family members SynCAM 1/2/3 equally well (Biederer et al., 2002) and biochemical fractionation (Fogel et al., 2007). Synaptic labeling was reduced by 84±4% in SynCAM 1 KO sections, validating antibody specificity (Figure S2A). At WT CA1 synapses, gold particles were most common at postsynaptic membranes (46±6% of total synaptic labeling) (Figure 2B). Less labeling was found in the pre- and post-synaptic cytomatrix and presynaptic membranes. To map SynCAM 1 distribution, we measured distances of the gold particles at postsynaptic membranes relative to the PSD center (Figure 2C). This determined that postsynaptic SynCAM 1 localizes to the cleft edge (Figure 2D). The less abundant SynCAM 1 particles at presynaptic membranes were distributed as two populations, one more central and the other peri-synaptic (Figure S2B). These results supported an enrichment of postsynaptic SynCAM 1 at the edge of excitatory synapses.
Figure 2. Postsynaptic SynCAM 1 marks the perimeter of excitatory synapses and EphB2 is enriched deeper within the postsynaptic area.
(A) Immuno-EM of SynCAM 1 in adult hippocampal CA1 area after high pressure freezing. Arrows mark 10 nm gold particles labeling SynCAM 1 at the postsynaptic membrane edge of an asymmetric synapse.
(B) majority of synaptic SynCAM 1 localizes to postsynaptic membranes (N=97 micrographs, 3 mice).
(C) Measurement of postsynaptic distances for quantification in (D). 0 marks the PSD center and 1 the edge.
(D) Postsynaptic membrane SynCAM 1 is enriched at the PSD edge. Particle distances to the PSD center were measured for each synapse and normalized as in (C) (N=97 micrographs, 3 mice).
(E) Hippocampal neurons were sequentially immunostained at 14 div to first detect surface SynCAM 1 (blue) and surface EphB2 (green), followed by permeabilization and staining for postsynaptic Homer (red), and confocal imaging. The box marks the synapse enlarged in the insets. Scale bar, 5 μm.
(F,G) Hippocampal neurons at 14 div were subjected to sequential immunostaining for (F) surface SynCAM 1 (magenta) and PSD-95 (green) or (G) surface EphB2 (magenta) and Homer (green), and imaged by 2-color STED microscopy. PSD borders based on STED image analysis are shown in center and right panels. Scale bars, 400 nm.
(H) Surface SynCAM 1 and EphB2 locations were determined by STED as in (F,G) and distances were measured from the PSD border defined by PSD-95 and Homer images, respectively. SynCAM 1 (red) reached maximum density at the PSD border (green). EphB2 (blue) was prominently located within PSD areas. Densities were normalized to the highest value. SynCAM 1 data from 696 PSD areas in 86 imaging fields; EphB2 data, 111 PSD areas in 10 fields.
See also Figure S2.
EphB2 and SynCAM 1 complexes mark distinct sub-synaptic areas
A second important player in the cleft is the postsynaptic receptor tyrosine kinase EphB2 that promotes excitatory synaptogenesis during the rapid phase of synapse addition before neurons mature (Kayser et al., 2008; Sheffler-Collins and Dalva, 2012). This role differs from SynCAM 1, which first induces and then maintains excitatory synapses (Robbins et al., 2010). We speculated that this functional difference may be reflected in distinct sub-synaptic localizations. We first addressed to what extent these proteins co-localize at excitatory synapses. Non-permeabilized hippocampal neurons were immunolabeled for endogenous, surface-expressed SynCAM 1 and EphB2 at 14 days in vitro (div) with antibodies against their extracellular domains, followed by permeabilization, immunostaining for the excitatory postsynaptic scaffold protein Homer, and confocal microscopy (Figure 2E). SynCAM 1 and EphB2 were detected at comparable densities along dendrites (data not shown). Automated three-channel co-localization analysis measured that 74±9% of Homer puncta contained SynCAM 1, EphB2 or both. Of the Homer-positive SynCAM 1 puncta, 88±1% co-localized with EphB2, while all synaptic EphB2 puncta were positive for SynCAM 1 (N=69 dendritic segments from 3 independent experiments).
We next analyzed the sub-synaptic distribution of surface SynCAM 1 and EphB2 by 2-channel STED super-resolution microscopy, applying the same sequential immunostaining (Figure 2F,G). This showed that SynCAM 1 has a high density at the PSD border while EphB2 resides within the bounds of the PSD proper (Figure 2H). Trans-synaptic complexes can therefore mark distinct synaptic zones.
3D localization shows differential EphB2 and SynCAM distribution and reveals cloud-like SynCAM ensembles
What is the three-dimensional distribution of synapse-organizing proteins? To address this question, we used 2-color 3D dSTORM (Huang et al., 2008) to analyze cultures of hippocampal neurons at 12-14 div. Endogenous, surface-expressed SynCAM 1 or EphB2 and postsynaptic Homer were visualized by sequential immunostaining and imaged by 3D dSTORM (Figure 3A,B). SynCAM 1 and EphB2 were found along dendrites, including near Homer-positive PSDs. The three-dimensional border of each PSD was defined with a convex hull around Homer localizations (Dani et al., 2010; MacGillavry et al., 2013), and the density of SynCAM localizations was determined within this border and outside it in 50 nm shells (Figure 3C). This revealed a prominent peak of SynCAM 1 density between 100 nm and 200 nm outside the PSD border. This agrees with the immuno-EM and STED localizations, though differences of synapses formed in vivo and by cultured neurons, maturation stage, and 2D STED and 3D STORM imaging need to be considered. We consider antibody penetration issues unlikely, as post-embedding immuno-EM which is not expected to be affected by access issues showed similarly restricted labeling (Figure 2D). In contrast to SynCAM 1, EphB2 localizations by 3D STORM were most prominent deeper within the postsynaptic area, with a smaller peak of EphB2 between 100 nm and 200 nm outside the PSD (Figure 3B,C)
Figure 3. Distinct 3D distribution of EphB2 and SynCAM 1 and presence of SynCAM 1 ensembles around the cleft.
(A,B) Left, hippocampal neurons at 12-14 div were subjected to sequential immunostaining for surface SynCAM 1 (A) or EphB2 (B) (red) and intracellular Homer (green), and imaged by 2-channel 3D STORM. Center and right, enlarged PSD with the calculated border outlined. Scale bar overview, 1 μm; enlarged panels, 400 nm.
(C) Surface SynCAM 1 and EphB2 localizations were determined by 3D STORM as in (A,B) and the PSD border was defined by super-resolved Homer localizations. SynCAM 1 localization density (red) reached a maximum around the PSD edge (green) and localizations within Homer hulls were rare. EphB2 (blue) was prominently localized within the area demarcated by the PSD and showed a smaller peak around the edge. SynCAM 1 data from 178 PSDs in 11 imaging fields; EphB2 data, 446 PSDs in 31 fields.
(D) Two 3D views of a convex, Homer-defined PSD hull (green; boxed in A) showing adjacent ensembles of SynCAM 1 (red). Scale bars, 200 nm in each axis.
(E) Distribution of SynCAM 1 ensemble volumes within 500 nm of the PSD. Gray graph, putative single molecules detected at low thresholds. Red, ensembles detected at a threshold excluding single molecules.
(F) PSDs marked by SynCAM 1 ensembles are larger. Cumulative frequency distribution of super-resolved Homer volumes in spines lacking SynCAM 1 ensembles (grey) or containing at least one SynCAM 1 ensemble (red) within 500 nm of the Homer hull. Ensembles were identified as in (E) (N=120 PSDs lacking SynCAM 1 ensembles, 31 PSDs with ensembles from 11 fields of imaging; Mann-Whitney test, p=0.026).
See also Figure S3.
Of all PSDs, 79% had SynCAM surface localizations within 500 nm of their border and a subset of SynCAM 1 localizations appeared in large ensembles (Figure 3D). To characterize these grouped localizations using automated analysis, we applied area and density criteria to select ensembles that likely consisted of multiple SynCAM 1 molecules (Figures 3E and S3). The volume of SynCAM 1 ensembles varied widely, averaging 8.4×106 nm3 with a standard deviation of 6.6×106 nm3. The median volume was 3.9×106 nm3, approximately 1/3 the volume of Homer-defined PSDs (median 9.4±1.5×106 nm3). 21% of PSDs had SynCAM 1 ensembles within 500 nm of their border, suggesting that they belonged to a unique subpopulation of synapses. To determine whether the presence of SynCAM 1 ensembles correlated with structural features, we measured Homer clusters (Figure 3F). This showed that synapses lacking SynCAM 1 ensembles had an average Homer volume of 7.2±0.8 ×106 nm3 (N=120), while Homer clusters had almost twice the volume when marked by SynCAM 1 ensembles (13.1±2.3 ×106 nm3; N=31; t-test, p=0.003). These ensembles thus revealed a novel macromolecular feature of trans-synaptic complexes.
The synaptic cleft is a dynamic compartment
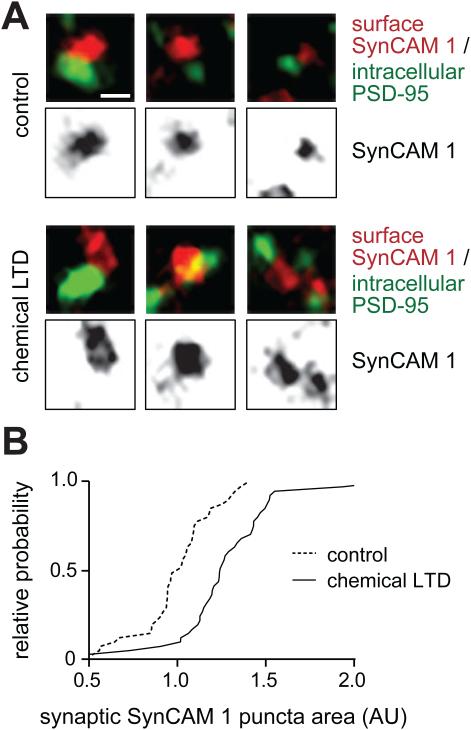
Neuronal activity and synaptic plasticity regulate the trafficking of N-cadherin and its interactions with β-catenins (Hirano and Takeichi, 2012). Can the distribution of synaptogenic proteins also change in an activity-dependent manner? SynCAM 1 was well-suited to test this question as the position of SynCAM complexes at the synaptic edge may allow them to be readily dispersed. We used a chemical LTD protocol because SynCAM 1 regulates LTD in vivo (Robbins et al., 2010). Sequential immunostainings were performed for surface SynCAM 1 and the excitatory postsynaptic scaffold protein PSD-95, and imaged by confocal microscopy (Figure 4A). Analysis was restricted to surface-labeled SynCAM 1 within 0.8 μm of the center of the nearest PSD-95 punctum to measure SynCAM 1 puncta within approximately one PSD diameter, which in our confocal analysis was 0.65±0.02 μm (N=41 neurites). LTD treatment enlarged synaptic SynCAM 1 puncta by 27±5% (t-test, p<0.0001; N=41 neurites each). Frequency distribution analysis supported a LTD-induced enlargement of SynCAM 1 puncta across all area ranges (Figure 4B). Synaptic SynCAM 1 proteins can therefore undergo dynamic changes in their surface distribution.
Figure 4. Activity-dependent area changes of synaptic SynCAM 1 complexes.
(A) Hippocampal neurons at 14 d.i.v. were subjected to chemical LTD. Surface SynCAM 1 was immunolabeled (red), followed by staining for postsynaptic PSD-95 (green) and confocal imaging. Three representative images show control synapses (top) or after chemical LTD treatment (bottom). Scale bar, 0.8 μm.
(B) LTD treatment enlarges the area of synaptic SynCAM 1 puncta. The graph shows the cumulative frequency distribution of SynCAM 1 puncta areas located within 0.8 μm of a PSD-95 punctum (Mann-Whitney test, p<0.0001; N=41 neurites each).
DISCUSSION
This study reports five findings about the cleft of excitatory synapses. First, the cleft is structurally patterned. Second, the cleft is molecularly organized, with SynCAM 1 marking the edge and EphB2 central areas. Third, SynCAM 1 shapes the edge of the cleft. Fourth, SynCAM 1 proteins can form cloud-like ensembles around the postsynaptic border. Fifth, our results support that synaptogenic proteins can undergo activity-dependent re-distribution.
Our cryo-ET analysis provides evidence that the excitatory synaptic cleft can be stratified into layers based on protein densities, reminiscent of the intra-cleft line (Gray, 1959; Hajos, 1980) prominent after phosphotungstic acid staining (Bloom and Aghajanian, 1968). This indicates that clusters of adhesion molecules are repetitively organized. Future studies may uncover effects of maturation stage and brain regions. The density and layer profile of the outer cleft column are shaped by SynCAM 1, consistent with its localization to the postsynaptic edge. We regularly observed synaptic vesicles opposite SynCAM 1-labeled PSD edges, and hence consider this zone unlikely to be puncta adhaerentia (Palay, 1967) but part of synaptic junctions. It is conceivable that SynCAM 1 participates in the conversion of nascent zones, specialized edge regions, into active zones (Bell et al., 2014). The shortened PSDs and active zones in SynCAM 1 KO mice (Robbins et al., 2010) may be due to SynCAM 1 loss in this outer column, consistent with our result that PSDs with SynCAM 1 ensembles are substantially larger than those without.
SynCAM 1 loss and overexpression selectively affect the structural organization of the cleft’s edge, supporting that differences in the adhesive makeup of synapses are a factor in shaping them. The central density along the cleft is unaffected by SynCAM 1 and is hence likely established by other proteins such as neuroligins/β-neurexins which form sheets at non-neuronal cell contacts with higher density closer to neuroligin (Tanaka et al., 2012). Indeed, one reason for the diversity of adhesion systems at excitatory synapses may be that they need to occupy distinct cleft nanodomains to instruct different characteristics of synapses. EphB2, acting during the initial period of synapse formation (Kayser et al., 2008), and SynCAM 1, which induces excitatory synapses and is then required to maintain this increase (Robbins et al., 2010), are differentially localized to areas deeper in the postsynaptic area and around the PSD. The mechanisms that position trans-synaptic organizers with such precision are currently unknown. Given the role of SynCAM 1 in synapse maintenance, it is conceivable that the stabilization of synapses involves interactions at their edge, possibly through links of SynCAM 1 to the postsynaptic cytoskeleton (Cheadle and Biederer, 2012). Selectively positioned trans-synaptic proteins may also affect synapse function and define specialized synaptic zones such as proposed for neurotransmitter release (Kavalali, 2015). The density profile of the cleft is relevant for transmission, too, as cleft geometry is predicted to shape synaptic currents (Savtchenko and Rusakov, 2007). Moreover, the density of the cleft’s edge could control access of extra-synaptic neurotransmitter receptors (Choquet and Triller, 2013) via presentation of binding sites or macromolecular crowding (Santamaria et al., 2010).
PSD size correlates with basal synaptic strength (Harris and Weinberg, 2012) and the presence of SynCAM 1 ensembles linked to larger PSDs may be regulated by the activity history of each synapse. Given the structural dynamics of synapses during plasticity, it is also of interest that postsynaptic SynCAM 1 complexes occupy a larger area after LTD. In contrast to SynCAM 1, neuroligin 1 is rapidly internalized upon LTD (Schapitz et al., 2010) and cleaved (Peixoto et al., 2012). Synapse-organizing proteins hence exhibit distinct activity-dependent dynamics, adding to the molecular diversity of the cleft.
Together, our study provides evidence that the synaptic cleft is organized into sub-compartments. This introduces the concept of nanodomains to the cleft and expands recent insights into active zones and postsynaptic sites (Choquet and Triller, 2013; MacGillavry et al., 2013; Sigrist and Sabatini, 2012). Differences in the nanoscale organization and dynamics of the cleft may be important parameters that specify synaptic properties.
EXPERIMENTAL PROCEDURES
Tomography
Cerebral cortex homogenates from mice were subjected to differential centrifugation, followed by Percoll gradient purification of synaptosomes and vitrification. Tilt series were collected using a Polara electron microscope (FEI), reconstructed, and segmented.
Immuno-EM
Acute hippocampal slices were prepared and high-pressure frozen CA1 samples were processed for post-embedding immuno-EM with monoclonal anti-SynCAM 1 antibodies (MBL Laboratories, clone 3E1; 1:50). Grids were observed with a FEI Tecnai Biotwin electron microscope.
Neuronal culture
Hippocampal neurons were cultured from E18 rat embryos for STORM imaging and confocal co-localization. Hippocampal cultures from P1 rats were used for STED imaging and LTD studies. Chemical LTD was induced at 14 div by treating neurons for 3 min with 20 μM NMDA.
Confocal imaging
Non-permeabilized neurons were subjected at 14 div to surface labeling with antibodies against the extracellular domain of SynCAM 1 (MBL, 3E1; 1:000) or EphB2 (R&D Systems, AF467; 1:100). Homer was immunodetected after permeabilization (Synaptic Systems, 160 003; 1:500), as was PSD-95 (NeuroMab, clone K28/43; 1:500). A Leica TCS SPE DM2500 microscope was used for confocal imaging.
STED imaging
Neurons were subjected to sequential immunostaining as above and imaged on a custom built gated detection, beam scanning, all pulsed laser STED system with a 100X oil objective (UPLAPO 100XO/PSF, Olympus). Fluorescence was detected by a single photon counting avalanche photodiode (SPCM-ARQ-13-FC, Perkin Elmer).
STORM imaging
Neurons were subjected to sequential immunostaining as above and imaged on an Olympus IX81 ZDC2 microscope with a 100X/1.49 TIRF oil objective using simultaneous excitation with 647 nm and 561 nm lasers. Stochastic blinking was detected with an iXon+ 897 EM-CCD camera (Andor). An astigmatic lens was added and Z-axis positions of localized molecules were deduced post hoc.
Statistical analyses
Data were analyzed using GraphPad Prism 6 and custom ImageJ (confocal) and MATLAB (Mathworks) scripts (STED, STORM) scripts. * denotes t-test p<0.05; ** p<0.01; *** p<0.001.
Animal procedures
Animal procedures in this study were approved by the Institutional Animal Care and Use Committees, in compliance with NIH guidelines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Graham for EM support, S. Asano and Z. Kochovski for help with cryo-ET, Dr. D. Lai for script development, Dr. T. Momoi for the SynCAM 1/RA175 KO line, Dr. W. Baumeister for supporting cryo-ET, and Dr. S. Chandra for discussions. We acknowledge support from the Tufts Center for Neuroscience Research under NIH grant P30 NS047243, as well as NIH grants F31 MH105105 (to S.W.R.M.), R01 MH080046 and R01 GM106000 (to T.A.B.) and R01 DA018928 (to T.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Data include 3 Figures, 2 Media Files, and detailed Experimental Procedures.
AUTHOR CONTRIBUTIONS
K.P. performed and analyzed EM and confocal imaging and performed STED, N.S. analyzed cryo-ET, S.M. performed and analyzed STORM, E.A. performed and analyzed STED, G.K. recorded cryo-ET, A.T. analyzed STORM, A.J.K. shared mice and supported cryo-ET, V.S. supervised A.J.K., X.L. supported EM, J.B. supervised E.A., T.A.B. designed STORM and supervised S.M. and A.T., V.L. designed cryo-ET, developed analysis and supervised N.S. and G.K., and T.B. designed the study with T.A.B and V.L., supervised K.P., and wrote the manuscript.
REFERENCES
- Bell ME, Bourne JN, Chirillo MA, Mendenhall JM, Kuwajima M, Harris KM. Dynamics of nascent and active zone ultrastructure as synapses enlarge during long-term potentiation in mature hippocampus. J Comp Neurol. 2014;522:3861–3884. doi: 10.1002/cne.23646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Bloom FE, Aghajanian GK. Fine structural and cytochemical analysis of the staining of synaptic junctions with phosphotungstic acid. J Ultrastruct Res. 1968;22:361–375. doi: 10.1016/s0022-5320(68)90027-0. [DOI] [PubMed] [Google Scholar]
- Cheadle L, Biederer T. The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 2012;199:985–1001. doi: 10.1083/jcb.201205041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Triller A. The dynamic synapse. Neuron. 2013;80:691–703. doi: 10.1016/j.neuron.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elste AM, Benson DL. Structural basis for developmentally regulated changes in cadherin function at synapses. J Comp Neurol. 2006;495:324–335. doi: 10.1002/cne.20876. [DOI] [PubMed] [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EG. Axo-somatic and axo-dendritic synapses of the cerebral cortex: an electron microscope study. J Anat. 1959;93:420–433. [PMC free article] [PubMed] [Google Scholar]
- Hajos F. The structure of cleft material in spine synapses of rat cerebral and cerebellar cortices. Cell Tissue Res. 1980;206:477–486. doi: 10.1007/BF00237976. [DOI] [PubMed] [Google Scholar]
- Harris KM, Weinberg RJ. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. 2012;4:a005587. doi: 10.1101/cshperspect.a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High B, Cole AA, Chen X, Reese TS. Electron microscopic tomography reveals discrete transcleft elements at excitatory and inhibitory synapses. Front Synaptic Neurosci. 2015;7:9. doi: 10.3389/fnsyn.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalali ET. The mechanisms and functions of spontaneous neurotransmitter release. Nat Rev Neurosci. 2015;16:5–16. doi: 10.1038/nrn3875. [DOI] [PubMed] [Google Scholar]
- Kayser MS, Nolt MJ, Dalva MB. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron. 2008;59:56–69. doi: 10.1016/j.neuron.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol. 2013;202:407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Yang T, Schweikert G, Forster F, Baumeister W. Morphological characterization of molecular complexes present in the synaptic cleft. Structure. 2005;13:423–434. doi: 10.1016/j.str.2005.02.005. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Südhof TC, Biederer T. Synaptic cell adhesion. In: Südhof TC, Sheng M, Sabatini B, editors. Synapses. Cold Spring Harbor Laboratory Press; 2012. pp. 31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortillo S, Elste A, Ge Y, Patil SB, Hsiao K, Huntley GW, Davis RL, Benson DL. Compensatory redistribution of neuroligins and N-cadherin following deletion of synaptic beta1-integrin. J Comp Neurol. 2012;520:2041–2052. doi: 10.1002/cne.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL. Principles of cellular organization in the nervous system. In: Quorton GC, Melnchuch T, Schmitt FO, editors. The Neurosciences. Rockefeller University Press; New York: 1967. pp. 24–31. [Google Scholar]
- Peixoto RT, Kunz PA, Kwon H, Mabb AM, Sabatini BL, Philpot BD, Ehlers MD. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron. 2012;76:396–409. doi: 10.1016/j.neuron.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees RP, Bunge MB, Bunge RP. Morphological changes in the neuritic growth cone and target neuron during synaptic junction development in culture. J Cell Biol. 1976;68:240–263. doi: 10.1083/jcb.68.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins EM, Krupp AJ, Perez de Arce K, Ghosh AK, Fogel AI, Boucard A, Sudhof TC, Stein V, Biederer T. SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron. 2010;68:894–906. doi: 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria F, Gonzalez J, Augustine GJ, Raghavachari S. Quantifying the effects of elastic collisions and non-covalent binding on glutamate receptor trafficking in the post-synaptic density. PLoS Comput Biol. 2010;6:e1000780. doi: 10.1371/journal.pcbi.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savtchenko LP, Rusakov DA. The optimal height of the synaptic cleft. Proc Natl Acad Sci U S A. 2007;104:1823–1828. doi: 10.1073/pnas.0606636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapitz IU, Behrend B, Pechmann Y, Lappe-Siefke C, Kneussel SJ, Wallace KE, Stempel AV, Buck F, Grant SG, Schweizer M, et al. Neuroligin 1 is dynamically exchanged at postsynaptic sites. J Neurosci. 2010;30:12733–12744. doi: 10.1523/JNEUROSCI.0896-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler-Collins SI, Dalva MB. EphBs: an integral link between synaptic function and synaptopathies. Trends Neurosci. 2012;35:293–304. doi: 10.1016/j.tins.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Sabatini BL. Optical super-resolution microscopy in neurobiology. Curr Opin Neurobiol. 2012;22:86–93. doi: 10.1016/j.conb.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Südhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci U S A. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Miyazaki N, Matoba K, Nogi T, Iwasaki K, Takagi J. Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin. Cell Rep. 2012;2:101–110. doi: 10.1016/j.celrep.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Herman JP, Sanes JR. Lamina-specific expression of adhesion molecules in developing chick optic tectum. J Neurosci. 1995;15:4556–4571. doi: 10.1523/JNEUROSCI.15-06-04556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber B, Nikonenko I, Klauser P, Muller D, Dubochet J. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc Natl Acad Sci U S A. 2005;102:19192–19197. doi: 10.1073/pnas.0509527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.