Abstract
Background
Panax L. is a medicinally important genus within family Araliaceae, where almost all species are of cultural significance for traditional Chinese medicine. Previous studies suggested two independent origins of the East Asia and North America disjunct distribution of this genus and multiple rounds of whole genome duplications (WGDs) might have occurred during the evolutionary process.
Results
We employed multiple chloroplast and nuclear markers to investigate the evolution and diversification of Panax. Our phylogenetic analyses confirmed previous observations of the independent origins of disjunct distribution and both ancient and recent WGDs have occurred within Panax. The estimations of divergence time implied that the ancient WGD might have occurred before the establishment of Panax. Thereafter, at least two independent recent WGD events have occurred within Panax, one of which has led to the formation of three geographically isolated tetraploid species P. ginseng, P. japonicus and P. quinquefolius. Population genetic analyses showed that the diploid species P. notoginseng harbored significantly lower nucleotide diversity than those of the two tetraploid species P. ginseng and P. quinquefolius and the three species showed distinct nucleotide variation patterns at exon regions.
Conclusion
Our findings based on the phylogenetic and population genetic analyses, coupled with the species distribution patterns of Panax, suggested that the two rounds of WGD along with the geographic and ecological isolations might have together contributed to the evolution and diversification of this genus.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0669-0) contains supplementary material, which is available to authorized users.
Keywords: Chloroplast, Disjunt distribution, nrITS, Nucleotide diversity, Panax, Single copy nuclear gene, Whole genome duplication
Background
Whole genome duplication (WGD) or polyploidy is thought to be central to the diversification of angiosperm plants [1, 2]. It is well recognized that all angiosperms are paleopolyploid [3, 4] and have experienced multiple rounds of WGD [5]. To date, about 30–70 % of the extant plant species are polyploidy [6]. The allopolyploid species reunite two or more sets of distinct genomes that entail a suite of genomic accommodations [7–9] which give rising to a variety of novel morphological and physiological phenotypes [10–12]. These observations have led to the hypothesis that polyploidy contributes to the diversification of angiosperm plants. Indeed, it has been demonstrated that 15 % of angiosperm speciation events are accompanied by ploidy increase [13]. In the grass tribe, for example, although no directly evidence indicates that the species diversification was accelerated by the allopolyploidy, at least one third of speciation events are associated with genetic allopolyploidy [14]. In addition, a series of studies from diverse plant taxa have documented that the “genomic shock” resulted from polyploidization has profound effects on the genetic architecture (e.g., gene loss), epigenetic modification (e.g., cytosine methylation) and gene expression (e.g., homeolog biased expression) [15–19], and some of these induced changes are linked to the phenotypic changes [20–22]. These attributes together suggested that polyploidy itself, as a mode of speciation and an avenue that generating novel variations, has indeed contributed to the evolution and diversification of plants.
Panax L. (Araliaceae) is a medicinally important genus in the East Asia and almost every species within the genus has cultural significance for traditional Chinese medicine [23]. The taxonomy of Panax has been controversial due to the circumscription of P. pseudoginseng and P. japonicus [24–26]. For example, all of the species from southwestern China have been treated as the varieties of P. pseudoginseng [27]. However, Zhou et al. [25] moved some P. pseudoginseng varieties (e.g., P. pseudoginseng var. bipinnatifidus) into the species P. japonicus based on their triterpenoids and seed morphologies. Thereafter, Wen and colleagues have reconstructed the phylogenetic trees of Panax based on nrITS and selected chloroplast genes [23, 28–30]. To date, seven well-recognized species and one species complex are defined according to their geographic distributions, chromosome numbers and phylogenetic relationships [30]. Based on the phylogenetic tree and chromosome number, Yi et al. [31] have proposed that at least two recent polyploidy events have occurred within the genus Panax, one of which has led to the formation of three geographically isolated tetraploid (2n = 48) species P. ginseng, P. japonicus and P. quinquefolius. The other recent polyploidy event had occurred within the P. bipinnatifidus species complex wherein both diploids (2n = 24) and tetraploids are identified. These previous studies provide a framework for understanding the evolutionary history of genus Panax. However, these phylogenetic analyses are mainly based on nrITS and selected chloroplast genes, the relationships and nucleotide variation patterns of diploid and tetraploid species remained uninvestigated. In addition, the fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH) analyses revealed the allotetraploid of P. ginseng [32, 33]. More importantly, recent investigations based on the expressed sequence tags (ESTs) suggested that the tetraploid species P. ginseng and P. quinquefolius have experienced two rounds of WGD and diverged to each other after the recent tetraploidization event [34, 35]. These features suggested that the evolutionary trajectories of Panax species are much more complicated than we thought.
In this study, we employed 12 chloroplast genomes of Panax and relative genera to address if the ancient WGD has occurred before the establishment of genus Panax. To further infer the evolutionary trajectories of the extant Panax species, we applied nrITS, four chloroplast and seven single copy nuclear genes to investigate the phylogenetic relationships of the diploid and tetraploid species. To evaluate the impacts of polyploidization on the genetic diversity, we investigated the nucleotide variation pattern of two tetraploids, P. ginseng and P. quinquefolius, and one diploid species P. notoginseng based on 36 single copy nuclear genes. In comparison with the other congeneric species, the three economically important species are well recognized and cultivated widely in East Asia. The tetraploid species P. ginseng and P. quinquefolius have been used as a tonic and fatigue-resistance medicine in East Asia for a long time. Likewise, the diploid species P. notoginseng is considered to be a remedy for preventing bleeding and recovering from injury for thousands of years [30]. We expect our study shed lights on how the polyploidization, geographic and ecological isolations contribute to the evolution and diversification of genus Panax.
Results
Phylogenetic analyses of panax
The geographic distributions and chromosome numbers of the Panax species are shown in Fig. 1. The lengths and informative characters of each alignment and detailed information of the specimens were presented in Additional file 1: Table S1 and Additional file 2: Table S2. In brief, the combined matrix of the four chloroplast genes includes 3031 characters, of which 211 (7.0 %) are variable sites. Similarly, the alignment of whole chloroplast genome of the 12 species contains a total of 144,303 bp in length and 11,506 (8.0 %) of which are polymorphic sites. In contrast, the percentages of informative characters in nrITS and single copy nuclear genes are apparently higher than those of chloroplast genes, which ranged from 8.2 % in Z8 to 25.6 % in nrITS (Additional file 2: Table S2). The numbers of haplotype of the seven nuclear genes were shown in Additional file 2: Table S2 and accession numbers of the DNA sequences downloaded from GenBank were listed in Additional file 3: Table S3.
Fig. 1.
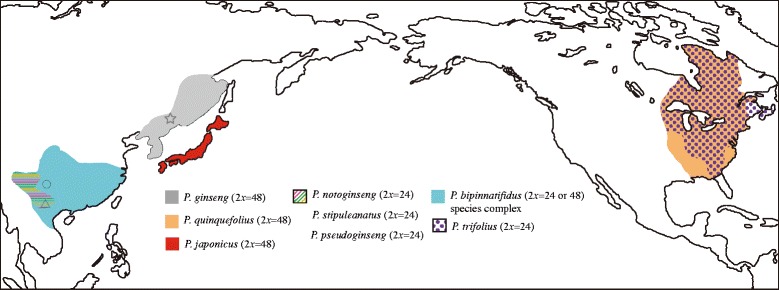
Geographic distributions and chromosome numbers of the extant diploid and tetraploid Panax species. The diploid species P. trifolius shows overlapped distribution with the tetraploid P. quinquefolius. The P. bipinnatifidus species complex covers the distribution ranges of P. notoginseng, P. stipuleanatus and P. pseudoginseng. Star, sampling locations of the species P. ginseng and P. quinquefolius in Jilin province of China; Circle, sampling location of P. bipinnatifidus species complex in Sichuan province of China; Triangle, sampling locations of P. notoginseng, P. stipuleanatus and P. bipinnatifidus species complex in Yunnan province of China. The original map was downloaded from Wikimedia Commons(https://commons.wikimedia.org/wiki/File:Map_of_the_Pacific_region.svg?uselang=zh-cn). The information of geographic distributions of Panax species was retrieved from the Natural Resources Conservation Service of USDA (www.plants.usda.gov) and Flora of China (www.efloras.org)
Phylogenetic reconstruction using Bayesian inference (BI) resulted in distinct topologies between the chloroplast and nuclear datasets (Fig. 2 and 3). In detail, the BI tree based on whole chloroplast genome revealed that species of genera Aralia and Panax grouped together as a clade, supporting previous observation that the two lineages are the closest genera within family Araliaceae (Fig. 2a). To this end, we employed the Aralia species as outgroup when we performed the phylogenetic analyses of the Panax species. As shown in the Fig. 2b, the North American diploid species P. trifolius was placed at the basal clade with high support value (poster prior value = 1.00). Likewise, the two Asiatic diploid species P. pseudoginseng and P. stipuleanatus formed a monophyletic clade and showed distinct phylogenetic positions to the other Asiatic species. It should be noted that the remaining species were separated into two distinct lineages, one of which contains the three tetraploid species P. ginseng, P. japonicus and P. quinquefolius, and the other clade includes the P. notoginseng and P. bipinnatifidus species complex. These features suggested that the two lineages shared the ancestral chloroplast genome and differed from the three basal diploid species.
Fig. 2.
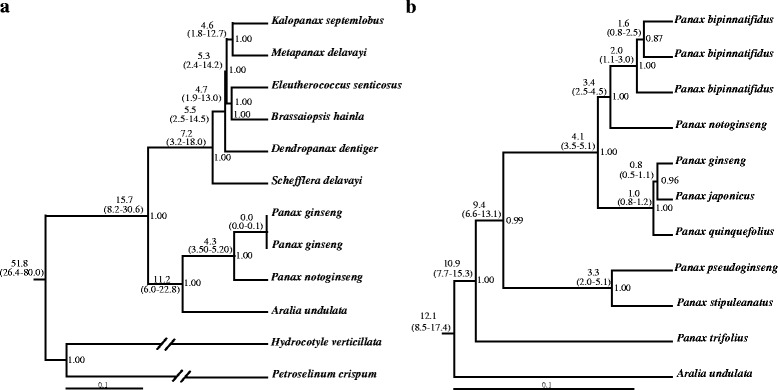
Divergence times and topologies of Bayesian trees based on whole chloroplast genome (a) and four selected chloroplast genes (b). The values on the left and right of each node are the divergence time (one million years ago) and poster prior support, respectively. Length of each branch is not shown in the two phylogenetic trees
Fig. 3.
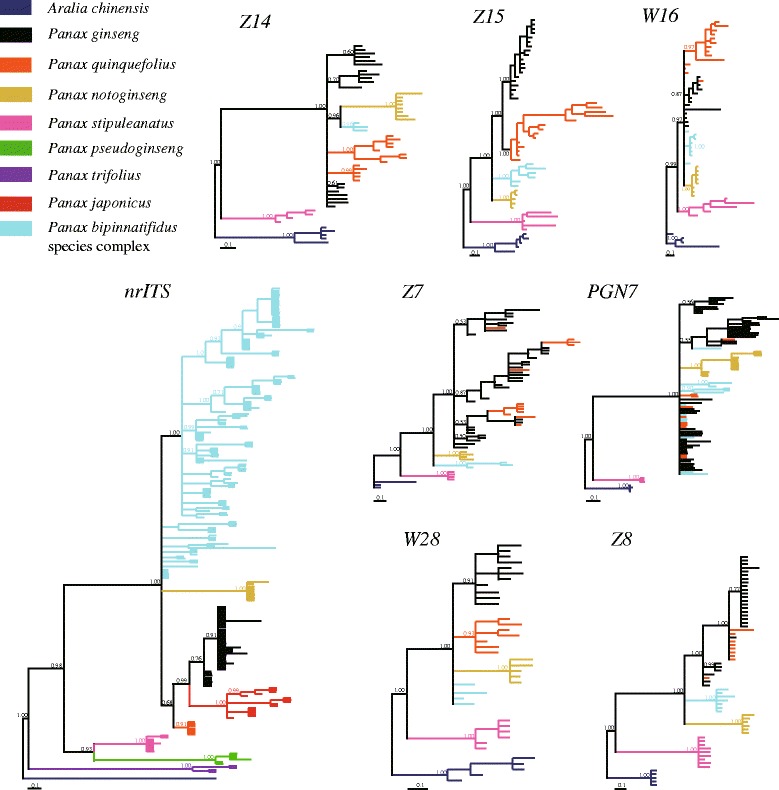
Topologies of Bayesian trees based on nrITS and seven single copy nuclear genes. Each branch represents one haplotype and identical sequences from the same species were removed. The color of branch stands for different species. Numbers of haplotypes for each gene were shown in Additional file 2: Table S2
In contrast, the BI trees of nrITS and seven nuclear genes revealed more complicated phylogenetic topologies for the Panax species (Fig. 3). For example, although the phylogenetic positions of the three basal species, P. trifolius, P. stipuleanatus and P. pseudoginseng, showed no significant differences between chloroplast and nrITS topologies, accessions of the P. bipinnatifidus species complex were not clustered together with P. notoginseng as a monophyletic clade in the BI tree of nrITS (Fig. 3). Instead, they exhibited polyphyletic pattern and then grouped together with the P. notoginseng and three tetraploid species P. ginseng, P. japonicus and P. quinquefolius. The P. bipinnatifidus accessions used in this study contains both diploids and tetraploids and cover their current distributions from southeastern and southwestern China. The polyphyletic pattern suggested the possibility of heterogeneous origins of this species complex. Similarly, topologies of the seven nuclear genes also revealed that P. stipuleanatus showed a distinct phylogenetic position to the other Asiatic species (Fig. 3). These findings suggested that the P. notoginseng and P. bipinnatifidus species complex are more close to the tetraploid species P. ginseng and P. quinquefolius than those of the basal diploid species (Fig. 3). However, we noted that the nrITS topology showed an autotetraploid pattern of the three tetraploid species. In contrast, topologies of the seven nuclear genes revealed that the haplotypes of P. ginseng and P. quinquefolius mixed together at most of these nuclear genes, clearly supporting the allotetraploid of the two species. Taken together, our results based on chloroplast and nuclear genes indicated that P. ginseng and P. quinquefolius are allotetraploid and all accessions of the P. bipinnatifidus species complex have the same maternal origin.
Whole genome duplication and divergence time
Previous investigations based on expression sequence tags (ESTs) have documented that the tetraploid (2n = 48) species P. ginseng and P. quinquefolius have experienced two rounds of WGD [34, 36]. To this end, we estimated the divergence times of the Panax species based on four chloroplast genes and whole chloroplast genome, respectively. Estimations of the divergence time showed that the genus Panax diverged from Aralia some 11.2 million years ago (MYA) (95 % confidence interval (CI): 6.0–22.8 MYA) for whole chloroplast genome data (Fig. 2a) and 12.1 MYA (CI: 8.5–17.4 MYA) for the four chloroplast genes (Fig. 2b), respectively. Thereafter, the basal species P. trifolius and the ancestor of P. stipuleanatus and P. pseudoginseng diverged from the remaining species before 9.4 MYA (CI: 6.6–13.1 MYA) (Fig. 2b). Notably, our results revealed that the three tetraploid species, P. ginseng, P. japonicus and P. quinquefolius, shared the same maternal donor and diverged to each other during 0.8–1.0 MYA (CI: 0.5–1.2 MYA) (Fig. 2b). In contrast, the divergence time between P. notoginseng and P. bipinnatifidus species complex is earlier than those of the three tetraploid species. It was suggested that the P. bipinnatifidus species complex have also experienced recent WGD [31]. In our study, the exact origins of the P. bipinnatifidus species complex can not be determined due to the limited sampling size used at the single copy nuclear genes and undetermined chromosome numbers. However, our phylogenetic results showed that the Asiatic diploid species P. notoginseng shared the maternal genome with the P. bipinnatifidus species complex (Fig. 2b) but showed independent phylogenetic position at the nrITS and nuclear genes (Fig. 3). Likewise, the Asiatic diploid species P. stipuleanatus was placed at the basal clade at the chloroplast, nrITS and nuclear genes, suggesting that it was not involved in the two recent WGD events. Similar phenomenon was also found in the two North American species where demonstrated that although both of the diploid P. trifolius and tetraploid P. quinquefolius are distributed in the North America, the two species fall into two distinct clades (Fig. 2b and 3).
Nucleotide diversity
To estimate if the orthologs showed heterogeneous evolutionary rates among the diploid and tetraploid species, we compared the nucleotide variation pattern of P. notoginseng, P. ginseng and P. quinquefolius based on 36 single copy nuclear genes. As shown in our results, the diploid species P. notoginseng harbored significantly lesser number of variations at total (St), synonymous (Ssyn) and nonsynonymous (Snon) sites than those of the two tetraploid species P. ginseng and P. quinquefolius (Fig. 4 and Additional file 4: Table S4, t-test, all p values < 0.003). For instance, the St of P. notoginseng ranged from 0 (locus W13, W31 and W59) to 58 (locus W48), while the St varied from 5 (locus W28 and W31) to 92 (locus Z63) and 4 (locus W28) to 94 (locus Z63) in P. ginseng and P. quinquefolius, respectively (Additional file 4: Table S4). Similar results were also observed at the parameter πT where most of the 36 genes showed obviously lower nucleotide diversity in P. notoginseng than those of P. ginseng and P. quinquefolius (Additional file 4: Table S4). In particular, we noted that the decreasing of nucleotide diversity at exon regions of the 36 nuclear genes is more apparent than that of the intron regions (Additional file 4: Table S4). For example, ten of the 36 nuclear genes in P. notoginseng showed no variations at the exon regions, but both synonymous and nonsynonymous muations were reported in the P. ginseng and P. quinquefolius. In addition, the P. notoginseng also showed significantly lower ka/ks values compared to the tetraploid species P. ginseng and P. quinquefolius (Additional file 4: Table S4, t-test, both p values < 0.03).
Fig. 4.
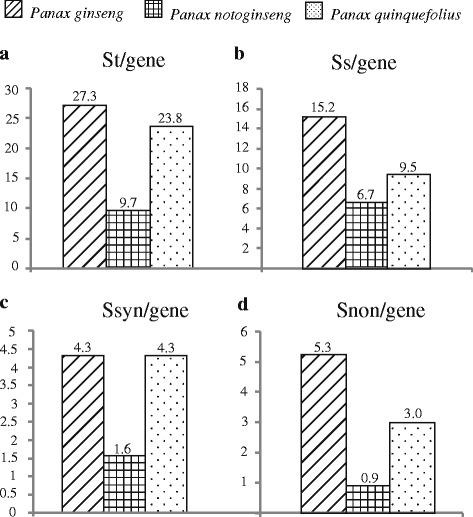
Quantification of segregating site per gene at total a, species-specific b synonymous c and nonsynonymous d sites for the 36 genes. The numbers above the vertical bars are the exact numbers of segregating sites per gene. Detailed information of the 36 nuclear genes was shown in Additional file 4: Table S4
To further evaluate the impacts of tetraploidization on the genetic constitution of tetraploid species, we compared the nucleotide variation pattern between the two tetraploids P. ginseng and P. quinquefolius. Our results revealed that the Asiatic tetraploid P. ginseng harbored slightly greater number of St than that of the North America tetraploid P. quinquefolius (Fig. 4 and Additional file 4: Table S4, t-test, p = 0.364). Notably, the two tetraploid species exhibited distinct nucleotide variation pattern at the exon regions (Fig. 4 and Additional file 4: Table S4). For instance, most of the 36 nuclear genes showed higher nonsynonymous mutation rates in the P. ginseng compared to P. quinquefolius (Fig. 4 and Additional file 4: Table S4, t-test, p = 0.02). Similarly, ka/ks values of the 36 nuclear genes also exhibited obviously different between P. ginseng and P. quinquefolius (Additional file 4: Table S4). It should be noted that each of the three species possessed high level of species-specific SNPs (Fig. 4 and Additional file 4: Table S4). For example, although the two tetraploid P. ginseng and P. quinquefolius diverged recently, 495 and 313 SNPs are specific to each of the two tetraploids.
Discussion
Ancient and recent polyploidy followed by geographic and ecological isolations
Polyploidy is a widespread feature of plant genomes and has played a crucial role in the evolution and diversification of plants [37]. In terms of time of origin, polyploidy can be broadly divided into paleopolyploidization (ancient WGD) and neopolyploidization (recent WGD) [38]. The recent polyploidy events are easily identified by the chromosome numbers, genome size, and gene copy number relative to progenitors. In contrast, the evidence of ancient polyploidy has mainly come from comparative genetic mapping, analysis of specific gene families or by the identification of duplicated genes in ESTs [39].
The contributions of ancient WGD on the evolution and diversification of plants are well-recognized [40–42]. In the case of legumes, for example, multiple independent polyploidy events had occurred in the early radiation stage and which might provide raw materials for the genetic innovations that resulted in the evolution of symbiotic nitrogen fixation [43–45]. In Panax, previous studies based on the ESTs indicated that the extant tetraploid species, P. ginseng and P. quinquefolius, have undergone two rounds of WGD, of which, the first round of WGD had occurred during 24.6–32.8 MYA [34, 35]. Here, our results based on phylogenetic and divergence time analyses suggested that the genus Panax have experienced both ancient and recent WGDs. Given that both of genera Panax and Aralia have the same basic chromosome number (n = 12) [31] and diverged to each other obviously later than that of the first round WGD, we proposed that the ancient WGD might have occurred before of the establishment of genus Panax. Under this hypothesis, it is tempting to predict that the extant diploid species of Panax are paleopolyploid, which is thought to be a common phenomenon in plants [46]. We noted that the genome sizes of extant diploid and tetraploid Panax species vary dramatically [47–49]. Similar observations were also reported in Gossypium and Arabidopsis where rapid genomic revolution during and/or soon after WGD and gradual process of diploidization are likely to result in variation and evolution in genome size [50–53]. Taken together, our findings indicated that the ancient WGD might have contributed to the evolution and diversification of Panax. In addition to the ancient WGD, recent polyploidy events were also revealed by our phylogenetic and divergence time analyses. However, we noted that the nrITS topology did not show the allotetraploid of the three species P. ginseng, P. japonicus and P. quinquefolius, which is not consistent with previous observations based on FISH and GISH [33]. The possible explanation might be that the orthologs from distinct genomes were homogenized through concerted evolution. As expected, topologies of the seven single copy nuclear genes confirmed the allotetraploid of the three species. It should be noted that both the four selected chloroplast genes and nrITS topologies suggested the single tetraploidization origin of the three tetraploid species. Similar phenomenon was also reported in the Gossypium where five extant tetraploid species (AADD) have derived from a single polyploidization event between G. raimondii (DD) and two extant A-genome species about 1–2 MYA [54–57]. In our case, however, the diploid species used in this study may not the direct progenitors of the three tetraploid species. Instead, our phylogenetic results suggested a possibility that the three tetraploids have the same maternal donor and might share the parental ancestor with P. notoginseng and P. bipinnatifidus specie complex. Similar phylogenetic patterns were also observed in the Panicum where the two teraploids P. miliaceum and P. repens shared the same parental genome but have distinct maternal donors [58]. To this end, it is possible that the direct donors of the three tetraploid species may not exist at present. It has also been suggested that recent WGD had occurred within the P. bipinnatifidus species complex [31]. In our study, although the diploid species P. notoginseng showed overlapped distributions with the P. bipinnatifidus species complex, phylogenetic analyses indicated that it might not be involved in the recent WGD of P. bipinnatifidus species complex. Instead, the observed polyphyletic pattern suggested that polyploids within the P. bipinnatifidus species complex might have formed through autopolyploidization. Together, our findings suggested that the recent WGDs have indeed promoted the diversification of Panax.
It has been demonstrated that the genus Panax shows a disjunct distribution between eastern Asia and eastern North America [59–61]. Here, our results confirmed previous hypothesis of two independent origins of the disjunct distributions of Panax [23, 28]. In particular, we noted that the diploid species P. trifolius was not involved in the tetraploidization of P. quinquefolius, although they showed overlapped distribution pattern in the eastern North America. In contrast, despite the three geographic isolated tetraploids, P. ginseng, P. quinquefolius and P. japonicus, are endemic to northeastern Asia (excluding Japan), North America and Japan, respectively, they have established through a single tetraploidization event and diverged almost simultaneous (0.5–1.2 MYA). These features suggested that geographic isolation is likely one of the underlying mechanisms that promoted the divergence of the three tetraploids. In addition, it has been reported that, in the P. bipinnatifidus species complex, tetraploids usually occur at high altitudes [28, 29]. Similar observations were also reported in the Alyssum montanum-A. repens complex in which the polyploidy provides raw materials for diversification and the geographic and ecological isolation have further stimulated speciation [62]. Under this hypothesis, our findings suggested that multiple rounds of ancient and recent polyploidization, along with geographic and ecological isolations, might have together played important roles in the evolution and diversification of Panax.
Nucleotide diversity of diploid and tetraploid species
It was widely recognized that WGD has profound effects on the genome constitution of plants [8, 11, 15, 17, 19, 63]. The notable feature of polyploidy is that it would increase the copy numbers of a given gene. As a result, orthologs in the polyploids would harbor relatively higher genetic diversity and heterozygosity compared to the diploids, mainly due to the relaxed selection and reuniting of multiple parental copies [16, 64–67]. In the case of Gossypium, for example, the population genetic analyses based on 48 nuclear genes showed that polyploidy in Gossypium has led to a modest enhancement in rates of nucleotide substitution [68]. Here, our study also demonstrated that the two tetraploid species, P. ginseng and P. quinquefolius, showed relatively higher nucleotide diversity at the total sites of the 36 nuclear genes than those of the diploid species P. notoginseng. The possible explanation might be that the two allotetraploids possessed two divergent genomes which would increase the heterozygous and nucleotide diversity at the genome-wide level. In addition, the limited sampling size of P. notoginseng might be also respossible for the low nucleotide diversity. Notably, we found that a vast of majority of SNPs is specific to each of the three species, suggesting that some of these SNPs might have accumulated after their divergent. Given the recent divergence and allopatric distributions of the three species, we propose that, in addition to the effects of recent WGD, geographic isolation might have also contributed to the distinct variation patterns of the three species.
Previous studies have suggested that gene duplication plays a crucial role in the coding sequence evolution [64, 69–71]. In the hexaploid wheat, duplicated orthologs that created by WGD can change the dynamic of coding sequence evolution through relaxing selection and then provide chances for the accumulation of new mutations which may impact gene function [72]. In our study, we found that, compared to the introns of the 36 nuclear genes, the deceasing in nucleotide diversity at exons in diploid species is more apparent than those of the two tetraploid species. In particular, the diploid species showed obviously lower ka/ks values at the 36 nuclear genes. In addition, we also noted that distinct variation pattern was also observed between the two tetraploid species. Taken the locus Z8 as an example, only two synonymous mutations were found in P. quinquefolius, yet eight and five synonymous and nonsynonymous mutations were identified in P. ginseng. These findings allow us to speculate that gene duplication might provide raw materials and natural selection favors different mutations between the diploid and tetraploid species.
Conclusion
WGD is thought to be a driving force that promoted the evolution and diversification of plants. Here, our phylogenetic analyses based on multiple chloroplast and nuclear genome markers demonstrated that the ancient and recent WGDs along with geographic and ecological isolations have together contributed to the diversification of Panax species. Through comparing the nucleotide variation patterns of the diploid and tetraploid species, we found that distinct selection pressures might have acted on these nuclear genes during their evolutionary processes.
Methods
Sampling and DNA extraction
The aims of this study are to infer the phylogenetic relationships of the extant Panax species and evaluate if the same ortholog exhibits heterogeneous evolutionary rates between the diploid and tetraploid species. To this end, 11 and 15 individuals of P. notoginseng (Burkill) Chen ex and P. ginseng were collected from the Yunnan and Jilin provinces of China, respectively. Similarly, seven and eight accessions of P. quinquefolius L. and P. stipuleanatus Tsai and Feng were collected from the Jilin and Yunnan provinces of China, respectively. Samples of these species were collected from a wide geographic area that several populations were included. In addition, four accessions sampled from Yunnan and Sichuan provinces of China were chosen to represent the P. bipinnatifidus Seem. species complex. The exact geographic locations of these samples were shown in Fig. 1. The four species, P. bipinnatifidus, P. stipuleanatus P. ginseng and P. notoginseng, are widely distributed in southwestern and northeastern China and no specific permissions are required for the specimen collection. The species P. quinquefolius is naturally distributed in North America and widely cultivated in North America and northeastern China. We collected seven cultivated accessions of P. quinquefolius from Jilin province of China with the owner’s permission. The remaining 13 accessions of P. quinquefolius were obtained from our collaborator who bought these samples from the market of the United States of America. The exact geographic location of these accessions is unclear. Detailed information of the specimens used in this study is listed in Additional file 1: Table S1. Genomic DNA was extracted from the silica-gel dried leaf material of each accession using Qiagen (Tiangen, Beijing) following the manufacturer’s instructions.
Chloroplast, nrITS and single copy nuclear gene selection
To infer the establishment and evolutionary process of Panax, we downloaded the whole chloroplast genomes of two Panax and nine relative genera species from GenBank (Panax ginseng, KF431956 and KC686332; Panax notoginseng, KJ566590; Aralia undulata, KC456163; Dendropanax dentiger, KP271241; Metapanax delavayi, KC456165; Kalopanax septemlobus, KC456167; Eleutherococcus senticosus, JN637765; Brassaiopsis hainla, KC456164; Schefflera delavayi, KC456166; Hydrocotyle verticillata, HM596070; Petroselinum crispum, HM596073). To further address the evolutionary trajectories of the extant diploid and tetraploid Panax species, we employed the nrITS and four chloroplast genes (trnD, psbK-psbI, rbcL and ycf1) to reconstruct the phylogenetic trees (Additional file 2: Table S2 and Additional file 3: TableS 3). The nrITS region is one of the most popular nuclear DNA regions in molecular phylogenetic studies, yet the intra-individual paralogy [73–75] and concerted evolution have largely limited its application in the phylogenetic work, especially in the polyploid species. Instead, single or low copy nuclear genes have been proposed to be particularly useful in resolving such problems and are an increasingly popular alternative to nrITS [76]. To this end, 53 single copy nuclear genes were selected according to our previous studies [77, 78], 36 of which were successfully amplified in the diploid species P. notoginseng and tetraploid species P. ginseng and P. quinquefolius (Additional file 5: Table S5). Seven genes that showed high transferability across the genera Panax and Aralia were used to construct the phylogenetic trees (Additional file 2: Table S2).
PCR, sequencing and phylogenetic analyses
Polymerase chain reactions (PCRs) of the single copy nuclear genes were performed in a 50 μL volume containing 0.2 mM of each dNTP, 1.5 mM MgCl2, 0.5 mM of each primer, 1U of rTaq polymerase (Takara, Dalian, China), and about 50 ng of DNA template under the following conditions: 5 min at 95 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at the annealing temperature of each primer combination (Additional file 5: Table S5), 60 s at 72 °C, and then a final 5 min extension at 72 °C. The amplifications of seven single copy nuclear genes were purified with Gel Band Purification Kit (Tiangen, Beijing, China) and cloned using pMD18 vector (Takara, Dalian, Liaoning) following the manufacturer’s instructions. To obtain different haplotypes of the seven nuclear genes, multiple accessions of each species were selected and 4–10 clones were sequenced for each accession studied.
The DNA sequences were aligned using the default parameters in Clustal [79] and edited manually using BioEdit [80] if necessary. To infer the phylogenetic relationships of the Panax species, the BI analyses for the nrITS, combined chloroplast and single copy nuclear genes were performed using MrBayes [81], separately. Model parameters for each data set were estimated using jModelTest [82]. The best-fit models for each data set were showed in Additional file 2: Table S2. For the Bayesian trees, two independent Markov chains were run and calculated simultaneously with 1,000,000 generations for each data set. The convergence of the two runs was evaluated by stopping the analysis when the average standard deviation was below 0.01. Bayesian posterior probabilities were estimated as the majority consensus of all sampled trees with the first 25 % discarded as burn-in. The divergence times of Panax and relative genera were calculated using mcmctree of PAML [83, 84]. The indepented rates and HKY85 were chosen as the molecular clock and nucleotide substitution model, respectively. The ambiguity characters were removed from alignments. The empirical divergence times of P. ginseng/P. quinquefolius (0.8–1.2 MYA) and P. ginseng/P. notoginseng (3.5–5.2 MYA) [35, 36] were assigned to constrain the age of the Panax. A Birth-Death prior on branching rates was employed and three independent analyses were run for 10,000 generations.
SNP recalibrating and nucleotide diversity
The references of the 36 single copy nuclear genes were obtained from our previous studies [77, 78]. The population data of P. ginseng, P. quinquefolius and P. notoginseng were sequenced using Illumina Hiseq 2000 (BGI, Shenzhen, China). The quality of raw reads was checked using FastQC [85] and low-quality (Phred < 30) reads were removed. Alignments of the clean reads were initially screened against the obtained references using Burrows-Wheeler Aligner [86]. The low quality single nucleotide polymorphisms (SNPs) (mapping quality < 30, depth < 10) and PCR duplicates were removed from the mapped reads using SAMtools [87]. The heterozygous and homozygous SNPs were reported according to our previous study [78]. The Perl scripts were applied to generate the alignment for each gene by replacing the references with reported SNPs. Insertions/deletions (INDELs) were excluded from the subsequent data analyses. Accordingly, a total of 0.55 million 100 bp paired-end reads (low quality reads and PCR duplicates were removed) were mapped to the references. We therefore obtained an average of ~80.6 × coverage for each gene per individual. The numbers of species-specific SNPs for each species were estimated based on the total segregating sites of the three species. The nucleotide diversity of the three Panax species was calculated using DnaSP v5 [88], including number of segregating sites (S), ration of nonsynonymous and synonymous site (Ka/Ks), nucleotide diversity π [89] for total, nonsynonymous and synonymous sites, respectively. The segregating sites that showed monomorphic within each of the three species were not included in the analyses of nucleotide diversity.
Availability of supporting data
All data generated from this study were submitted to GenBank under the accession number KT593555-KT593862 and PRJNA291547.
Acknowledgments
We thank Haifei Yan for providing the materials of Panax species. This work was financially supported by the National Natural Science Foundation of China (31470010).
Additional files
Accessions of Panax species and outgroups sampled from the 36 single copy nuclear genes. (DOCX 43 kb)
Detailed information of the chloroplast and nuclear genes used in this study. (DOCX 58 kb)
Accession numbers of the Panax species and outgroups used in this study. (DOCX 23 kb)
Nucleotide variation patterns of the 36 single copy nuclear genes used in this study. (DOCX 34 kb)
Detailed information of the 36 single copy nuclear genes used in this study. (DOCX 22 kb)
Footnotes
Feng-Xue Shi, Ming-Rui Li and Ya-Ling Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FXS and MRL carried out the molecular genetic studies, performed the phylogenetic analyses and drafted the manuscript. YLL analyzed the high throughput data of the nuclear genes and performed the population genetic analyses. PJ and CZ developed the single copy nuclear genes and tested their transferability. HXX and YZP collected the samples and helped to draft the manuscript. LFL, HXX and BL participated in the design and coordination of this study. LFL and BL wrote this manuscript. All authors read and approved the final manuscript.
Contributor Information
Feng-Xue Shi, Email: shifx816@nenu.edu.cn.
Ming-Rui Li, Email: limr145@nenu.edu.cn.
Ya-Ling Li, Email: liyl378@nenu.edu.cn.
Peng Jiang, Email: jiangp446@nenu.edu.cn.
Cui Zhang, Email: zhangc034@nenu.edu.cn.
Yue-Zhi Pan, Email: panyuezhi@mail.kib.ac.cn.
Bao Liu, Email: baoliu@nenu.edu.cn.
Hong-Xing Xiao, Phone: +86043185099126, Email: xiaohx771@nenu.edu.cn.
Lin-Feng Li, Phone: +86043185099367, Email: lilf241@nenu.edu.cn.
References
- 1.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 2.Wall PK, Soltis PS, DePamphilis CW, Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 3.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–U113. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 5.Renny-Byfield S, Wendel JF. Doubling down on genomes: polyploidy and crop plants. Am J Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- 6.Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 1998;29:467–501. doi: 10.1146/annurev.ecolsys.29.1.467. [DOI] [Google Scholar]
- 7.Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee H-S, et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/S0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 8.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. In: Annu Rev Plant Biol; 2007. pp. 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams KL. Evolution of duplicate gene expression in polyploid and hybrid plants. J Hered. 2007;98:136–141. doi: 10.1093/jhered/esl061. [DOI] [PubMed] [Google Scholar]
- 11.Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 13.Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estep MC, McKain MR, Diaz DV, Zhong J, Hodge JG, Hodkinson TR, et al. Allopolyploidy, diversification, and the Miocene grassland expansion. Proc Natl Acad Sci U S A. 2014;111:15149–15154. doi: 10.1073/pnas.1404177111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, et al. Evolutionary genetics of genome merger and doubling in plants. In: Annu Rev Genet; 2008. pp. 443–461. [DOI] [PubMed] [Google Scholar]
- 16.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 17.Adams KL, Wendel JF. Dynamics of Duplicated Gene Expression in Polyploid Cotton. In: Chen ZJ, Birchler JA, editors. Polyploid and Hybrid Genomics. Oxford, UK: John Wiley & Sons, Inc.; 2013. [Google Scholar]
- 18.Jiao Y, Paterson AH. Polyploidy-associated genome modifications during land plant evolution. Philos Trans R Soc Lond B Biol Sci. 2014;369(1648):20130355. doi: 10.1098/rstb.2013.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soltis DE, Visger CJ, Soltis PS. The polyploidy revolution then…and now: Stebbins revisited. Am J Bot. 2014;101:1057–1078. doi: 10.3732/ajb.1400178. [DOI] [PubMed] [Google Scholar]
- 20.Pires JC, Zhao J, Schranz M, LEON EJ, Quijada PA, Lukens LN, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linn Soc. 2004;82:675–688. doi: 10.1111/j.1095-8312.2004.00350.x. [DOI] [Google Scholar]
- 21.Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni Z, Kim E-D, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–U327. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi H-K, Wen J. A phylogenetic analysis of Panax (Araliaceae): Integrating cpDNA restriction site and nuclear rDNA ITS sequence data. Plant Syst Evol. 2000;224:109–120. doi: 10.1007/BF00985269. [DOI] [Google Scholar]
- 24.Hara H. On the Asiatic species of the genus Panax. J Japanese botany. 1970;45(7):197–212. [Google Scholar]
- 25.Zhou J, Huang W, Wu M, Yang C, Feng K, Wu Z. Triterpenoids from Panax Linn. and their relationship with taxonomy and geographical distribution. Acta Phytotaxon Sin. 1975;13:29–45. [Google Scholar]
- 26.Hoo G, Tseng Cj, Tsai SC. Flora reipublicae popularis Sinicae delectis florae reipublicae popularis Sinicae agendae academiae Sinicae edita: Tom 54. Angiospermae. Dicotyledoneae. Araliaceae. Facultas Biologica Universitatis Amoiensis; Beijing, 1978.
- 27.Ho C, Tseng C. On the Chinese species of Panax Linn. Acta Phytotaxonom Sinica. 1973. [Google Scholar]
- 28.Wen J, Zimmer EA. Phylogeny and biogeography of Panax L. (the ginseng genus, araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol. 1996;6:167–177. doi: 10.1006/mpev.1996.0069. [DOI] [PubMed] [Google Scholar]
- 29.Lee C, Wen J. Phylogeny of Panax using chloroplast trnC-trnD intergenic region and the utility of trnC-trnD in interspecific studies of plants. Mol Phylogen Evol. 2004;31:894–903. doi: 10.1016/j.ympev.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Zuo Y, Chen Z, Kondo K, Funamoto T, Wen J, Zhou S. DNA barcoding of Panax species. Planta Med. 2011;77:182–187. doi: 10.1055/s-0030-1250166. [DOI] [PubMed] [Google Scholar]
- 31.Yi T, Lowry PP, Plunkett GM. Chromosomal evolution in Araliaceae and close relatives. Taxon. 2004;53:987–1005. doi: 10.2307/4135565. [DOI] [Google Scholar]
- 32.Choi HW, Koo DH, Bang KH, Paek KY, Seong NS, Bang JW. FISH and GISH analysis of the genomic relationships among Panax species. Genes Genom. 2009;31:99–105. doi: 10.1007/BF03191143. [DOI] [Google Scholar]
- 33.Choi HI, Waminal NE, Park HM, Kim NH, Choi BS, Park M, et al. Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J. 2014;77:906–916. doi: 10.1111/tpj.12441. [DOI] [PubMed] [Google Scholar]
- 34.Kim NH, Choi HI, Kim KH, Jang W, Yang TJ. Evidence of genome duplication revealed by sequence analysis of multi-loci expressed sequence tag-simple sequence repeat bands in Panax ginseng Meyer. J Ginseng Res. 2014;38:130–135. doi: 10.1016/j.jgr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi HI, Kim NH, Lee J, Choi BS, Do Kim K, Park JY, et al. Evolutionary relationship of Panax ginseng and P. quinquefolius inferred from sequencing and comparative analysis of expressed sequence tags. Genet Resour Crop Evol. 2013;60:1377–1387. doi: 10.1007/s10722-012-9926-3. [DOI] [Google Scholar]
- 36.Choi HI, Kim NH, Kim JH, Choi BS, Ahn I-O, Lee JS, et al. Development of reproducible EST-derived SSR markers and assessment of genetic diversity in panax ginseng cultivars and related species. J Ginseng Res. 2011;35:399–412. doi: 10.5142/jgr.2011.35.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soltis PS, Soltis DE. Polyploidy and genome evolution. New York: Springer; 2012. pp. 225-249.
- 38.Hilu K. Polyploidy and the evolution of domesticated plants. Am J Bot. 1993;80:1494–1499. doi: 10.2307/2445679. [DOI] [Google Scholar]
- 39.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solds DE, Bell CD, Kim S, Soltis PS. Origin and early evolution of angiosperms. Year in Evol Biol. 2008;1133:3–25. doi: 10.1196/annals.1438.005. [DOI] [PubMed] [Google Scholar]
- 42.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 43.Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/480162a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li QG, Zhang L, Li C, Dunwell JM, Zhang YM. Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae. Mol Biol Evol. 2013;30:2602–2611. doi: 10.1093/molbev/mst152. [DOI] [PubMed] [Google Scholar]
- 45.Cannon SB, McKain MR, Harkess A, Nelson MN, Dash S, Deyholos MK, et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 2015;32:193–210. doi: 10.1093/molbev/msu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tate JA, Joshi P, Soltis KA, Soltis PS, Soltis DE. On the road to diploidization? Homoeolog loss in independently formed populations of the allopolyploid Tragopogon miscellus (Asteraceae) BMC Plant Biol. 2009;9:80. doi: 10.1186/1471-2229-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan YZ, Zhang YC, Gong X, Li FS. Estimation of genome size of four Panax species by flow cytometry. Plant Diversity Res. 2014;36:233–236. [Google Scholar]
- 48.Hong C, Lee S, Park J, Plaha P, Park Y, Lee Y, et al. Construction of a BAC library of Korean ginseng and initial analysis of BAC-end sequences. Mol Genet Genomics. 2004;271:709–716. doi: 10.1007/s00438-004-1021-9. [DOI] [PubMed] [Google Scholar]
- 49.Obae GS. Nuclear DNA, content and genome size of American ginseng. J Med Plants Res. 2012;6:4719–4723. doi: 10.5897/JMPR11.221. [DOI] [Google Scholar]
- 50.Cronn RC, Small RL, Haselkorn T, Wendel JF. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am J Bot. 2002;89:707–725. doi: 10.3732/ajb.89.4.707. [DOI] [PubMed] [Google Scholar]
- 51.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 52.Hendrix B, Stewart JM. Estimation of the nuclear DNA content of gossypium species. Ann Bot. 2005;95:789–797. doi: 10.1093/aob/mci078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf DE, Steets JA, Houliston GJ, Takebayashi N. Genome size variation and evolution in allotetraploid Arabidopsis kamchatica and its parents, Arabidopsis lyrata and Arabidopsis halleri. . AoB Plants. 2014;6:plu025. doi: 10.1093/aobpla/plu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wendel JF. New world tetraploid cottons contain old world cytoplasm. Proc Natl Acad Sci U S A. 1989;86:4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- 56.Grover CE, Grupp KK, Wanzek RJ, Wendel JF. Assessing the monophyly of polyploid Gossypium species. Plant Syst Evol. 2012;298:1177–1183. doi: 10.1007/s00606-012-0615-7. [DOI] [Google Scholar]
- 57.Grover CE, Gallagher JP, Jareczek JJ, Page JT, Udall JA, Gore MA, et al. Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol Phylogenet Evol. 2015;92:45–52. doi: 10.1016/j.ympev.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 58.Hunt HV, Badakshi F, Romanova O, Howe CJ, Jones MK, Heslop-Harrison JS. Reticulate evolution in Panicum (Poaceae): the origin of tetraploid broomcorn millet. P miliaceum J Exp Bot. 2014;65:3165–3175. doi: 10.1093/jxb/eru161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li HL. Floristic relationships between eastern Asia and eastern North America. Trans Am Philos Soc. 1952;42:371–429. doi: 10.2307/1005654. [DOI] [Google Scholar]
- 60.Zhengyi W. On the significance of Pacific intercontinental discontinuity. Ann Mo Bot Gard. 1983;70:577–590. doi: 10.2307/2398977. [DOI] [Google Scholar]
- 61.Wen J, Nowicke JW. Pollen ultrastructure of Panax (the ginseng genus, Araliaceae), an eastern Asian and eastern North American disjunct genus. Am J Bot. 1999;86:1624–1636. doi: 10.2307/2656799. [DOI] [PubMed] [Google Scholar]
- 62.Zozomová-Lihová J, Marhold K, Španiel S. Taxonomy and evolutionary history of Alyssum montanum (Brassicaceae) and related taxa in southwestern Europe and Morocco: Diversification driven by polyploidy, geographic and ecological isolation. Taxon. 2014;63:562–591. doi: 10.12705/633.18. [DOI] [Google Scholar]
- 63.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 64.Ohno S. The enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Trans Am Fish Soc. 1970;99:120–130. doi: 10.1577/1548-8659(1970)99<120:TEDIGS>2.0.CO;2. [DOI] [Google Scholar]
- 65.Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. doi: 10.1023/A:1006392424384. [DOI] [PubMed] [Google Scholar]
- 67.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 68.Senchina DS, Alvarez I, Cronn RC, Liu B, Rong J, Noyes RD, et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol. 2003;20:633–643. doi: 10.1093/molbev/msg065. [DOI] [PubMed] [Google Scholar]
- 69.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 70.Lin J-Y, Stupar RM, Hans C, Hyten DL, Jackson SA. Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell. 2010;22:2545–2561. doi: 10.1105/tpc.110.074229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Adams KL. Duplicate gene divergence by changes in microRNA binding sites in Arabidopsis and Brassica. Genome Biol Evol. 2015;7:646–655. doi: 10.1093/gbe/evv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akhunov ED, Sehgal S, Liang H, Wang S, Akhunova AR, Kaur G, et al. Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 2013;161:252–265. doi: 10.1104/pp.112.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailey C. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol Phylogen Evol. 2003;29:435–455. doi: 10.1016/j.ympev.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 74.Nieto Feliner G, Rossello JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 75.Baldwin BG, Sanderson MJ, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard. 1995;82:247–277. doi: 10.2307/2399880. [DOI] [Google Scholar]
- 76.Zimmer EA, Wen J. Reprint of: using nuclear gene data for plant phylogenetics: progress and prospects. Mol Phylogenet Evol. 2013;66:539–550. doi: 10.1016/j.ympev.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 77.Li MR, Wang XF, Zhang C, Wang HY, Shi FX, Xiao HX, et al. A simple strategy for development of single nucleotide polymorphisms from non-model species and its application in Panax. Int J Mol Sci. 2013;14:24581–24591. doi: 10.3390/ijms141224581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li MR, Shi FX, Zhou YX, Li YL, Wang XF, Zhang C, et al. Genetic and epigenetic diversities shed light on the domestication of cultivated ginseng (Panax ginseng) Mol Plant. 2015;8:1612–1622. doi: 10.1016/j.molp.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 79.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symp Ser; 1999. pp. 95–98. [Google Scholar]
- 81.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 84.Xu B, Yang Z. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- 85.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. Genome project data processing S. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 89.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]


