Abstract
Activation of the ERK pathway is a hallmark of cancer and targeting of upstream signalling partners led to the development of approved drugs. Recently SCH772984 has been shown to be a selective and potent ERK1/2 inhibitor. Here we report the structural mechanism for its remarkable selectivity. In ERK1/2, SCH772984 induced a so far unknown binding pocket that accommodated the piperazine-phenyl-pyrimidine decoration. This novel binding pocket was created by an inactive conformation of the phosphate binding loop and an outward tilt of helix αC. In contrast, structure determination of SCH772984 with the off-target haspin and JNK1 revealed canonical but two distinct type-I binding modes. Intriguingly, the novel binding mode with ERK1/2 was associated with slow binding kinetics in vitro as well as in cell based assay systems. The described binding mode of SCH772984 with ERK1/2 enables the design of a new type of specific kinase inhibitors with prolonged on-target activity.
Introduction
The Ras-Raf-MEK-ERK cascade constitutes a central signalling pathway that tightly controls key cellular functions such as cell proliferation. Aberrant activation of this pathway has been extensively targeted for the development of cancer therapeutics, best exemplified by clinical B-RAF and MEK inhibitors1,2. In particular the RAF inhibitor vemurafenib (PLX4032) has demonstrated excellent efficacy treating patients with BRAFV600E mutated melanoma which led to the recent approval of this drug3. However, response to vemurafenib is often temporary due to the rapid development of drug resistance by a number of diverse mechanisms3-5.
Vemurafenib strongly attenuates ERK signalling in BRAFV600E mutated melanoma but not in cancer types harbouring other mutations that activate the ERK pathway6. Surprisingly, in wild type or non-BRAF mutated cancers, ATP competitive RAF inhibitors lead to increased ERK signalling, an unexpected finding that has been attributed to a drug activated dimerization mechanism of RAF kinases7,8.
Most identified resistant mechanisms to RAF inhibitors results in strong reactivation of the ERK pathway by a large variety of different mechanisms9-11. This observation led to a number of clinical studies combining RAF and MEK inhibitors which have demonstrated a significant increase in progression free survival in BRAFV600E melanoma12. The strong activation of ERK in RAF inhibitor resistant tumours and other MAPK activated cancers suggests direct targeting of ERK as an attractive strategy for the cancer treatment4,13. To date, only few ERK1/2 inhibitors have been reported. Initial inhibitor development has been focussed on pyrazolo-pyridazines such as FR180204, a modest ERK inhibitor which has not been profiled comprehensively14. Further development led to the discovery of the pyrimidyl-pyrrole-based ERK inhibitor VTX-11e, a potent ERK inhibitor with oral bioavailability15.
Two main strategies are currently employed developing kinase inhibitors: ATP mimetic inhibitors that target the kinase active state (type-I inhibitors) and inhibitors that target a structurally more diverse inactive state, usually characterized by an “out” conformation of the ATP/Mg2+ coordinating DFG motif (type-II inhibitors)16. However, selectivity remains the major challenge also for type-II inhibitors. In contrast, non-ATP competitive allosteric inhibitors are usually highly selective as demonstrated by inhibitors that target an allosteric pocket in MEK1/22 or the myristyl binding site of ABL17. However, most allosteric inhibitors have been discovered coincidentally as strategies that would lead to the systematic development of these inhibitors are largely lacking. The binding mode of representative type-I, type-II and allosteric inhibitor binding modes are summarized in Figure 1.
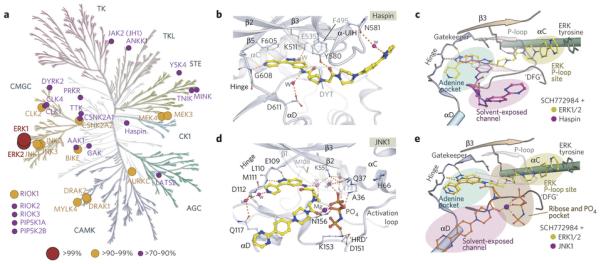
Figure 1. Illustration of inhibitor modes of kinases Inhibitors.
a) Schematic representation of a type-I binding mode (left, p38α/SB220225 complex, pdb id:4LOO), a type-II binding mode (middle, p38 α/BIRB796 complex, pdb id:1KV2) and an allosteric non-ATP competitive binding mode (MEK1/ATP/Mg2+/PD318088 complex, pdb id:1S9J). The main structural elements are labelled. b) Superimposition of all three inhibitors. Unique binding pockets targeted by each inhibitor class are indicated by coloured ellipsoids.
ERK1/2 has a low propensity for the “DFG-out” conformation due to the presence of residues in the catalytic domain that stabilize the “DFG-in” state18. Indeed, ERK1/2 co-crystal structures exclusively revealed type I binding modes15 and to date VTX-11e remains the only available potent, type-I ERK1/2 inhibitor4,13,15. Interestingly, the highly potent and selective ERK1/2 inhibitor SCH772984 of unknown binding mode has been reported recently19. SCH772984 contains a putative indazole hinge binding moiety and an elongated linear scaffold suggesting a possible type-II binding mode.
Here we report the crystal structures of SCH772984 with human ERK1 and ERK2. The structural data unravelled a novel induced allosteric pocket located adjacent to the ATP site that accommodated the SCH772984 piperazine-phenyl-pyrimidine decoration while the indazole moiety acted as a hinge binding motif. Kinetic measurements using biolayer interferometry (BLI) showed that the unexpected binding mode of SCH772984 was associated with slow inhibitor off-rates. In contrast, SCH772984 off-target activity showed fast off-rates suggesting that inhibitor specificity in cellular systems is additionally enhanced by prolonged target engagement. Indeed, wash-out experiments confirmed sustained ERK1/2 and ERK pathway inhibition in cellular systems. In the present paper we discuss the molecular mechanisms leading to slow binding kinetics of SCH772984 and how the identified binding pocket can be explored for the development of new generations of selective kinase inhibitors.
Results
SCH772984 adopts a unique kinase binding mode in ERK1/2
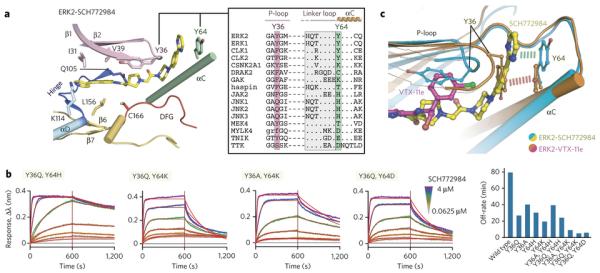
SCH772984 is a novel pyridine-indazole inhibitor with an unusual extended piperazine-phenyl-pyrimidine decoration19 (Fig. 2a). To understand the molecular mechanisms of SCH772984 selectivity we determined crystal structures of this compound with ERK1 and ERK2. Both structures were refined to high resolution (Supplementary Results, Supplementary Table 1), and the bound inhibitor was well defined by electron density in both structures (Fig. 2b). The binding mode of SCH772984 was conserved in ERK1/2 and revealed an intricate network of interaction across the ATP binding site. Consistent with the lack of propensity of ERK1/2 to adopt a ‘DFG’ out conformation, the extended linear decoration of the inhibitor did not interact with the type-II binding pocket but with a so far unseen induced binding pocked located between helix αC and the phosphate binding loop (P-loop) (Fig. 2c). Analysis of the ERK1/2 structures suggested that tight binding of the inhibitor was due to three key interactions (Fig. 2d). First, the indazole acted as a hinge binding scaffold forming two hydrogen bonds with the hinge backbone while the pyridine nitrogen formed a hydrogen bond with lysine K114 (ERK2 numbering). Second, the pyrrolidine linker was positioned in proximity to the conserved active site salt bridge (K54-E71) forming a network of direct and water mediated hydrogen bonds involving also the gatekeeper Q105 and the ‘DFG’ motif which adopted an ‘in’ conformation. Intriguingly, the P-loop tyrosine Y36 flipped into the ATP site and stacked onto the pyrrolidine ring, leading to a strong distortion of the P-loop and opening of the P-loop binding pocket. Third, the linker between the pyrrolidine and the piperazine produced a sharp kink that oriented the phenyl-pyrimidine moiety towards the P-loop pocket flanked also by the αC helix. Interactions between the kinase and the phenyl-pyrimidine rings were limited to π-stacking interaction with the αC Y64 and water-mediated hydrogen bonds to the pyrimidine group.
Figure 2. SCH772984 has a unique binding mode in ERK1/2.
a) Chemical structure of SCH772984. b) |2FO|-|FC| omitted SCH772984 electron density map contoured at 1σ. c) Surface representation of the ERK2-SCH772984 complex. SCH772984 spans across the ATP binding site and induces the P-loop pocket. d) Details of key interactions between SCH772984 and ERK1/2. Amino acid numbering is according to ERK2. e) Schematic illustration summarizing key structural features of SCH772984 binding to ERK1/2.
Structural comparison with the unphosphorylated, inactive20 and phosphorylated, active conformations of ERK21 demonstrated that the allosteric pocket induced by SCH772984 does not exist in either states of the kinase (Supplementary Fig. 1). The formation of the P-loop binding pocket in ERK1/2 involved a tilt of the αC and significant structural distortion of the P-loop while other key structural elements such as the conserved VIAK/αC salt bridge as well as the DFG motif assumed active conformations (Fig. 2e). However, the P-loop conformation with the tyrosine Y36 oriented towards the ATP site would not be compatible with ATP binding and must therefore be considered an inactive state of ERK1/2.
SCH772984 has high selectivity for ERK1/2
Next we asked if this unique binding mode confers a high degree of selectivity. Morris et al. screened SCH772984 against a panel of kinases using enzymatic assays, identifying only few additional kinases that were inhibited with considerably weaker potency19. Here we used a comprehensive KINOMEscan panel22 to assess selectivity against 456 kinases, which confirmed high specificity of SCH772984 for ERK1/2 detecting few off-targets of significantly weaker affinity (Fig. 3a and Supplementary Fig. 2). We then performed enzymatic assays to determine IC50 values for ERK1/2 revealing low nanomolar inhibition (8.3 and 2.7 nM for ERK1 and ERK2, respectively), similar to previous reports19. In addition, the assays confirmed considerably weaker IC50 values for the most relevant off-targets identified from the KINOMEscan, exemplified by ~40-70 weaker inhibitions of the most potently inhibited kinases such as Cdc2-like kinase 2 (CLK2), DAP-kinase-related apoptosis-inducing protein kinase (DRAK1) and TTK/MPS1 (monopolar spindle 1 kinase) (Supplementary Fig. 3). Correlation between the KINOMEscan binding assay and enzyme kinetic data was high considering the significant differences of these two assay systems. The most notable exceptions were CLK2 and CSNK2A2 with similar inhibitions at 1 μM in KINOMEscan but remarkably different IC50 values in the enzymatic assay (104 nM for the former and no activity in the enzymatic assay for the latter).
Figure 3. Selectivity and off-target binding modes of SCH772984.
a) Inhibitor selectivity determined using KinomeScan22. Binding affinities of the inhibitor are represented as spheres mapped onto the kinase phylogenetic tree. The sphere radius corresponds to inhibitor affinities as indicated in the figure. b) Detailed interactions of SCH772984 with the haspin ATP binding pocket. c) Schematic illustration for superimposition of ERK2 and haspin structures reveals the distinct accommodations of the inhibitor. d) Detailed interactions of SCH772984 with JNK1. e) Superimposition of the ERK2 and JNK1 complexes.
SCH772984 binding modes in off-targets haspin and JNK1
To explore whether the ERK1/2 binding mode is conserved in off-targets, we determined the structures of SCH772984 with the atypical kinase haspin (IC50: 398 nM) as well as the MAPK C-JUN kinase 1 (JNK1; IC50: 1080 nM). The co-crystal structure of haspin revealed a dramatically altered binding mode (Fig. 3b and Supplementary Fig. 4). The inhibitor rotated in the binding site positioning the pyridine nitrogen at the hinge region and the indazole towards the back pocket forming a water-mediated hydrogen bond to the active site lysine (K511). The piperazine-phenyl-pyrimidine ring system was oriented towards solvent-exposed space interacting with the haspin specific insertion23. Superimposition of the ERK and haspin complexes demonstrated the dramatic differences of the observed binding modes. In haspin, SCH772984 interacted with the active state of the kinase in a typical type-I binding mode (Fig. 3c).
Haspin is a highly diverse protein kinase which shares only weak sequence homology with MAPKs. We were therefore interested if the binding mode of SCH772984 is conserved in kinases that are structurally more related to ERK1/2. MAP kinases of the JNK family (JNK1-JNK3) were weakly inhibited by SCH772984 with IC50 values ranging from 632 nM (JNK3) to 1080 nM (JNK1). However, due to the failure to obtain co-crystals of the inhibitor with either JNK1 or JNK2 we therefore employed crystal soaking which led to a high resolution model of the JNK1-SCH772984 complex (Supplementary Table 1). The electron density map allowed us to unambiguously determine the binding mode of SCH772984. However, additional electron density was visible in the ATP binding pocket that was interpreted as a Mg2+ ion and a (β,γ-imido)triphosphate group from the hydrolysed AMP-PNP, that were present in the initial crystallization (Supplementary Fig. 4). Interestingly, SCH772984 bound with a diverse type-I binding mode to JNK1 in which, similar to the ERK1/2 binding mode, the indazole interacted with the hinge region while the pyridine nitrogen formed water-mediated interactions with Q117 and with the main chain of the hinge residue D112 (Fig. 3d, 3e). In contrast to ERK1/2 the remaining parts of the inhibitor engaged different interactions, potentially dictated by the gatekeeper M108. Unlike the polar glutamine gatekeeper in ERK1/2 which attracted the protruding of the linker into the back pocket, the bulky, hydrophobic methionine gatekeeper in JNK1 forced the pyrrolidine linker to provide a sharp kink orienting the piperazine-phenyl-pyrimidine ring system towards the solvent. The resulting space in the back pocket of the ATP site was thus mainly occupied by water molecules, which together with triphosphate/Mg2+ bridged interactions between the inhibitor and the conserved active site lysine/glutamate salt bridge as well as several other residues of the kinase. The few direct interactions explain the weak activity of SCH772984 for JNK. However, due to the presence of the triphosphate/Mg2+ group we cannot rule out that these ions influence the binding mode of the inhibitor. We therefore synthesized a small series of SCH772984 analogues with modifications in the pyridine ring to probe hinge interactions of this moiety and screened these derivatives using a temperature shift binding assay (ΔTm)24,25. As expected, removal of the pyridine nitrogen or a ring substitution at this position resulted in inactivity of the inhibitors for haspin (Supplementary Fig. 4c). In contrast, in JNK1 these substitutions were well tolerated consistent with the experimental binding mode envisaged from the crystal structure. Other off-targets (CLK1, MEK4) were also not affected by the introduced modifications in the pyridine ring, suggesting that similar to JNK1 the indazole and not the pyridine likely interacts with the kinase hinge backbone.
The SCH772984-ERK1/2 complex has slow dissociation rates
Inhibitor binding thermodynamics and kinetics have developed into important design criteria. In particular, slow binding off-rates have been associated with improved inhibitor efficacies due to prolonged target engagement in vivo26. This prompted us to characterize the biophysical properties of the SCH772984 interaction with ERK1/2. First, the thermodynamic signature of interaction of SCH772984 with ERK1/2 and haspin was investigated using isothermal titration calorimetry (ITC) (Fig. 4a, Supplementary Fig. 5). Binding of the inhibitor was associated with large favourable binding enthalpy changes of about −20 kcal/mol while interaction with ERK1/2 was strongly opposed by entropic forces, suggesting induction of unfavourable conformations in either the ligand or the protein. However, also haspin exhibited the similar thermodynamic signature, probably due to its large and polar interaction surface with this kinase. Interestingly, the measured KD values for ERK1 and ERK2 of ~200 nM were significantly larger than the determined IC50 values of ~2.7-8.3 nM, while this was not the case for haspin where ITC data and IC50 values were similar (Supplementary Fig. 5). Careful inspection of the titration curves suggested that unlike haspin, the normalized binding heats were not well represented by a single site fitting model. This was even more evident in ITC experiments carried out at higher temperature where normalized binding enthalpies clearly showed a biphasic binding behaviour (Supplementary Fig. 5). It is therefore likely that non-equilibrium binding of SCH772984 leads to determination of incorrect binding constants. In support of this notion, this unusual behaviour was not observed using fast off-rate inhibitors such as for the haspin inhibitor 5-iodotubercidin23 that also inhibits ERK1/2, albeit with weaker affinity27,28 (Supplementary Fig. 6).
Figure 4. Slow kinetics of SCH772984 both in vitro and in cell based systems.
a) ITC binding isotherms for the interactions of SCH772984 with ERK2 and haspin. Shown are raw titration heats (insets) as well as normalized binding heats. b) BLI data showing the association and dissociation sensograms at different inhibitor concentrations for the interaction of SCH772984 with ERK2 and haspin and ERK2 with VTX-11e. c) Comparison of cellular activities of the ERK inhibitors in MDA-MB 231 cells. Cells were treated with 100nM inhibitors for 4 hours and lysates were analysed using Western blotting before and after inhibitor wash out. Western blots band intensities were quantified and normalized relative to DMSO-treated cells. Quantification of Western blot signals for indicated phospho-proteins relative to loading controls is shown in the extrapolated recovery curves (lower panels) and reflects the recovery rates of MAPK pathway specific phosphorylation events. Western Blots show representative experiments of two repeats.
In addition, we studied the kinetic aspects of the interactions using biolayer interference measurements (BLI). Interestingly, we observed slow binding kinetics affecting association and dissociation rates of SCH772984 when binding to ERK1/2, but not for the interaction of this inhibitor with haspin (Fig. 4b, Supplementary Fig. 5). In general, slower kinetics has been described as a distinguishing characteristic of some type-II and allosteric inhibitors possibly due to kinetic constrains of the necessary structural rearrangements29,30, and slow off-rates have also been observed for the ‘out’-DFG-sensitive ERK mutant interacting with a type-II compound18. Slow inhibitor off-rates have emerged as an important parameter for the sustained efficacies of kinase inhibitors and other target families but the structural mechanisms that influence inhibitor binding kinetics are poorly understood and it is likely that slow-off rates are not unique features of the type-II binding mode. For comparison, we further assessed the kinetic characteristics of other type-I ERK inhibitors, including VTX-11e, FR18020414 and 5-iodotubercidin (5-iTU). Interestingly, while FR180204 and 5-iTU exhibited fast kinetic behaviours, the similar slow association and dissociation patterns to SCH772984 were also observed for VTX-11e (Fig. 4b). The observation that VTX-11e has slow dissociation rates was rather surprising, nonetheless potentially not too unusual features for type-I class since slow kinetics was also demonstrated for the interaction between 5-iTU and its main target haspin (Supplementary Fig. 6). Dissociation half-lifes for the SCH772984-ERK1/2 interactions were estimated to be between 25 and 80 minutes but we were unable to determine these values with high accuracy due signal instability using BLI. The half-life of SCH772984 was comparable to that of VTX-11e, which was estimated to be ~35-46 minutes. Both off-rates were considerably longer when compared to FR180204 and 5-iTU which showed off-rates of less than one minute (Supplementary Fig. 5 and 6).
SCH772984 has prolonged ERK inhibitory activity in cells
Given the observed slow kinetics of SCH772984 in vitro, we investigated whether treatment of cells with this inhibitor would result in sustained ERK repression after the inhibitor was removed from the culture medium. We treated human MDA-MB-231 metastatic adenocarcinoma breast cancer cells with SCH772984 and monitored the recovery of ERK signalling after inhibitor removal by Western blotting in a time-dependent manner. ERK1/2 activation, as well as the phosphorylation of the downstream ERK targets, RSK1 (ribosomal protein S6 kinase) and ETS1 (v-ets avian erythroblastosis virus E26 oncogene homolog 1), was monitored using phosphorylation site specific antibodies. We found that phosphorylation of pERKs, pRSK1 and pETS1 recovered to levels similar to the ones before inhibitor treatment within 1 to 2 hours after inhibitor wash-out, consistent with the estimated off-rates determined by BLI in vitro (Fig. 4c). Interestingly, SCH772984 also inhibited ERK1/2 phosphorylation, a phenomenon observed also previously19, and this phosphorylation inhibition also showed similar time dependence after the inhibitor washout in a synchronized manner to the time-dependent phosphorylation levels of the down-stream ERK1/2 substrates. It is likely that the significant structural rearrangement induced by SCH772984 inhibits ERK1/2 phosphorylation and activation by MEK1/2 (Supplementary Fig. 7). Consistent with its type-I binding mode, VTX-11e did not inhibit activating phosphorylation of ERK1 and ERK2. As predicted by its slow in vitro off-rates, this inhibitor showed also prolonged down-stream inhibition after inhibitor washout as demonstrated by the levels of pRSK1 and pETS1. For comparison, FR180204 did similar to VTX-11e not inhibit ERK1/2 phosphorylation and showed as expected rapid recovery after the inhibitor washout which restored rapidly pETS1 levels. Surprisingly, we did however not see a strong effect on pRSK1 inhibition using this inhibitor (Supplementary Fig. 6), potentially a consequence of its fast off-rate.
Structural requirements for SCH772984 slow-off rates
Based on the binding mode of SCH772984 with ERK1/2, the complex structures with the off-target haspin and JNK1 and available structural data on other kinases that are inhibited by SCH772984 including CLK131, GAK (cyclin G associated kinase)32, JAK233, TNIK (pdb id: 2×7f) and TTK34, we sought to identify the structural requirements facilitating this novel binding mode as well as the mechanisms leading to slow off-rates. Comparison of 14 available crystal structures, the four structures determined here as well as structure-based sequence alignments revealed conservation of key residues lining the ERK1/2-SCH772984 interface (Fig. 5a and Supplementary Fig. 8 and 9). While the residues within the type-I adenine mimetic pocket were quite conserved, we hypothesized that the SCH772984 binding mode in ERK1/2 depends on a strong structural rearrangement of the P-loop as well as an aromatic stacking interaction between Y64 located in helix αC. 11 of the 16 off-targets harbour an aromatic residue at the tip of the P-loop that may facilitate stabilizing the distorted P-loop conformation necessary for the opening of the P-loop pocket. Furthermore, only two kinases, CLK1 and CLK2, contain also an aromatic residue in an analogues position to ERK1/2 αC tyrosine that would allow a stacking interaction with SCH772984. To assist our structural comparison, we analysed the binding kinetics of SCH772984 on the off-targets CLK1, CLK2, JNK2, CSNK2A1/A2, DRAK1 and TTK. Interestingly, BLI binding data revealed that all studied off-targets exhibited fast association and dissociation rates (Supplementary Fig. 10). Thus, for the use of SCH772984 in cellular systems and in vivo the unique slow off-rate for ERK1/2 will additionally increase inhibitor efficacy and selectivity.
Figure 5. Structural analysis of inhibitor interactions and mutagenesis of key residues that modulate binding kinetics.
a) Analysis of the residues located within the P-loop pocket in ERK2. The inset shows the structure-based sequence alignment of ERK1/2 with some off-targets focused mainly on the two tyrosine residues from P-loop and αC. A full alignment is shown in supplementary Fig. 9. b) Binding kinetics of ERK2 mutants with SCH772984 using BLI with sensograms and global fits for selected double mutants shown. Experiments show representative data of two identical repeats. The bar diagram on the right illustrates dissociation half-life for all tested mutants. See Supplementary Fig. 10b for the summary of BLI kinetics for the interaction between ERK2 mutants with SCH771984 and VTX-11e. c) Structural comparison of the VTX-11e and SCH772984 binding modes in ERK2 with a detailed view of inhibitor accommodations and the key aromatic residues located in the P-loop and αC highlighted. Dashed lines indicate potential π-stacking interactions in vicinity to the ERK P-loop pocket.
Importance of aromatic stacking interactions
Based on our structural comparison we hypothesized that two aromatic tyrosine residues located within the P-loop and αC could be the keys for stabilization of the binding mode of SCH772984 in ERK1/2 and its slow binding kinetics (Fig. 5a). To test this hypothesis, we mutated the P-loop and αC tyrosine residues to the corresponding residues present in off-targets that all showed fast off-rates, and studied the binding kinetics of SCH771984 with these mutants. Interestingly, all four single-site mutants tested (Y36Q, Y36A, Y64H and Y64K) retained slow association and dissociation kinetics, albeit with reduced binding affinities. Also the double mutants Y36A/Y64H and Y36Q/Y64H still showed slow binding kinetics suggesting that H64 can compensate the loss of the αC tyrosine through a similar stacking interaction with the inhibitor. In support of this notion, the other double mutants with the substitutions of Y64 to lysine or aspartate strongly reduced inhibitor off-rates (Fig. 5b, Supplementary Fig. 10). The mutagenesis study suggested that both tyrosine residues are required for tight binding and slow inhibitor binding kinetics in agreement with our prediction based on the analysed crystal structures.
Since we also observed slow binding kinetics of the type-I VTX-11e, we used the generated mutants to investigate if the slow binding kinetics of this inhibitor is also affected by these substitutions (Supplementary Fig. 10). Surprisingly, the two tested single mutants (Y36Q and Y64K) significantly reduced inhibitor off-rates. In particular, the strong effect of Y64K on the binding kinetics was unexpected since this residue is distantly located to the ATP binding site. Mutations of both tyrosine residues (Y36Q/Y64K and Y36Q/Y64D) increase inhibitor off-rates further while also reducing binding affinities. Thus, interactions mediated by the two aromatic residues were also important for slow binding kinetics for VTX-11e. Since an experimental structure of the VTX-11e complex was not available, we determined the co-crystal structure of ERK2 with VTX-11e and performed a structural comparison (Fig. 5c). As expected, the typical type-I binding mode of VTX-11e did not occupy the P-loop binding pocket, however the chlorobenzene group was located in a similar position as Y36 in the ERK1/2-SCH772984 complexes leading to a distortion of the P-loop. As a result, the P-loop tyrosine, Y36, was oriented towards the αC Y64 in the VTX-11e complex, forming a stacking interaction mimicking that of the SCH772984 tail and Y64 (Fig. 5c). Thus, despite the fundamentally different binding modes of VTX-11e and SCH772984, aromatic moieties in VTX-11e and interactions formed by Y36 and Y64 mimicked key aromatic stacking interactions of the SCH772984 complex, explaining why the mutated aromatic residues in ERK2 affected binding kinetics of both inhibitor types.
Effects of ERK inhibition in BRCA2-deficient cells
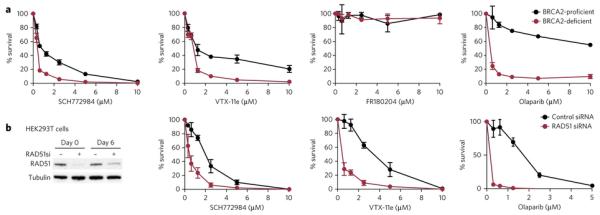
ERK1 has been recently described as a kinase required for the proliferation of BRCA2-deficient cells, suggesting potential benefits of targeting ERK1 for the treatment of BRCA2-deficient tumours35. To test if selective inhibition of ERK1/2 will affect survival of BRCA2-deficient cells we used the hamster cell line V-C8, which lacks detectable BRCA2 expression36. Exposure of V-C8 cells to SCH772984 led to a significant reduction in cell survival compared to the corresponding cells complemented with wild type BRCA2 (Fig 6a). Significant differences in cell survival between BRCA2-deficient and proficient cells were also observed following treatment with VTX-11e, but not to FR180204. BRCA2 deficiency has been shown to significantly sensitize cells and tumours to PARP (poly ADP ribose polymerase) inhibitors37,38. We used therefore the PARP inhibitor olaparib as a control compound based on its established ability to preferentially kill BRCA2-deficient cells (Fig 6a). Furthermore, similar growth suppression by ERK inhibitors was also observed in the human cell line MDA-MB 231 in which BRCA2 shRNA-mediated depletion was induced with doxycycline, as well as in human HEK293T cells in which BRCA2 expression was repressed using siRNA (Supplementary Fig. 11 and 12). In these cell lines, BRCA2 knockdown also sensitized cells to SCH772984 and/or VTX-11e.
Figure 6. Effects of ERK and PARP inhibitors on BRCA2-deficient cell survival.
a) Effect of SCH772984, VTX-11e, FR180204 and the PARP inhibitor olaparib on the survival of BRCA2-deficient and -proficient V-C8 hamster cells, following three-day exposure to each drug at the concentrations indicated. b) Human HEK239T cells were transfected with control or RAD51 siRNAs twice (day 0 and day 3) over a six day period. Cell extracts were prepared 12 hours and six days after the first transfection and analysed by Western blotting as indicated. Survival of HEK293T cells lacking RAD51 relative to control cells in the presence of SCH772984, VTX-11e and the PARP inhibitor olaparib, following six-day exposure to each drug at the concentrations indicated. Data represented are the means of two biological repeats and the standard deviations (SD).
BRCA2 is a central regulator of the homologous recombination DNA repair pathway by promoting the assembly of the RAD51 recombinase at sites of DNA double stranded breaks or stalled replication forks and protecting them from degradation39-41. We tested therefore if RAD51 deficiency also sensitizes cells to ERK1/2 inhibition. Indeed, siRNA-mediated depletion of RAD51 in human 293T cells led to a significant increase in cell killing upon exposure to SCH772984 as well as VTX-11e (Fig 6b), similarly to V-C8. However, the effects of the studied ERK inhibitors on RAD51 deficient cells were less pronounced when compared to olaparib, which was used as a control for specific elimination of RAD51-depleted cells as previously reported42. Nonetheless, consistent with genetic studies35, treatment with the ERK1/2 inhibitor eliminates specifically cells lacking BRCA2 or RAD51.
Discussion
In this study we suggest targeting of a novel binding pocket created by an inactive P-loop conformation as well as an outward movement of helix αC. The tip of the P-loop frequently contains an aromatic residue which may flip into the ATP binding site forming aromatic π-π stacking interaction inhibitors. The “in” or “folded” P-loop conformation has been described for a number of protein kinases including major drug targets such as ABL1, Aurora A, FGFR1, cMET, p38 and MAP4K4 and folded P-loop conformations have been associated with favourable inhibitor selectivity43,44. However, in contrast to the binding mode of SCH772984 in ERK1/2, the P-loop tyrosine usually interacts in previously described structures in a “side on” aromatic stacking conformation which blocks at least partly access of the generated binding site between the P-loop and αC. The presence of the P-loop binding pocket in a large diversity of kinases suggests that this site can be targeted by optimizing current inhibitor chemistry to this new binding mode adding a novel design strategy for the development of kinase inhibitors. The P-loop pocket may also be targeted independently by removing or substituting adenine mimetic moieties. Due to the diversity of the P-loop pocket this would lead to a considerable expansion of kinase inhibitor chemical space and commercialization opportunities.
Drug residence time has been recently recognized as an important parameter for the development of efficient inhibitors45,46 but the lack of our understanding of the “structure/kinetic relationships” precludes rational optimization of this property. Only few reports have analysed potential mechanisms of drug binding kinetics including electrostatic steering which modulates inhibitor binding on-rates47 or water-shielded hydrogen bonds48. In proteins kinases, the coupling of ligand binding to presumably slow structural transitions such as the DFG-out movement has been associated with slow binding kinetics26,30. Here we found that also type-I inhibitors such as VTX-11e and 5-iodotubercidin may have slow off-rates and identified that aromatic stacking interactions seem to play a key role in determining binding kinetics.
We also reported that chemical inhibition of ERK1/2 preferentially affects cell survival of BRCA2 deficient breast cancer cells potentially expanding future therapeutic applications of ERK inhibitors to this difficult to treat patient population. The described structural mechanism of SCH772984 selectivity and its slow binding kinetics will facilitate the development of further highly selective inhibitors for ERK1/2 and other kinases in the future.
Online methods
Protein expression and purification
Full length ERK1, ERK2 and the kinase domains of haspin (aa 465-798) and JNK1 (aa 1-363) cloned into pNIC28-Bsa4 were co-expressed with λ-phosphatase in E. coli BL21 (DE3)-R3 cells, which were initially cultured in terrific broth (TB) media to OD600 of ~1.6 at 37 °C followed by an additional growth while cooling to 18 °C to OD600 of ~3 prior to an induction with 0.5 mM IPTG overnight. Cells were harvested by centrifugation, and were lysed using sonication method. The N-terminal His6-tagged recombinant proteins were purified using Ni-affinity chromatography. For haspin, the eluted protein was passed directly onto Superdex s200 column, and the resulting pure protein incorporating the His6-tag was stored at −80 °C in 50 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 mM TCEP and 5% glycerol. For ERK1, ERK2 and JNK1, the eluted proteins from Ni-affinity purification were treated with TEV protease for the histidine tag removal. The cleaved proteins were separated by passing through Ni-Sepharose resin, and further purified using size-exclusion chromatography (Superdex S200). The resulting pure recombinant ERKs and JNK1 were stored at −80 °C in a buffer containing 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 5% glycerol.
For generation of biotinylated proteins, the same constructs were cloned into pNIC-Bio2, incorporating an N-terminal His10-tag and a C-terminal AviTag. All recombinant proteins were co-expressed with λ-phosphatase and biotin-protein ligase BirA in E. coli BL21 (DE3)-R3 cells, cultured in TB media using similar protocol as above, albeit with supplementation of 0.3 mM biotin upon IPTG addition for in vivo biotinylation as previously recommended49. After cell lysis, the biotinylated recombinant proteins were purified via two steps, Ni-affinity and size-exclusion chromatography. The pure proteins were stored at −80 °C in 50 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 mM TCEP and 5% glycerol.
Commercial inhibitors
SCH772984 was purchased from Selleckchem (Cat. No. S7101). VX11e was purchased from ChemieTek (Cat. No. CT-VX11e). FR180204 was purchased from Tocris (Cat. No 3706).
Crystallization
All crystallization experiments were performed using sitting-drop vapour diffusion method at 4 °C. For ERK1 and ERK2, the proteins were buffer exchanged into 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM TCEP, and concentrated up to 10-15 mg/mL. For ERK2, the protein was incubated with 1 mM ligands, and crystallized using the reservoir solutions containing 30% PEG4000 and 0.2 M ammonium sulphate for the SCH772984 complex or 25% PEG smears (PEG2000, PEG3350, PEG4000 and PEG5000MME), 0.1 M cacodylate pH 5.5 and 0.2 M ammonium sulphate for the VTX-11e complex.
For ERK1, ligand substitution soaking was employed due to a failure to achieve the co-crystals of the protein with the compound. The ERK1 crystals were initially obtained with 5-iodotubercidin inhibitor using a similar condition (33% PEG4000, 0.1 M Tris pH 8.0 and 0.2 M lithium sulphate) as previously described28. The ERK1 crystals were soaked with 4 mM SCH772984 in a stabilization solution containing 27% PEG 4000, 0.03 M Tris pH 8.0, 0.03 M lithium sulphate and 20% ethylene glycol for 5 days.
For haspin, without a requirement of buffer exchange step the protein at 15 mg/ml in the storage buffer was mixed with 1 mM inhibitor prior setting up crystallization. The viable co-crystals grew using a condition containing 51% MPD and 0.1 M SPG pH 6.0.
For JNK1, the protein was buffer exchanged into 25 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM DTT, and concentrated to ~11-13 mg/ml. The initial crystals were achieved by co-crystallizing with 3 mM AMP-PNP and 5 mM MgCl2 using the reservoir solution containing 12-15% PEG3350 and 0.1 M HEPES pH 6.8-7.8. Ligand substitution soaking was performed using similar procedure as that employed for ERK1 with the soaking solution containing 4 mM SCH772984, 15-18% PEG3350, 0.1 M HEPES pH 6.8 and 20% ethylene glycol.
Data collection and structural determination
Viable crystals of ERK2 were briefly transferred into a cryo-protectant made from the crystallization mother liquor supplemented with 22% ethylene glycol, while this step was not required for the soaked ERK1 and JNK1 crystals and haspin co-crystals. All crystals were flash-cooled in liquid nitrogen, and diffraction data were collected at Diamond Light Source and processed with MOSFLM50 before subsequent scaling using SCALA51 from CCP4 suite52. Molecular replacement was performed for structure solutions using Phaser program53 and the kinase coordinates of ERK1-5-iodotubercidin complex28, inactive ERK220, haspin-AMP complex23 and JNK1-inhibitor complex54 as search models for ERK1, ERK2, haspin and JNK1, respectively. All structures were subjected to iterative cycles of manual model building in COOT55 alternated with refinement using REFMAC56. TLS definitions used in the late refinement step were calculated using TLSMD server57. Geometric correctness of all kinase-SCH772984 complexes was validated with MOLPROBITY58. Statistics for data collection and structure refinement are summarized in Supplementary Table 1.
Selectivity screening
Competitive binding assays of SCH772984 were performed against a panel of human kinases using the KINOMEscan method as previously described22. Selected kinases were further profiled for their inhibitor binding and the IC50 values were determined using either Z’LYTE®, Adapta® or LanthaScreen® assays from Life Technologies (www.lifetechnologies.com).
Thermal stability shift assays
The kinases at 2 μM were mixed with 10 μM inhibitors. The assays and data evaluation for melting temperatures were performed using a Real-Time PCR Mx3005p machine (Stratagene) and the protocols previously described25.
Isothermal titration calorimetry
All calorimetric titration experiments were carried out on VP-ITC (MicroCal) at 15 °C. The buffer condition used for ERK1 and ERK2 was 20 mM HEPES, pH 7.5, 150 mM NaCl and 0.5 mM TCEP, while that for haspin was 20 mM HEPES, pH 7.5, 250 mM NaCl and 0.5 mM TCEP due to instability of the protein at low salt concentration. Titration was performed by injecting the proteins (100 μM) into a reaction cell containing the inhibitors (7 μM). The same experimental protocol was also used for the 5-iodotubercidin-kinase titrations. Integrated heat of the titrations after corrected for the heat of dilution were analysed using the Origin program. The corrected data were fitted to a single binding site model using a nonlinear least-square minimization algorithm, and the binding parameters including reaction enthalpy changes (ΔH), reaction enthalpy changes (TΔS), equilibrium dissociation constants (KD), stoichiometry (n) were calculated.
Bio-Layer Interferometry binding assays
Binding kinetics of inhibitor to kinases was determined by the bio-layer inferometry (BLI) method using Octet RED384 system (FortéBio). All experiments were performed at 25 °C under a buffer condition containing 20 mM HEPES, pH 7.5, 150 mM NaCl and 0.5 mM TCEP. Biotinylated proteins were immobilized onto Super Streptavidin biosensors, which were subsequently used in association and dissociation measurements each performed for a time window of 600 seconds. The interference patterns from the protein-coated biosensors with no compound and the uncoated biosensors with compound at corresponding concentrations were measured as two sets of controls. After double referencing corrections, the subtracted binding interference data were applied to the calculations of binding constants using the FortéBio analysis software provided with the instrument.
Monitoring ERK signalling in cells
Human MDA-MB-231 breast cancer cells were cultivated in monolayers in DMEM medium (Sigma Aldrich) supplemented with 10% foetal bovine serum (Clontech), penicillin and streptomycin (Sigma Aldrich). ERK1/2 inhibitors SCH772984 and VTX-11e were added at 100 nM final concentration for 4 hours, followed by two washes in PBS and release into fresh media for 0.5, 1 and 2 hours. Cells were harvested by trypsinization, washed once with cold PBS, re-suspended in SDS-PAGE loading buffer and sonicated. Equal amounts of protein were analysed by gel electrophoresis followed by Western blotting. NuPAGE-Novex 10% Bis-Tris gels (Invitrogen) were run according to manufacturer’s instructions. The following antibodies were used in immunoblotting: anti-pERK (Cell Signaling Technology), anti-pRSK (Abcam) and anti-pETS1 (Immunoway). Anti-ERK1/2 (Cell Signaling Technology), anti-GAPDH (Novus Biologicals) or anti-α-tubulin (Cancer Research UK Monoclonal Antibody Service) antibodies were used as loading controls.
siRNAs
293T cells were transfected using Dharmafect 1 (Dharmacon Research). Briefly, 2 × 106 cells were transfected with 40 nM siRNA/plate by reverse transfection in 10 cm plates. Following overnight incubation, cells were were seeded at 1 × 104 cells cells per well in 24-well plates. Three days after the first transfection, cells were transfected again as above. siRNA duplexes were 21 base pairs long with 2-base deoxynucleotide overhang. The sequence of BRCA2 siRNA was UCA GCU GGC UUC AAC UCC AUU and RAD51 siRNA was ordered from Qiagen. GFP siRNA with the sequence GCU GAC CCU GAA GUU CAU CUU was used as a control.
Cell viability assays
Human HEK293T and MDA-MB-231 cells, as well as BRCA2-mutated V-C8 hamster cells transduced with empty vector or wild type BRCA2 were seeded at 2500 cells per well in 96-well plates and grown in monolayers in DMEM medium (Sigma Aldrich) supplemented with 10% foetal bovine serum (Life Technologies), penicillin and streptomycin (Sigma Aldrich). ERK1/2 inhibitors SCH772984 and VTX-11e were added at the indicated concentrations ranging between 0.3-10 mM. Cell number was determined by incubating cells with media containing 10 mg/mL of resazurin for 2 hours. Fluorescence was measured at 590 nm using a plate reader (POLARstar, Omega one). Cells exposed to the indicated treatments were collected at the indicated timepoints and immunoblotted with the following antibodies: anti-BRCA2 (Calbiochem), anti-SMC1 (Bethyl Laboratories), anti-pERK (Cell Signaling Technology) and sheep polyclonal antibody raised against mouse BRCA2.
Supplementary Material
Acknowledgements
SK is supported by the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Bayer, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z]. AC is supported by the European Union FP7 Grant No. 278568 “PRIMES” (Protein interaction machines in oncogenic EGF receptor signalling). Work in M.T. laboratory is supported by Cancer Research UK, EMBO Young Investigator Program and The Royal Society. We thank the staffs at Diamond Light Source for assistance during data collection at the synchrotron.
Footnotes
Competing financial interests: The authors declare no competing financial interests
The coordinates and structure factors have been deposited in the protein data bank with the accession codes: 4QTA, 4QTB, 4QTC, 4QTD, 4QTE.
References
- 1.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 2.Hatzivassiliou G, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501:232–6. doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whittaker SR, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107:14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 9.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady DC, et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–496. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G, et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol Cancer Ther. 2012;11:1143–54. doi: 10.1158/1535-7163.MCT-11-1010. [DOI] [PubMed] [Google Scholar]
- 14.Ohori M, et al. Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex. Biochem Biophys Res Commun. 2005;336:357–63. doi: 10.1016/j.bbrc.2005.08.082. [DOI] [PubMed] [Google Scholar]
- 15.Aronov AM, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem. 2009;52:6362–8. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, et al. Exploration of Type II Binding Mode: A Privileged Approach for Kinase Inhibitor Focused Drug Discovery? ACS Chem Biol. 2014;9:1230–1241. doi: 10.1021/cb500129t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–6. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hari SB, Merritt EA, Maly DJ. Sequence determinants of a specific inactive protein kinase conformation. Chem Biol. 2013;20:806–15. doi: 10.1016/j.chembiol.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–50. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–11. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 21.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–69. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 22.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 23.Eswaran J, et al. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A. 2009;106:20198–203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedorov O, Niesen FH, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol. 2012;795:109–18. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- 25.Fedorov O, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007;104:20523–8. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood ER, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–9. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 27.Fox T, et al. A single amino acid substitution makes ERK2 susceptible to pyridinyl imidazole inhibitors of p38 MAP kinase. Protein Sci. 1998;7:2249–55. doi: 10.1002/pro.5560071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T, et al. Crystal structure of human mono-phosphorylated ERK1 at Tyr204. Biochem Biophys Res Commun. 2008;377:1123–7. doi: 10.1016/j.bbrc.2008.10.127. [DOI] [PubMed] [Google Scholar]
- 29.Gruenbaum LM, et al. Inhibition of pro-inflammatory cytokine production by the dual p38/JNK2 inhibitor BIRB796 correlates with the inhibition of p38 signaling. Biochem Pharmacol. 2009;77:422–32. doi: 10.1016/j.bcp.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Pargellis C, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–72. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 31.Bullock AN, et al. Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation. Structure. 2009;17:352–62. doi: 10.1016/j.str.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaikuad A, et al. Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies. Biochem J. 2014;459:59–69. doi: 10.1042/BJ20131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–83. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–68. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlos AR, et al. ARF triggers senescence in Brca2-deficient cells by altering the spectrum of p53 transcriptional targets. Nat Commun. 2013;4:2697. doi: 10.1038/ncomms3697. [DOI] [PubMed] [Google Scholar]
- 36.Kraakman-van der Zwet M, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22:669–79. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evers B, et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res. 2008;14:3916–25. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 38.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 39.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–11. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCabe N, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 43.Guimaraes CR, et al. Understanding the impact of the P-loop conformation on kinase selectivity. J Chem Inf Model. 2011;51:1199–204. doi: 10.1021/ci200153c. [DOI] [PubMed] [Google Scholar]
- 44.Hari SB, Perera BG, Ranjitkar P, Seeliger MA, Maly DJ. Conformation-selective inhibitors reveal differences in the activation and phosphate-binding loops of the tyrosine kinases Abl and Src. ACS Chem Biol. 2013;8:2734–43. doi: 10.1021/cb400663k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–9. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 46.Swinney DC. The role of binding kinetics in therapeutically useful drug action. Curr Opin Drug Discov Devel. 2009;12:31–9. [PubMed] [Google Scholar]
- 47.Selzer T, Albeck S, Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat Struct Biol. 2000;7:537–41. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 48.Schmidtke P, Luque FJ, Murray JB, Barril X. Shielded hydrogen bonds as structural determinants of binding kinetics: application in drug design. J Am Chem Soc. 2011;133:18903–10. doi: 10.1021/ja207494u. [DOI] [PubMed] [Google Scholar]
- 49.Keates T, et al. Expressing the human proteome for affinity proteomics: optimising expression of soluble protein domains and in vivo biotinylation. N Biotechnol. 2012;29:515–25. doi: 10.1016/j.nbt.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oza V, et al. Discovery of checkpoint kinase inhibitor (S)-5-(3-fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by structure-based design and optimization of thiophenecarboxamide ureas. J Med Chem. 2012;55:5130–42. doi: 10.1021/jm300025r. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 57.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 58.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.