Abstract
Quaternary climatic factors have played a significant role in population divergence and demography. Here we investigated the phylogeography of Osteomeles schwerinae, a dominant riparian plant species of the hot/warm-dry river valleys of the Hengduan Mountains (HDM), Qinling Mountains (QLM) and Yunnan-Guizhou Plateau (YGP). Three chloroplast DNA (cpDNA) regions (trnD-trnT, psbD-trnT, petL-psbE), one single copy nuclear gene (glyceraldehyde 3-phosphate dehydrogenase; G3pdh), and climatic data during the Last Interglacial (LIG; c. 120–140 ka), Last Glacial Maximum (LGM; c. 21 ka), and Current (c. 1950–2000) periods were used in this study. Six cpDNA haplotypes and 15 nuclear DNA (nDNA) haplotypes were identified in the 40 populations of O. schwerinae. Spatial Analysis of Molecular Variance, median-joining networks, and Bayesian phylogenetic trees based on the cpDNA and nDNA datasets, all suggested population divergence between the QLM and HDM-YGP regions. Our climatic analysis identified significant heterogeneity of the climatic factors in the QLM and HDM-YGP regions during the aforementioned three periods. The divergence times based on cpDNA and nDNA haplotypes were estimated to be 466.4–159.4 ka and 315.8–160.3 ka, respectively, which coincide with the time of the weakening of the Asian monsoons in these regions. In addition, unimodal pairwise mismatch distribution curves, expansion times, and Ecological Niche Modeling suggested a history of population expansion (rather than contraction) during the last glaciation. Interestingly, the expansion times were found being well consistent with the intensification of the Asian monsoons during this period. We inferred that the divergence between the two main lineages is probably caused by disruption of more continuous distribution because of weakening of monsoons/less precipitation, whilst subsequent intensification of the Asian monsoons during the last glaciation facilitated the expansion of O. schwerinae populations.
Introduction
Exploring factors associated with population processes (e.g. population divergence and demography) is an important issue in ecology and evolution [1–4]. Divergent selection and adaptation associated with different climatic factors have increasingly become recognized to have major influence on population demography, divergence and/or speciation in plants [4–7]. In recent years, a considerable number of phylogeographic studies have been conducted in the Hengduan Mountains (HDM) and adjacent regions [e.g. the Qinghai-Tibetan Plateau (QTP); Eastern Himalayas (EH); Yunnan-Guizhou Plateau (YGP); Qinling Mountains (QLM); Fig 1]. These studies provide robust molecular evidence that (sub)alpine plants presently occurring in these regions are strongly affected by glacial-interglacial climatic fluctuations during the Quaternary [8–14].
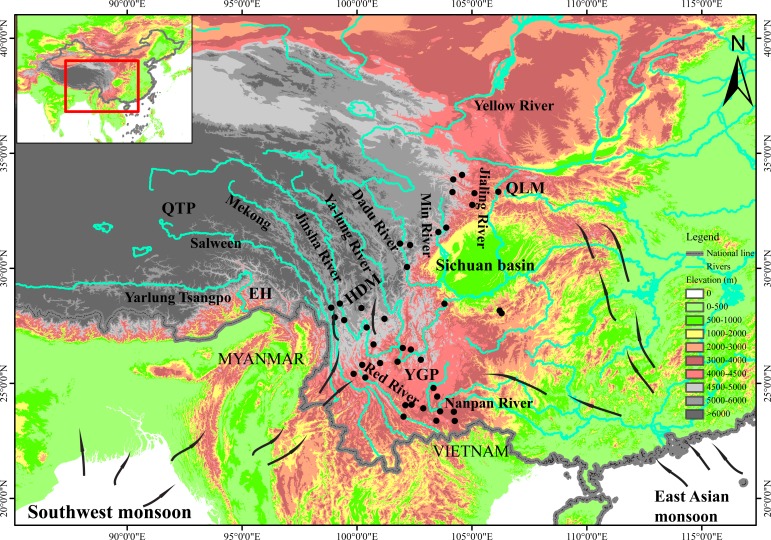
Fig 1. Topography, monsoons, and locations of sampled Osteomeles schwerinae populations in the involved regions.
Hot/warm-dry river valleys are one of the most prominent natural landscape features in the HDM and adjacent regions [15]. In comparison to studies of (sub)alpine plants, however, few riparian plants of hot/warm-dry river valleys in these regions have been studied. The phylogeographic studies of riparian plants conducted so far have focused on two species: Terminalia franchetii (Combretaceae) and Buddleja crispa (Buddlejaceae), both of which indicated that the discontinuous distribution patterns of these plants are strongly linked to historical drainage re-organization events [16, 17]. More phylogeographic studies of riparian plants in the HDM and adjacent regions (particularly the QLM) are required to obtain a better understanding of their recent population history; and in particular, to detect whether some riparian species in these regions have been affected by climatic factors (i.e. heterogeneity and changes) during the Quaternary.
River valleys in different regions probably differ in climate because of different monsoon systems. The QLM, which is located on the north-eastern margin of the HDM (Fig 1), is dominated by the East Asian monsoon, originating from the Pacific Ocean [18, 19]. The east–west orientated mountains form a natural obstacle, preventing the wet summer East Asian monsoon from penetrating northern China, but provide good corridors for it to penetrate into the western QLM or even HDM. In contrast, The HDM is affected by both the southwest monsoon from the Indian Ocean and the East Asian monsoon from the Pacific Ocean, and the YGP is mainly affected by the East Asian monsoon [20]. Similarly, the south–north orientated river valleys in the HDM could also provide convenient corridors for the warm, moist air from the Indian and Pacific Oceans to penetrate into the northern and/or north-eastern HDM [21, 22]. During the Quaternary, the Asian monsoons appear to have fluctuated with the alternating interglacial-glacial periods, with more intense summer monsoons during interglacial periods and weaker monsoons during glaciations [23–25]. However, some strong summer monsoons seem to have also occurred during glacial periods (e.g. the last glaciation) in addition to their anticipated occurrence during interglacial periods. Conversely, some significantly weak summer monsoons have also been found to have occurred during interglacial periods (e.g. the last interglacial period) in addition to their occurrence during glacial periods [26–28]. Given the river valleys’ role as corridors for monsoons, it seems plausible that such climatic heterogeneity and changes due to Asian monsoons would have an impact on the population divergence and demography of riparian plants.
Osteomeles schwerinae Schneid. (Rosaceae) is an iconic riparian plant species endemic to the dry river valleys of the HDM, YGP, and QLM [29–31] (Fig 1), and probably ideal to study such impacts. It is a small tree species, characterized by imparipinnate leaves with 5 to 10 pairs of elliptic or elliptic-oblong leaflet blades, a campanulate hypanthium possessing sepals, and stony pomes [32]. According to the Flora of China [32] and our field observations, it commonly grows on the slopes of dry river valleys. It includes two varieties (O. schwerinae var. schwerinae and O. schwerinae var. microphylla Rehder & E. H. Wilson). The former is mainly confined to the river valleys of the HDM-YGP and has larger leaflet blades (5–10 mm), while the latter is mostly present in the river valleys of the QLM and has smaller leaflet blades (3–5 mm) [32]. Although there is now much molecular evidence, as mentioned above, that the population divergence and demographic history of taxa (e.g. alpine plants) in the HDM and adjacent regions were closely linked with historical climatic events [9, 11, 20], very little climatic data have been explicitly used to examine these inferences. In order to understand better how climatic factors contributed to the present-day distribution and population divergence of plants in the HDM and adjacent regions, we constructed the phylogeography of O. schwerinae based on molecular approaches and paleo-and current climatic data. We had the following objectives: (1) to quantify whether there is genetic divergence between the populations of O. schwerinae; (2) to explore whether the genetic divergence coincide with the regional morphological differentiation of O. schwerinae in the QLM and HDM-YGP regions; (3) to date divergences and determine whether these dates relate to geological events (e.g. historical drainage re-organization) or climatic changes (e.g. the monsoon fluctuation and/or the glacial-interglacial alternation); and (4) to unravel the demographic history of O. schwerinae.
Material and Methods
Ethics Statement
Osteomeles schwerinae is not protected nor endangered in the sampled areas, and all samples were collected by researchers following current Chinese regulations. None of the sampled locations are privately owned or protected by law.
Sample Collection, DNA Extraction, and Sequencing
We sampled 304 individuals from 40 populations of O. schwerinae covering its geographical range; out of these, eight populations originated from the QLM region, and 32 populations from the HDM-YGP region (Table 1). Individuals were randomly sampled and were at least 100 m apart within each population. Fresh green leaves were collected from each individual and dried in silica gel for DNA extraction.
Table 1. Details of sampled populations of Osteomeles schwerinae, haplotypes (num. of individuals) detected per population, and the assignment of each population to the identified regions and drainage basins.
| Code | Regions | Drainages | Latitude (°N) | Longitude (°E) | Altitude (m) | cpDNA haplotypes (num. of individuals) | nDNA haplotypes (num. of individuals) |
|---|---|---|---|---|---|---|---|
| 1 | QLM | Jialing River | 33.87 | 104.18 | 1484 | H1(7) H4(1) | Hap3(2) Hap4(3) Hap5(3) Hap11 (4) Hap13 (4) |
| 2 | QLM | Jialing River | 34.07 | 104.57 | 1595 | H1(8) | Hap2(1) Hap4(4) Hap11(6) Hap13(5) |
| 3 | QLM | Jialing River | 33.27 | 105.11 | 1005 | H1(6) H4(2) | Hap2(1) Hap3(2) Hap4(1) Hap5(3) Hap11(6) Hap13(3) |
| 4 | QLM | Jialing River | 32.76 | 105.01 | 1668 | H1(6) H4(2) | Hap2(2) Hap3(4) Hap5(4) Hap11(6) |
| 5 | QLM | Jialing River | 33.33 | 106.15 | 663 | H1(3) H4(5) | Hap3(4) Hap4(5) Hap13(6) Hap14(1) |
| 6 | QLM | Jialing River | 33.32 | 104.15 | 1480 | H1(8) | Hap1(1) Hap2(2) Hap3(6) Hap4(5) Hap5(2) |
| 7 | QLM | Min River | 31.78 | 103.89 | 1578 | H1(4) H6(3) | Hap2(2) Hap4(1) Hap5(2) Hap11(6) Hap13(5) |
| 8 | QLM | Min River | 31.58 | 103.54 | 1476 | H1(6) H6(2) | Hap2(2) Hap4(4) Hap5(1) Hap11(9) |
| 9 | HDM-YGP | Dadu River | 31.08 | 101.87 | 2018 | H1(1) H4(5) H5(2) | Hap2(6) Hap3(7) Hap5(3) |
| 10 | HDM-YGP | Dadu River | 30.06 | 102.17 | 1410 | H4(8) | Hap5(3) Hap7(2) Hap11(5) Hap12(6) |
| 11 | HDM-YGP | Dadu River | 31.01 | 102.31 | 1860 | H2(2) H4(6) | Hap5(16) |
| 12 | HDM-YGP | Ya-lung River | 27.81 | 101.2 | 2268 | H2(1) H4(6) | Hap9(10) Hap10(4) |
| 13 | HDM-YGP | Jinsha River | 26.54 | 102.08 | 1091 | H2(6) H4(1) | Hap3(6) Hap10(8) |
| 14 | HDM-YGP | Jinsha River | 28.47 | 99.25 | 2125 | H2(8) | Hap3(7) Hap6(9) |
| 15 | HDM-YGP | Jinsha River | 27.75 | 99.44 | 1964 | H2(5) H4(3) | Hap3(10) Hap6(6) |
| 16 | HDM-YGP | Jinsha River | 28.28 | 100.19 | 2328 | H2(8) | Hap3(16) |
| 17 | HDM-YGP | Jinsha River | 27.43 | 100.42 | 2322 | H2(6) H3(2) | Hap6(12) Hap7(4) |
| 18 | HDM-YGP | Jinsha River | 26.7 | 100.72 | 2015 | H2(8) | Hap3(8) Hap10(8) |
| 19 | HDM-YGP | Jinsha River | 25.94 | 101.76 | 1335 | H2(7) H4(1) | Hap10(16) |
| 20 | HDM-YGP | Jinsha River | 25.89 | 101 | 1485 | H2(8) | Hap3(12) |
| 21 | HDM-YGP | Jinsha River | 26.48 | 102.34 | 1756 | H2(6) H4(2) | Hap3(8) Hap10(8) |
| 22 | HDM-YGP | Jinsha River | 26.03 | 102.78 | 1958 | H2(5) | Hap3(10) |
| 23 | HDM-YGP | Jinsha River | 28.46 | 103.8 | 926 | H2(7) | Hap3(7) Hap10(7) |
| 24 | HDM-YGP | Jinsha River | 28.17 | 106.21 | 923 | H4(8) | Hap3(16) |
| 25 | HDM-YGP | Jinsha River | 28.05 | 106.29 | 184 | H4(8) | Hap3(16) |
| 26 | HDM-YGP | Red River | 25.81 | 100.23 | 1977 | H2(5) H4(2) | Hap3(10) Hap8(2) Hap9(2) |
| 27 | HDM-YGP | Red River | 25.42 | 99.85 | 2165 | H2(6) H4(2) | Hap3(11) Hap10(5) |
| 28 | HDM-YGP | Red River | 25.26 | 100.36 | 1359 | H2(8) | Hap3(7) Hap6(6) Hap10(3) |
| 29 | HDM-YGP | Red River | 24.06 | 102.1 | 1194 | H4(8) | Hap9(8) Hap10(8) |
| 30 | HDM-YGP | Red River | 23.56 | 102.02 | 508 | H2(8) | Hap3(16) |
| 31 | HDM-YGP | Red River | 23.92 | 102.88 | 1399 | H2(2) H4(5) | Hap10(14) |
| 32 | HDM-YGP | Red River | 23.38 | 103.45 | 1356 | H2(8) | Hap3(7) Hap10(9) |
| 33 | HDM-YGP | Nanpan River | 24.8 | 103.33 | 1354 | H2(7) H4(1) | Hap3(10) Hap10(6) |
| 34 | HDM-YGP | Nanpan River | 24.43 | 103.49 | 1538 | H2(6) | Hap10(12) |
| 35 | HDM-YGP | Nanpan River | 24.08 | 102.37 | 1609 | H4(7) | Hap3(2) Hap10(14) |
| 36 | HDM-YGP | Nanpan River | 23.79 | 103.61 | 1467 | H2(6) | Hap3(5) Hap10(7) |
| 37 | HDM-YGP | Nanpan River | 23.37 | 104.25 | 1505 | H2(7) | Hap3(6) Hap10(8) |
| 38 | HDM-YGP | Nanpan River | 23.77 | 104.21 | 1565 | H2(7) | Hap10(14) |
| 39 | HDM-YGP | Mekong | 28.3 | 98.88 | 2351 | H4(8) | Hap10(6) Hap15(10) |
| 40 | HDM-YGP | Mekong | 27.87 | 99.03 | 1935 | H4(8) | Hap10(13) Hap15(3) |
Total genomic DNA was extracted from approximately 15 mg dried leaf using an Axygen Plant Mini Kit (www.axygen.com). Universal primers and methods described in previous studies [33, 34] were used to amplify three chloroplast DNA (cpDNA) intergenic spacers (trnD-trnT, psbD-trnT, petL-psbE). Nuclear DNA (nDNA), namely the single copy nuclear gene (glyceraldehyde 3-phosphate dehydrogenase; G3pdh), was amplified following the PCR procedure in Olsen and Schaal (1999) [35], using one self-designed primer combination (forward: 5′-TTCAGCCACCCAGAAGAC-3′, reverse: 5′- CGACAACGGATAACAATAC -3′). All PCR products were purified and sequenced by the Sangon Corporation, Shanghai, China.
Molecular Data Analysis
Haplotypes of the combined three cpDNA sequences were produced in DnaSP v5.0 [36]. Method of Clark (1990) was used to identify the haplotypes for G3pdh heterozygotes. In this method, sequences of heterozygotes were compared with those of homozygotes until we could account for the observed combination of double-banded sites. ArcMap v9.3 (ESRI, Inc.) was used to plot the distribution of haplotypes on a relief map coving the range. Original map in the figures was downloaded from http://www.diva-gis.org/Data. The elevation dataset in the figures was derived from NASA’s SRTM data (http://www2.jpl.nasa.gov/srtm).
Spatial Analysis of Molecular Variance (SAMOVA) was conducted to analyze the spatial genetic structure of O. schwerinae using SAMOVA v1.0 [37]. This program uses a simulated annealing approach to define K groups of populations that are geographically homogenous and maximally differentiated from each other. In this analysis, we assumed that K varied from 2 to 13, and computed a corresponding F CT index, which measures the proportion of genetic differentiation among the K groups [38]. The configuration with the F CT value that achieves a plateau first is usually considered the optimal grouping of populations. An analysis of molecular variance (AMOVA) was conducted within ARLEQUIN v3.5 [39] to partition variation within and among defined groups and populations.
Genealogical relationships among haplotypes were inferred from an unrooted statistical parsimony network within Network v4.6.1.2 [40] (available at http://www.fluxus-engineering.com) using a median-joining method. Indels (gaps) were treated separately as binary states, namely single events, following Caicedo and Schaal (2004) [41]. Mononucleotide repeats and duplicated indels of sequences in this study were removed as they introduced homoplasious reticulations into the network, due to their high levels of bidirectional mutation [42]. Observed and inferred haplotypes were nested following the rules in Posada and Crandall (2001) [43].
Phylogenetic relationships among haplotypes were reconstructed by Bayesian inference. This phylogenetic analysis was conducted in MrBayes v3.1.2 [44] using substitution models (GTR+G) explored by Modeltest v3.7 [45]. Four independent Markov chain Monte Carlo analyses with runs of 10 million generations were carried out, sampling at every 1000 generations. The first 20% of generations was discarded as burn-in, while the remaining data was used to construct a 50% majority rule consensus tree. The posterior probabilities found in this consensus tree were given to evaluate the robustness of the trees.
The divergence time of haplotypes was estimated via a molecular dating method, and estimated under a strict molecular clock in BEAST v1.7.5 [46], using the substitution rate range for angiosperm species (cpDNA: 1.01–3.0×10−9 substitutions per site per year (s/s/y) [47, 48]; nDNA: 1.58–3.15×10−8 s/s/y [48]). Results from BEAST were compiled and visualized in the program Tracer v1.5 [49].
Pairwise mismatch distribution was carried out using ARLEQUIN to infer the demographic history of O. schwerinae. In this analysis, an expected distribution was generated from 1000 parametric bootstrap replicates to test ‘demographic expansion’ and ‘spatial expansion’ model, respectively [2]. Generally, a unimodal mismatch distribution curve indicates a recent population expansion, whereas a multimodal distribution usually suggests that populations are at demographic equilibrium. The populations for which the hypothesis of expansion was not rejected, the value for the mode of the mismatch distribution (Tau) was assessed by 1000 parametric bootstrap replicates in ARLEQUIN, and was converted into estimates of time since expansion (t, in years BP) using t = Tau/2μkg [2]. In this formula, μ is the substitution rate, and was assumed to be based on the substitution rate range for cpDNA mentioned above; k is the average sequence length used for analysis (2039 bp in this study); and g is the generation time in years, and was assumed to be 6 years here based on personal observations of on the age of first reproduction of O. schwerinae in cultivation at Kunming Botanical Garden.
Climatic Data Analysis
Climatic data [50] that summarized temperature and precipitation information (S1 Table), were downloaded from the WorldClim database (http://www.worldclim.org/), and were used to identify climatic factors potentially associated with the genetic structure and demographic history identified in this study. DIVA-GIS v7.5 [51] was used to extract the 19 climatic factors during the LIG, LGM, and Current periods (S2 Table). The mean values and standard errors of these factors for populations of the QLM and HDM-YGP regions were calculated with SPSS v18.0.0 [52], and were further analyzed using independent-Samples t-tests to validate the impact of the climatic factors on the genetic differentiation between the QLM and HDM-YGP regions.
Ecological Niche Modeling (ENM) can simulate past species distributions, and further elucidate demographic history through time [53]. Therefore, ENM was also performed in this study to validate potential present and past climate envelopes for this species. Assuming the species has not changed with respect to its climatic preference, we re-constructed the range of O. schwerinae during the LIG, LGM, and Current periods. Species occurrence data for O. schwerinae were obtained from collection records from the three main herbaria in China (IBSC, KUN and PE; S3 Table) and sampling records collected in this study (Table 1). ENM based on the 19 BioClim factors and species occurrence data was performed using MAXENT v3.3.3k [54] with the default parameters for number of iterations (500) and convergence threshold (10−5). We set up 10 replicate runs in each analysis in order to ensure more reliable results. Meanwhile, in order to test the performance of each model, 25% of the data in each run was randomly chosen by MAXENT and compared with the model output created with the remaining data. To reduce the effects of spatial autocorrelation, duplicate records from the same locality were also removed. The area under the receiver operating characteristic curves (AUC) was used to compare model performance. An AUC value of 0.5 indicates that the performance of the model is no better than random, while values closer to 1.0 indicate better model performance [54].
Results
Chloroplast and Nuclear DNA Sequencing Data
Variations in the length of two sequenced regions of cpDNA were detected (953–958 bp, trnT-trnD; 658–697 bp, psbD-trnT), while the petL-psbE was uniformly 449 bp long in our samples. Removal of mononucleotide repeats and duplicated indels for the alignment of three cpDNA regions from the 304 samples resulted in an overall alignment length of 2039 bp, and produced six cpDNA haplotypes (designated H1-6; S4 Table). The aligned sequences of the G3pdh gene were 550 bp long, and produced 15 nDNA haplotypes (designated Hap1-15) based on 18 nucleotide substitutions (S5 Table). These sequences were deposited in the GenBank database (accession numbers KT724331–KT724335, KT750890–KT750920).
Genetic Divergence and Demography
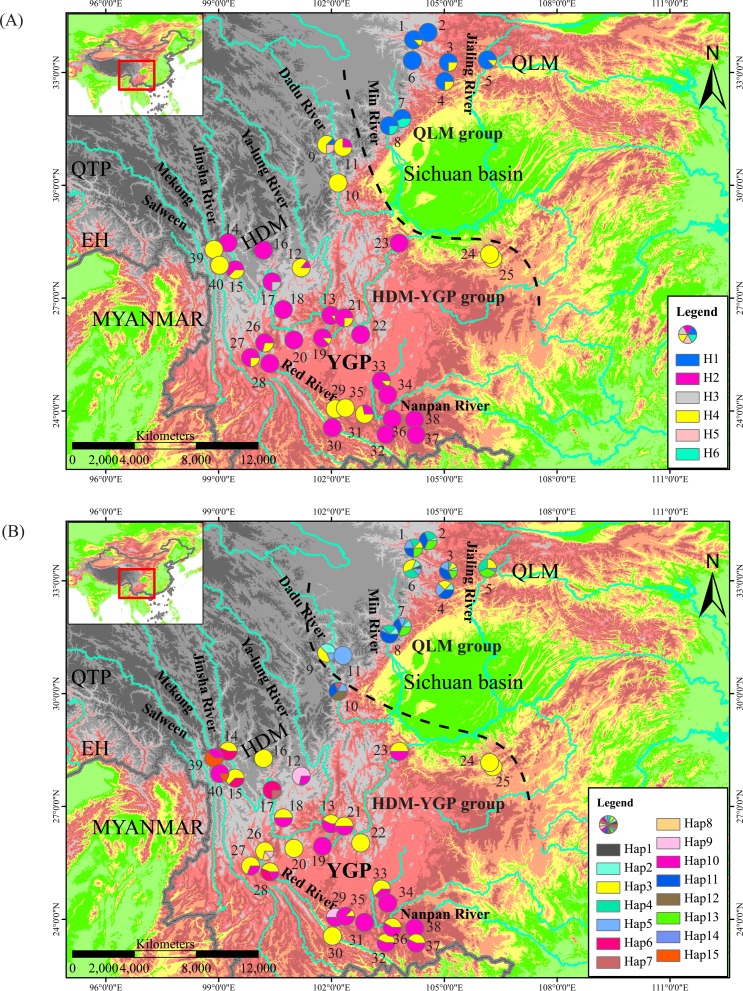
In the SAMOVA analysis based on cpDNA sequences, two groups corresponding to the QLM and HDM-YGP regions were well defined when K = 2. Although the F CT value was not the highest, the F CT values reached a plateau, and fluctuated little with an increasing number of groups (S1 Fig). In addition, when divided into two groups, the AMOVA revealed high genetic variation partitioned between the groups (68.57%), and low variations among populations within groups (18.60%) as well as within populations. As shown in Fig 2A, 8 populations located in the QLM region were contained in one group, whilst the remaining 32 populations from the HDM-YGP were included in the other group. The SAMOVA analysis based on nDNA sequences showed relatively low F CT values, and fluctuated little with the increase of K (S2 Fig). Nonetheless, when divided into two groups, the AMOVA revealed that still 30.55% of the total nDNA variation was due to differences among the two groups identified, comparing with 22.61% among populations and 46.84% within populations. In this scenarios of grouping, 8 populations from the QLM (pops. 1–8) and 2 populations from the HDM-YGP (pops. 9 and 11 of the Dadu River) formed one group, whereas the remaining 30 populations from the HDM-YGP were included in the other group (Fig 2B).
Fig 2. Geographical distributions of (A) six cpDNA haplotypes (H1-6) and (B) 15 nDNA haplotypes (Hap1-15) identified in 40 populations of Osteomeles schwerinae.
Dashed black line delineates the boundary between the two groups derived from SAMOVA.
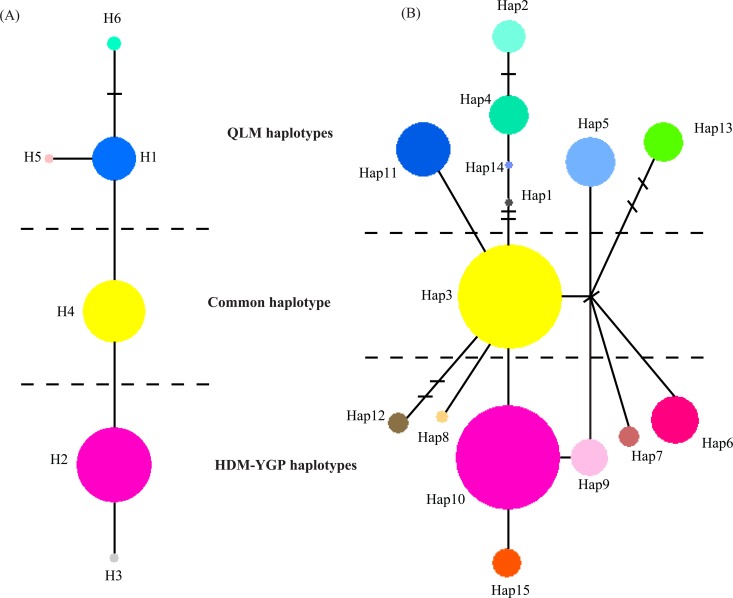
Genealogical analysis of cpDNA haplotypes identified a common haplotype (H4) shared by populations along all rivers surveyed in QLM, HDM, and YGP regions (Fig 3A). Apart from this common haplotype, the remaining haplotypes were distinctly regional, and could be divided into two clusters from the QLM and HDM-YGP regions (Fig 3A). In addition, it is worth noting that the intermediate zone between these two regions (namely the drainage basin of the Dadu River) possess high cpDNA haplotype diversity, containing the common haplotype (H4), the main regional haplotypes from the QLM (H1) and HDM-YGP (H2), as well as its own unique haplotype (H5) (Table 1; Fig 2A). Genealogical analysis of nDNA haplotypes showed similar structure, which also identified a common haplotype (Hap3) and two clusters of regional haplotypes from the QLM and HDM-YGP, respectively (Fig 3B). Simultaneously, the intermediate zone between QLM and HDM-YGP (the Dadu River basin) also showed high nDNA haplotype diversity, possessing the common haplotype (Hap3), regional haplotypes both from the QLM (Hap2, 5, 11) and HDM-YGP (Hap7), as well as its own unique haplotype (Hap12; Table 1; Fig 2B).
Fig 3. Median-joining networks of (A) six cpDNA haplotypes (H1-6) and (B) 15 nDNA haplotypes (Hap1-15) identified in this study.
The size of circles corresponds to the frequency of each haplotype. Short lines represent haplotypes missing in the dataset and each branch represents one mutation.
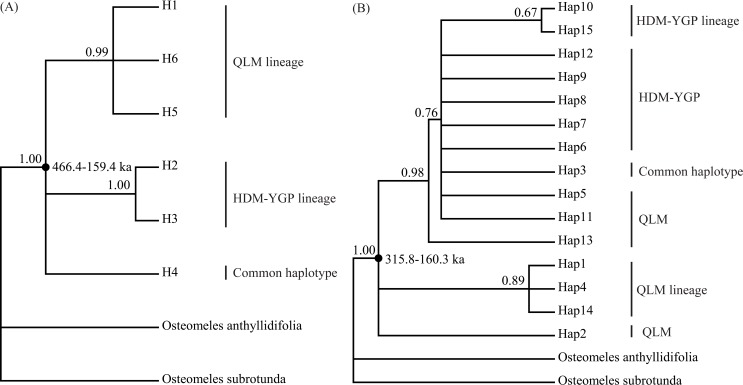
The Bayesian phylogenetic tree clustered five of the cpDNA haplotypes (all except H4) into two lineages, respectively from the QLM and HDM-YGP regions (Fig 4A). In the phylogenetic tree based on nDNA haplotypes, one haplotype (Hap15) and the dominated haplotype (Hap10) in HDM-YGP region fell in one lineage, while three haplotypes (Hap 1, 4, 14) from QLM in another lineage (Fig 4B). In addition, this tree showed several polytomies, however, it is not surprising since the nuclear gene is two parents–inherited. Based on molecular dating, the divergence times of the cpDNA and nDNA haplotypes were estimated to be 466.4–159.4 ka and 315.8–160.3 ka, respectively.
Fig 4. Bayesian phylogenetic trees of (A) six cpDNA haplotypes (H1-6) and (B) 15 nDNA haplotypes (Hap1-15) identified in this study.
Numbers above the branch indicate posterior probabilities. Numbers next to the nodes indicate the divergence times.
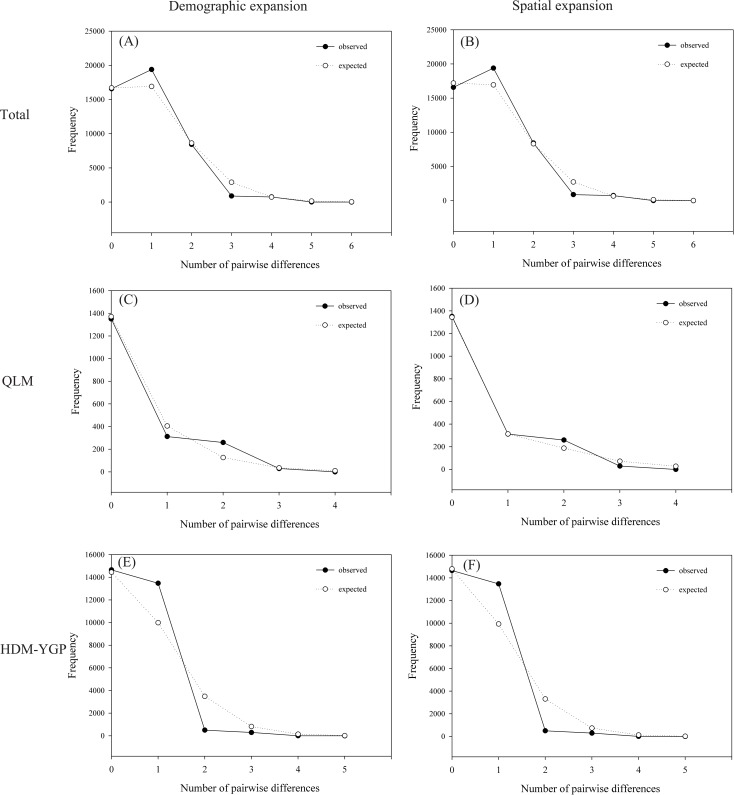
The mismatch distribution curves for the total, QLM and HDM-YGP populations were clearly unimodal (Fig 5). Therefore, the hypothesis of population expansion could not be rejected. Based on the Tau value, the demographic and spatial expansion times for the total, QLM and HDM-YGP populations fell within the last glacial period (Table 2).
Fig 5. Mismatch distribution curves of the total (A, B) populations, QLM (C, D) populations, and HDM-YGP (E, F) populations.
Left: demographic expansion; right: spatial expansion.
Table 2. Results of the expansion time for the Osteomeles schwerinae populations.
| Populations | Expansion types | Tau | Time since expansion began (ka) |
|---|---|---|---|
| Total | Demographic expansion | 0.98438 | 13.41–39.83 |
| Spatial expansion | 0.98510 | 13.42–39.86 | |
| QLM region | Demographic expansion | 3.00000 | 40.87–121.39 |
| Spatial expansion | 1.20902 | 16.47–48.93 | |
| HDM-YGP region | Demographic expansion | 0.66992 | 9.13–27.11 |
| Spatial expansion | 0.65968 | 8.98–26.69 |
Climatic Data
Out of the 19 bioclimatic factors, all but five (bio2, 5, 8, 10, 15) during the LGM and the current periods showed highly significant differences in mean values between populations of the QLM and HDM-YGP regions (Table 3). When comparing with the LGM and Current periods, two more bioclimatic factors (bio8 and 15) during the LIG exhibited highly significant differences in mean values between the two regions (Table 3). As shown in Table 4, there was reduction in temperature during the LGM. For instance, the annual average temperature (bio1) in the QLM and HDM-YGP regions decreased 2.3 and 2.6°C during the LGM than the LIG, respectively. In contrast, the majority of the bioclimatic factors relating to the precipitation (6/8 = 75%) for both the QLM and HDM-YGP during the LGM period were higher than during the LIG, with most variables (5/6 = 83.33%) being significantly higher (Table 4). It suggested a more moist environment in the QLM and HDM-YGP regions during the LGM, whereas a more drought environment in the LIG.
Table 3. Means, standard errors (SE) and results of independent-Sample t-test on the 19 BioClim factors of the LIG, LGM, and Current periods for the QLM and HDM-YGP regions surveyed in this study.
| Independent-Sample t-test | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (°C) | (°C) | (°C) | (SD*100) | (°C) | (°C) | (°C) | (°C) | (°C) | (°C) | (°C) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | ||
| LIG | ||||||||||||||||||||
| QLM | Mean | 10.4 | 10.5 | 2.7 | 985.1 | 29.8 | -8.4 | 38.3 | 14.3 | 1.0 | 22.8 | -2.8 | 289.4 | 162.3 | 0.0 | 198.9 | 231.0 | 0.0 | 40.3 | 0.0 |
| SE | 1.3 | 0.5 | 0.2 | 36.9 | 1.7 | 1.3 | 0.8 | 1.0 | 0.9 | 1.7 | 0.9 | 46.7 | 12.9 | 0.0 | 13.4 | 26.7 | 0.0 | 23.0 | 0.0 | |
| HDM-YGP | Mean | 14.3 | 11.3 | 3.6 | 652.5 | 29.2 | -1.6 | 30.8 | 20.7 | 8.5 | 21.6 | 5.5 | 894.8 | 247.9 | 0.0 | 115.7 | 620.3 | 5.6 | 440.7 | 18.3 |
| SE | 0.5 | 0.3 | 0.1 | 15.8 | 0.5 | 0.7 | 0.3 | 0.5 | 0.9 | 0.5 | 0.6 | 27.2 | 7.2 | 0.0 | 2.6 | 19.2 | 1.2 | 25.0 | 3.4 | |
| P | 0.002 | 0.167 | 0.000 | 0.000 | 0.644 | 0.000 | 0.000 | 0.000 | 0.000 | 0.535 | 0.000 | 0.000 | 0.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| LGM | ||||||||||||||||||||
| QLM | Mean | 8.1 | 11.9 | 3.7 | 695.7 | 23.5 | -8.9 | 32.3 | 16.0 | -1.4 | 16.7 | -1.4 | 541.1 | 103.6 | 2.5 | 80.9 | 282.4 | 10.5 | 273.1 | 10.5 |
| SE | 1.1 | 0.6 | 0.1 | 22.1 | 1.3 | 1.3 | 0.8 | 1.2 | 0.9 | 1.4 | 0.9 | 46.8 | 8.8 | 0.5 | 1.3 | 23.7 | 1.8 | 24.5 | 1.8 | |
| HDM-YGP | Mean | 11.7 | 11.2 | 4.5 | 486.6 | 22.6 | -2.3 | 24.9 | 17.1 | 5.1 | 17.3 | 4.9 | 1098.5 | 229.5 | 10.6 | 85.1 | 618.8 | 40.1 | 608.2 | 40.7 |
| SE | 0.6 | 0.3 | 0.1 | 14.9 | 0.5 | 0.8 | 0.5 | 0.5 | 0.8 | 0.5 | 0.8 | 41.6 | 9.3 | 1.1 | 1.7 | 25.4 | 3.5 | 26.9 | 3.4 | |
| P | 0.012 | 0.311 | 0.001 | 0.000 | 0.478 | 0.000 | 0.000 | 0.358 | 0.000 | 0.648 | 0.000 | 0.000 | 0.000 | 0.000 | 0.226 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Current | ||||||||||||||||||||
| QLM | Mean | 11.9 | 10.8 | 3.4 | 714.7 | 27.0 | -4.7 | 31.7 | 19.9 | 2.2 | 20.8 | 2.2 | 696.6 | 135.8 | 3.5 | 81.5 | 368.0 | 14.8 | 356.8 | 14.8 |
| SE | 1.3 | 0.4 | 0.1 | 24.8 | 1.5 | 1.4 | 0.5 | 1.4 | 1.1 | 1.6 | 1.1 | 55.4 | 10.0 | 0.6 | 1.9 | 27.5 | 2.6 | 28.8 | 2.6 | |
| HDM-YGP | Mean | 16.3 | 10.9 | 4.4 | 488.3 | 27.0 | 2.5 | 24.6 | 21.7 | 9.6 | 21.8 | 9.5 | 945.8 | 195.9 | 9.3 | 84.3 | 530.1 | 35.3 | 516.3 | 36.2 |
| SE | 0.5 | 0.3 | 0.1 | 15.2 | 0.5 | 0.6 | 0.4 | 0.4 | 0.6 | 0.4 | 0.6 | 19.4 | 4.7 | 0.8 | 1.7 | 12.6 | 2.7 | 15.3 | 2.7 | |
| P | 0.001 | 0.879 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.255 | 0.000 | 0.52 | 0.000 | 0.000 | 0.000 | 0.000 | 0.424 | 0.000 | 0.000 | 0.000 | 0.000 | |
Significantly different: P<0.05; significant P values were given in bold print.
-, cannot be compared.
Table 4. Means, standard errors (SE) and results of independent-Sample t-test on the 19 BioClim factors of the two regions (the QLM, HDM-YGP) for the LIG and LGM periods.
| Independent-Sample t-test | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (°C) | (°C) | (°C) | (SD*100) | (°C) | (°C) | (°C) | (°C) | (°C) | (°C) | (°C) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | ||
| QLM | ||||||||||||||||||||
| LIG | Mean | 10.4 | 10.5 | 2.7 | 985.1 | 29.8 | -8.4 | 38.3 | 14.3 | 1.0 | 22.8 | -2.8 | 289.4 | 162.3 | 0.0 | 198.9 | 231.0 | 0.0 | 40.3 | 0.0 |
| SE | 1.3 | 0.5 | 0.2 | 36.9 | 1.7 | 1.3 | 0.8 | 1.0 | 0.9 | 1.7 | 0.9 | 46.7 | 12.9 | 0.0 | 13.4 | 26.7 | 0.0 | 23.0 | 0.0 | |
| LGM | Mean | 8.1 | 11.9 | 3.7 | 695.7 | 23.5 | -8.9 | 32.3 | 16.0 | -1.4 | 16.7 | -1.4 | 541.1 | 103.6 | 2.5 | 80.9 | 282.4 | 10.5 | 273.1 | 10.5 |
| SE | 1.1 | 0.6 | 0.1 | 22.1 | 1.3 | 1.3 | 0.8 | 1.2 | 0.9 | 1.4 | 0.9 | 46.8 | 8.8 | 0.5 | 1.3 | 23.7 | 1.8 | 24.5 | 1.8 | |
| P | 0.186 | 0.087 | 0.001 | 0.000 | 0.010 | 0.826 | 0.000 | 0.302 | 0.079 | 0.015 | 0.332 | 0.002 | 0.002 | 0.001 | 0.000 | 0.172 | 0.001 | 0.000 | 0.001 | |
| HDM-YGP | ||||||||||||||||||||
| LIG | Mean | 14.3 | 11.3 | 3.6 | 652.5 | 29.2 | -1.6 | 30.8 | 20.7 | 8.5 | 21.6 | 5.5 | 894.8 | 247.9 | 0.0 | 115.7 | 620.3 | 5.6 | 440.7 | 18.3 |
| SE | 0.5 | 0.3 | 0.1 | 15.8 | 0.5 | 0.7 | 0.3 | 0.5 | 0.9 | 0.5 | 0.6 | 27.2 | 7.2 | 0.0 | 2.6 | 19.2 | 1.2 | 25.0 | 3.4 | |
| LGM | Mean | 11.7 | 11.2 | 4.5 | 486.6 | 22.6 | -2.3 | 24.9 | 17.1 | 5.1 | 17.3 | 4.9 | 1098.5 | 229.5 | 10.6 | 85.1 | 618.8 | 40.1 | 608.2 | 40.7 |
| SE | 0.6 | 0.3 | 0.1 | 14.9 | 0.5 | 0.8 | 0.5 | 0.5 | 0.8 | 0.5 | 0.8 | 41.6 | 9.3 | 1.1 | 1.7 | 25.4 | 3.5 | 26.9 | 3.4 | |
| P | 0.002 | 0.889 | 0.000 | 0.000 | 0.000 | 0.508 | 0.000 | 0.000 | 0.006 | 0.000 | 0.539 | 0.000 | 0.122 | 0.000 | 0.000 | 0.963 | 0.000 | 0.000 | 0.000 | |
Significantly different: P<0.05; significant P values were given in bold print.
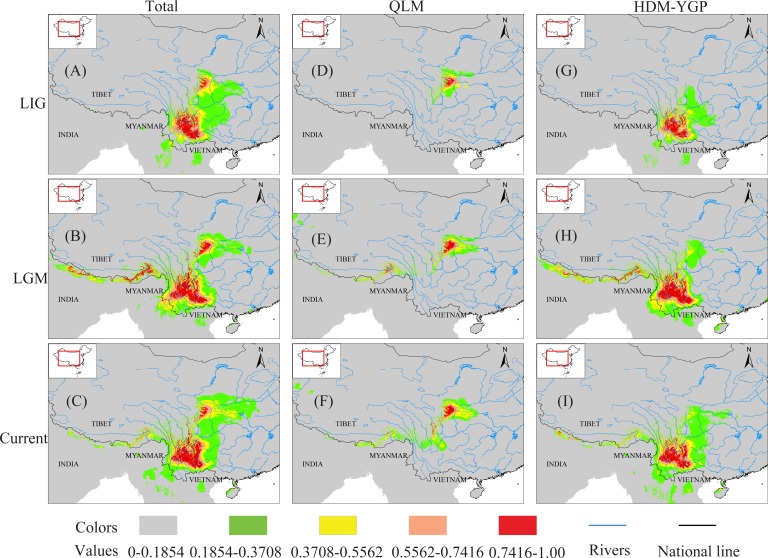
The AUCs of the ENM for the total, QLM and HDM-YGP populations under all climate scenarios were ≥ 0.987, indicating quite better model performance of ENM. The current distribution predictions in this study generally represented the actual distributions of O. schwerinae. Paleo-distribution modeling for the total, QLM and HDM-YGP populations, all indicated a much more restricted distribution during the LIG than the Current, with subsequent expansion during the LGM. This predicted subsequent expansion covered a slightly greater range than that predicted based on the current climatic factors (Fig 6).
Fig 6. Ecological Niche Modeling (ENM) using bioclimatic factors of the three periods (LIG; LGM; Current) based on the total, QLM, and HDM-YGP populations.
Colors with higher values show areas with more suitable predicted conditions. ENM for the total, QLM, and HDM-YGP populations indicated a much wider distribution during the LGM.
Discussion
Genetic and Morphological Divergence Relative to Climatic Heterogeneity and Changes
In this study, the SAMOVA, median-joining networks, and Bayesian phylogenetic trees, all suggested population genetic divergence between the QLM and HDM-YGP regions (Figs 2, 3 and 4), which coincide with the morphological divergence of the two varieties of O. schwerinae in the QLM and HDM-YGP regions. Climatically, the QLM has been primarily dominated by the East Asian monsoon, and is situated near the northern limit of this monsoon system [19] (Fig 1). However, the HDM-YGP has been affected both by the East Asian monsoon from the Pacific and the southwest monsoon from the Indian Ocean [20] (Fig 1). In addition, the HDM-YGP has experienced more intense monsoons than the QLM owing to its more southerly latitude. Therefore, the climate in the HDM-YGP is generally more warm and moist than that in the QLM. Our climate data further confirmed this, demonstrating that the temperature and precipitation values in the QLM were significantly lower than that in the HDM-YGP during the LIG, LGM, and Current periods (Table 3). Considering the two varieties of O. schwerinae, the leaflet blades of the one growing in the QLM (3–5 mm) are smaller than the one (5–10 mm) growing in the HDM-YGP. Since the sizes and shapes of leaves are quite sensitive to moisture, with leaves in dry climates generally smaller than in wet climates [55], it is quite likely that the difference in morphology of O. schwerinae between the two regions is the result of long-term adaptation to the significant climatic heterogeneity between the two regions.
It has been speculated that drainage re-organization was a main cause of genetic divergence for riparian plants in the HDM and adjacent regions [16, 17], but we did not detect its effects on O. schwerinae. When using the same substitution rate range for cpDNA (1.01–3.0×10−9 s/s/y) as previous studies on riparian plants [16, 17] in these regions, the genetic divergence time in this study was estimated to be 466.4–159.4 ka (Fig 4A). In addition, the divergence time based on nDNA was calculated to be 315.8–160.3 ka (Fig 4B). They are much more recent than the timings found in previous riparian plants’ studies (4.24–0.4 Ma, Terminalia franchetii [17]; 3.683–1.228 Ma, Buddleja crispa [16]) and the geological drainage re-organization events (no later than the later Pliocene; 5.332–2.588 Ma) [56–58]. However, our divergence times largely coincide with the time of the weakening of the monsoons in these regions. According to paleo-climatic analysis, the southwest monsoon was sharply weakened from 342–118 ka [59]. Coincidentally, the East Asian monsoon was also found to decrease during the period 200–130 ka [23, 27, 60]. Because of this, weaker airflows associated with monsoons would reach the HDM-YGP as well as the QLM, which probably led to a decrease in precipitation in both regions. On one hand, reduced precipitation from 342-118/200-130 ka would cause the contraction of the O. schwerinae populations in both regions (e.g. Fig 6A, 6D and 6G), and result in the loss/reduction of gene flow between them. On the other hand, the weakening of monsoons probably further increased the climatic differences between the two regions. For instance, the LIG (c. 140–120 ka) falls in the aforementioned time range of the weakening of monsoons (342-118/200-130 ka), and, as expected, displayed two more significantly different bioclimatic factors between the QLM and HDM-YGP than the LGM and the Current periods (Table 3). Finally, these two reasons together led to the genetic divergence between the QLM and HDM-YGP regions.
Demographic History of Osteomeles schwerinae
Usually, the demographic scenarios reported for species from the HDM and adjacent regions suggest distribution contraction during glacial periods, and expansions during interglacial periods [8–11]. However, our results revealed an unusual demographic history of O. schwerinae populations: the opposite pattern to most demographic scenarios in the HDM and adjacent regions. Before the period “342-118/200-130 ka” (including the LIG; c. 140–120 ka), the O. schwerinae populations probably had a more continuous or wider distribution linking the two geographic regions because of stronger monsoons/more precipitation in both regions. However, during the LIG, the O. schwerinae populations probably had a less continuous or more narrow distribution because of weaker monsoons/less precipitation. The ENM analysis further confirmed this, showing more narrow distribution during the LIG, but larger distribution ranges during the LGM period (Fig 6), which implied the “contraction during interglacial periods, and expansions during glacial periods” demographic pattern of O. schwerinae populations. Unimodal pairwise mismatch distribution curves of O. schwerinae (Fig 5) were identified in this study, which fit the sudden expansion model particularly well. Moreover, the expansion times for the total, QLM and HDM-YGP populations of O. schwerinae fell into the last glacial period (Table 2), which further supported this pattern.
This pattern is similar to the “phalanx” model [61], which has only been sporadically found in a few cold-adapted species at high-elevation in (sub)tropical areas [62–65]. These plants were found to expand to lower elevations during glacial periods, allowing isolated patches to join together, because of the retreat of the (sub)tropical forests occupying lower altitudes. In contrast, these plants would be forced back to ‘refugia’ at higher elevations during interglacial periods as a result of the re-expansion of the (sub)tropical forests at low elevations [66]. In the HDM and adjacent regions, the majority of plants have been found to retreat to refugia because of cooling during the glacial periods, which probably left a large space for O. schwerinae populations to expand during the last glacial period. Meanwhile, because of the Foehn effect [67, 68], the climate in river valleys is warmer than in neighboring areas [15]. Therefore, the cooling during the last glacial period might have had a limited impact on the populations of O. schwerinae. For instance, the annual global-mean continental cooling during the LGM has been found to have been 4.3–9.8°C [69, 70]. In contrast, the climatic data for our sampling sites reveals a lower annual average temperature reduction during the LGM (only 2.3 and 2.6°C lower than the LIG for the QLM and HDM-YGP, respectively; Table 4), which probably reflects the warm climate in river valleys. In addition, owing to the intensification of the Asian monsoons affecting precipitation during the last glacial period in these regions [23, 27, 28, 59], the precipitation at our sampling sites was significantly increased during the LGM compared to the LIG (Table 4). This may have relieved or even eliminated the impact of the drought since 342-118/200-130 ka [23, 27, 59, 60].
In this context, the river valleys could have provided warm and moist habits for O. schwerinae to expand during the last glacial period, resulting in a second contact between the populations from the QLM and HDM-YGP. This second contact probably just explains the high cpDNA and nDNA haplotype diversity within their intersecting distributions in the Dadu River basin, where there is the common haplotype (cpDNA: H4; nDNA: Hap3), regional haplotypes both from the QLM (cpDNA: H1; nDNA: Hap2, 5, 11) and HDM-YGP (cpDNA: H2; nDNA: Hap7), as well as its own exclusive haplotypes (cpDNA: H5; nDNA: Hap12) (Table 1; Fig 2A and 2B).
Conclusions
Our analyses of O. schwerinae, based on molecular phylogeography and climatic data, reveal a major population divergence between the QLM and HDM-YGP regions, and its correlation to the climatic heterogeneity and changes within both regions since the late Pleistocene. We inferred that weakening of Asian monsoons since 342-118/200-130 ka increased the climatic differences between the two regions, caused the loss/reduction of gene flow, and triggered the genetic divergence between them. Subsequent intensification of the monsoons during the last glaciation facilitated the expansion of O. schwerinae populations, contributed to the second contact between the populations from the QLM and HDM-YGP, and finally led to the development of the current distribution pattern of O. schwerinae populations. This study provides a new example in which riparian plant species in the HDM and adjacent regions are shown to be affected by climatic heterogeneity and changes since the late Pleistocene. However, more studies of riparian plants in these regions are needed to explore this hypothesis.
Supporting Information
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors thank Jinlong Dong, Drs. Liangliang Yue and Xinxin Zhu, Profs. Shih-Wen Chung from Taiwan, and Shinobu Akiyama from Japan for helping with the field survey and leaf collection. We also thank Minshu Song, and Drs. Guodong Li, Deli Peng, and Dong Luo for assistance with the molecular techniques and data analysis. In addition, we are grateful to Sees-editing Ltd for English editing.
Data Availability
All relevant data are within the paper and its Supporting Information files except the DNA sequences. They can be available from the GenBank database (accession numbers KT724331–KT724335, KT750890–KT750920).
Funding Statement
The study was supported by the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB03030112 to HS), NSFC-Yunnan Natural Science Foundation Combining Program (U1136601 to HS), the Hundred Talents Program of the Chinese Academy of Sciences (2011312D11022 to HS) and the CAS/SAFEA International Partnership Program for Creative Teams. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Avise JC. Phylogeography: the history and formation of species Cambridge: Harvard university press; 2000. [Google Scholar]
- 2. Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9(3):552–569. [DOI] [PubMed] [Google Scholar]
- 3. Schaal B, Hayworth D, Olsen K, Rauscher J, Smith W. Phylogeographic studies in plants: problems and prospects. Molecular Ecology. 1998;7(4):465–474. [Google Scholar]
- 4. Wang IJ, Glor RE, Losos JB. Quantifying the roles of ecology and geography in spatial genetic divergence. Ecology Letters. 2013;16(2):175–182. 10.1111/ele.12025 [DOI] [PubMed] [Google Scholar]
- 5. Andrew RL, Wallis IR, Harwood CE, Foley WJ. Genetic and environmental contributions to variation and population divergence in a broad-spectrum foliar defence of Eucalyptus tricarpa . Annals of Botany. 2010;105(5):707–717. 10.1093/aob/mcq034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist. 2013;199(4):1093–1108. 10.1111/nph.12336 [DOI] [PubMed] [Google Scholar]
- 8. Wang F-Y, Gong X, Hu C-M, Hao G. Phylogeography of an alpine species Primula secundiflora inferred from the chloroplast DNA sequence variation. Journal of Systematics and Evolution. 2008;46(1):13–22. [Google Scholar]
- 9. Yang FS, Li YF, Ding X, Wang XQ. Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Molecular Ecology. 2008;17(23):5135–5145. 10.1111/j.1365-294X.2008.03976.x [DOI] [PubMed] [Google Scholar]
- 10. Cun Y-Z, Wang X-Q. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Molecular Phylogenetics and Evolution. 2010;56(3):972–982. 10.1016/j.ympev.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 11. Chen S, Wu G, Zhang D, Gao Q, Duan Y, Zhang F, et al. Potential refugium on the Qinghai–Tibet Plateau revealed by the chloroplast DNA phylogeography of the alpine species Metagentiana striata (Gentianaceae). Botanical Journal of the Linnean Society. 2008;157(1):125–140. [Google Scholar]
- 12. Wang L, Abbott RJ, Zheng W, Chen P, Wang Y, Liu J. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Molecular Ecology. 2009;18(4):709–721. 10.1111/j.1365-294X.2008.04055.x [DOI] [PubMed] [Google Scholar]
- 13. Li Y, Zhai S-N, Qiu Y-X, Guo Y-P, Ge X-J, Comes HP. Glacial survival east and west of the ‘Mekong–Salween Divide’ in the Himalaya–Hengduan Mountains region as revealed by AFLPs and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Molecular Phylogenetics and Evolution. 2011;59(2):412–424. 10.1016/j.ympev.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 14. Opgenoorth L, Vendramin GG, Mao K, Miehe G, Miehe S, Liepelt S, et al. Tree endurance on the Tibetan Plateau marks the world’s highest known tree line of the Last Glacial Maximum. New Phytologist. 2010;185(1):332–342. 10.1111/j.1469-8137.2009.03007.x [DOI] [PubMed] [Google Scholar]
- 15. Tang Y, Xie J, Sun H. Revisiting sustainable development of dry valleys in Hengduan Mountains Region. Journal of Mountain Science. 2004;1(1):38–45. [Google Scholar]
- 16. Yue L-L, Chen G, Sun W-B, Sun H. Phylogeography of Buddleja crispa (Buddlejaceae) and its correlation with drainage system evolution in southwestern China. American Journal of Botany. 2012;99(10):1726–1735. 10.3732/ajb.1100506 [DOI] [PubMed] [Google Scholar]
- 17. Zhang T-C, Comes HP, Sun H. Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Molecular Phylogenetics and Evolution. 2011;60(1):1–12. 10.1016/j.ympev.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 18. Yang F, Wang N, Shi F, Ljungqvist FC, Wang S, Fan Z, et al. Multi-proxy temperature reconstruction from the West Qinling Mountains, China, for the past 500 years. PloS One. 2013;8(2):e57638 10.1371/journal.pone.0057638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garfin GM, Hughes MK, Yu L, Burns JM, Touchan R, Leavitt SW, et al. Exploratory temperature and precipitation reconstructions from the Qinling Mountains, north-central China. Tree-Ring Research. 2005;61(2):59–72. [Google Scholar]
- 20. Fan DM, Yue JP, Nie ZL, Li ZM, Comes HP, Sun H. Phylogeography of Sophora davidii (Leguminosae) across the ‘Tanaka‐Kaiyong Line’, an important phytogeographic boundary in Southwest China. Molecular Ecology. 2013;22(16):4270–4288. 10.1111/mec.12388 [DOI] [PubMed] [Google Scholar]
- 21. Wang R, Wang JF, Qiu ZJ, Meng B, Wan FH, Wang YZ. Multiple mechanisms underlie rapid expansion of an invasive alien plant. New Phytologist. 2011;191(3):828–839. 10.1111/j.1469-8137.2011.03720.x [DOI] [PubMed] [Google Scholar]
- 22. Wan YX, Zhang WC, Xiao ZN. The variations of atmospheric precipitable water over Longitudinal Range-Gorge Region (LRGR) in southwestern China. Journal of Natural Resources. 2008;23(4):657–664. [Google Scholar]
- 23. Hao Q, Guo Z. Spatial variations of magnetic susceptibility of Chinese loess for the last 600 kyr: implications for monsoon evolution. Journal of Geophysical Research. 2005; 110:B12101. [Google Scholar]
- 24. An Z, Liu T, Lu Y, Porter S, Kukla G, Wu X, et al. The long-term paleomonsoon variation recorded by the loess-paleosol sequence in central China. Quaternary International. 1990;7:91–95. [Google Scholar]
- 25. Ding Z, Liu T, Rutter NW, Yu Z, Guo Z, Zhu R. Ice-volume forcing of East Asian winter monsoon variations in the past 800,000 years. Quaternary Research. 1995;44(2):149–159. [Google Scholar]
- 26. Ao H, Dekkers MJ, Qin L, Xiao G. An updated astronomical timescale for the Plio-Pleistocene deposits from South China Sea and new insights into Asian monsoon evolution. Quaternary Science Reviews. 2011;30(13–14):1560–1575. [Google Scholar]
- 27. Wang Y, Cheng H, Edwards RL, Kong X, Shao X, Chen S, et al. Millennial- and orbital-scale changes in the East Asian monsoon over the past 224,000 years. Nature. 2008;451(7182):1090–1093. 10.1038/nature06692 [DOI] [PubMed] [Google Scholar]
- 28. Ao H, Dekkers MJ, Xiao G, Yang X, Qin L, Liu X, et al. Different orbital rhythms in the Asian summer monsoon records from North and South China during the Pleistocene. Global and Planetary Change. 2012;80:51–60. [Google Scholar]
- 29. Shui Y. Seed plants of Honghe region in SE Yunnan, China Kunming: Yunnan Science and Technology Press; 2003. [Google Scholar]
- 30. Jin Z. Study on the floristic elements of seed plant in the dry-warm valleys of Yunnan and Sichuan. Guihaia. 1998;18(4):313–321. [Google Scholar]
- 31. Jin Z, Ou X, Ou P, Chen J. A preliminary study on the floristic characteristics of seed plants in the dry-hot river valley of Jinshajiang. Acta Botanica Yunnanica. 1993;16(1):1–16. [Google Scholar]
- 32. Wu ZY, Raven PH. Flora of China Gu CZ, Spongberg SA, editors. Beijing; St. Louis: Science Press; Missouri Botanical Garden Press; 2003. [Google Scholar]
- 33. Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92(1):142–166. 10.3732/ajb.92.1.142 [DOI] [PubMed] [Google Scholar]
- 34. Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94(3):275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- 35. Olsen KM, Schaal BA. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proceedings of the National Academy of Sciences. 1999;96:5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 37. Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11(12):2571–2581. [DOI] [PubMed] [Google Scholar]
- 38. Wright S. Variability within and among natural populations Chicago: University of Chicago Press; 1978. [Google Scholar]
- 39. Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 40. Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16(1):37–48. [DOI] [PubMed] [Google Scholar]
- 41. Caicedo AL, Schaal BA. Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Molecular Ecology. 2004;13(7):1871–1882. [DOI] [PubMed] [Google Scholar]
- 42. Xia K, Su T, Liu Y-SC, Xing Y-W, Jacques FM, Zhou Z-K. Quantitative climate reconstructions of the late Miocene Xiaolongtan megaflora from Yunnan, southwest China. Palaeogeography, Palaeoclimatology, Palaeoecology. 2009;276(1):80–86. [Google Scholar]
- 43. Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology & Evolution. 2001;16(1):37–45. [DOI] [PubMed] [Google Scholar]
- 44. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- 45. Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. [DOI] [PubMed] [Google Scholar]
- 46. Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293(5538):2242–2245. [DOI] [PubMed] [Google Scholar]
- 48. Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences. 1987;84(24):9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A, Drummond A. Tracer version 1.5. 0. 2009. Available: http://wwwbeastbioedacuk/software/Tracer/.
- 50. Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International journal of climatology. 2005;25(15):1965–1978. [Google Scholar]
- 51. Hijmans R, Guarino L, Cruz M, Rojas E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genetic Resources Newsletter. 2001:15–19. [Google Scholar]
- 52.Statistics P. Statistical program for the social sciences (Version 18.0. 0). SPSS, Chicago, IL, USA. 2009.
- 53. Wang YH, Jiang WM, Comes HP, Hu FS, Qiu YX, Fu CX. Molecular phylogeography and ecological niche modelling of a widespread herbaceous climber, Tetrastigma hemsleyanum (Vitaceae): insights into Plio–Pleistocene range dynamics of evergreen forest in subtropical China. New Phytologist. 2015;206(2):852–867. 10.1111/nph.13261 [DOI] [PubMed] [Google Scholar]
- 54. Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190(3):231–259. [Google Scholar]
- 55. Peppe DJ, Royer DL, Cariglino B, Oliver SY, Newman S, Leight E, et al. Sensitivity of leaf size and shape to climate: global patterns and paleoclimatic applications. New Phytologist. 2011;190(3):724–739. 10.1111/j.1469-8137.2010.03615.x [DOI] [PubMed] [Google Scholar]
- 56. Schoenbohm LM, Burchfiel BC, Liangzhong C, Jiyun Y. Miocene to present activity along the Red River fault, China, in the context of continental extrusion, upper-crustal rotation, and lower-crustal flow. Geological Society of America Bulletin. 2006;118(5–6):672–688. [Google Scholar]
- 57. Clark MK, House M, Royden L, Whipple K, Burchfiel B, Zhang X, et al. Late Cenozoic uplift of southeastern Tibet. Geology. 2005;33(6):525–528. [Google Scholar]
- 58. Clift PD, Blusztajn J, Nguyen AD. Large-scale drainage capture and surface uplift in eastern Tibet–SW China before 24 Ma inferred from sediments of the Hanoi Basin, Vietnam. Geophysical Research Letters. 2006;33(19): L19403. [Google Scholar]
- 59. Xiao X, Shen J, Wang S, Xiao H, Tong G. Climate change and evolution of the southwest monsoon revealed by pollen records in the Heqing deep drilling core. Acta Palaeontologica Sinica. 2009;48:185–193. [Google Scholar]
- 60. Boulay S, Colin C, Trentesaux A, Frank N, Liu Z. Sediment sources and East Asian monsoon intensity over the last 450 ky. Mineralogical and geochemical investigations on South China Sea sediments. Palaeogeography, Palaeoclimatology, Palaeoecology. 2005;228(3):260–277. [Google Scholar]
- 61. Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58(3):247–276. [Google Scholar]
- 62. Hewitt G. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2004;359(1442):183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jaramillo-Correa JP, Aguirre-Planter E, Khasa DP, Eguiarte LE, Pinero D, Furnier GR, et al. Ancestry and divergence of subtropical montane forest isolates: molecular biogeography of the genus Abies (Pinaceae) in southern México and Guatemala. Molecular Ecology. 2008;17(10):2476–2490. 10.1111/j.1365-294X.2008.03762.x [DOI] [PubMed] [Google Scholar]
- 64. Jaramillo-Correa JP, Beaulieu J, Ledig FT, Bousquet J. Decoupled mitochondrial and chloroplast DNA population structure reveals Holocene collapse and population isolation in a threatened Mexican-endemic conifer. Molecular Ecology. 2006;15(10):2787–2800. [DOI] [PubMed] [Google Scholar]
- 65. Tian S, López-Pujol J, Wang H-W, Ge S, Zhang Z-Y. Molecular evidence for glacial expansion and interglacial retreat during Quaternary climatic changes in a montane temperate pine (Pinus kwangtungensis Chun ex Tsiang) in southern China. Plant Systematics and Evolution. 2010;284(3–4):219–229. [Google Scholar]
- 66. Qian H, Ricklefs RE. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 2000;407(6801):180–182. [DOI] [PubMed] [Google Scholar]
- 67. Li F, Bao W. Elevational trends in leaf size of Campylotropis polyantha in the arid Minjiang River valley, SW China. Journal of Arid Environments. 2014;108:1–9. [Google Scholar]
- 68. Steinhoff DF, Bromwich DH, Monaghan A. Dynamics of the Foehn mechanism in the McMurdo Dry Valleys of Antarctica from Polar WRF. Quarterly Journal of the Royal Meteorological Society. 2013;139(675):1615–1631. [Google Scholar]
- 69. Bush ABG, Philander SGH. The climate of the Last Glacial Maximum: Results from a coupled atmosphere-ocean general circulation model. Journal of Geophysical Research. 1999;104:24509–24525. [Google Scholar]
- 70. Schneider von Deimling T, Ganopolski A, Held H, Rahmstorf S. How cold was the last glacial maximum? Geophysical Research Letters. 2006;33(14):L14709. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files except the DNA sequences. They can be available from the GenBank database (accession numbers KT724331–KT724335, KT750890–KT750920).