Abstract
Toll‐like receptor (TLR) 13 and TLR2 are the major sensors of Gram‐positive bacteria in mice. TLR13 recognizes Sa19, a specific 23S ribosomal (r) RNA‐derived fragment and bacterial modification of Sa19 ablates binding to TLR13, and to antibiotics such as erythromycin. Similarly, RNase A‐treated Staphylococcus aureus activate human peripheral blood mononuclear cells (PBMCs) only via TLR2, implying single‐stranded (ss) RNA as major stimulant. Here, we identify human TLR8 as functional TLR13 equivalent that promiscuously senses ssRNA. Accordingly, Sa19 and mitochondrial (mt) 16S rRNA sequence‐derived oligoribonucleotides (ORNs) stimulate PBMCs in a MyD88‐dependent manner. These ORNs, as well as S. aureus‐, Escherichia coli‐, and mt‐RNA, also activate differentiated human monocytoid THP‐1 cells, provided they express TLR8. Moreover, Unc93b1 −/−‐ and Tlr8 −/−‐THP‐1 cells are refractory, while endogenous and ectopically expressed TLR8 confers responsiveness in a UR/URR RNA ligand consensus motif‐dependent manner. If TLR8 function is inhibited by suppression of lysosomal function, antibiotic treatment efficiently blocks bacteria‐driven inflammatory responses in infected human whole blood cultures. Sepsis therapy might thus benefit from interfering with TLR8 function.
Keywords: bacteria, human TLR8, mitochondrial, ribosomal, RNA
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction
Introduction
Toll‐like receptors (TLRs) mediate recognition of pathogen‐ and host‐derived “danger associated molecular patterns” (P‐/DAMPs) driving severe inflammation such as in sepsis, which is mostly induced by Gram‐positive and Gram‐negative bacterial infection 1. Anti‐inflammatory interventions in the acute sepsis phase are thus promising therapeutic approaches 2. Indeed, blockade of TLR2 and TLR4 at the start of antibiotic therapy inhibited human peripheral blood mononuclear cell (PBMC) activation and also saved mice upon Gram‐negative but not Gram‐positive bacterial infection 3. Accordingly, the Sa19 segment of bacterial 23S ribosomal (r) RNA is a murine (m) TLR13 ligand strongly operative primarily in Gram‐positive bacterial infection 4, 5, 6, 7. Sa19 sequence modification, as is frequently found in methicillin‐resistant Staphylococcus aureus (MRSA) strains that are also macrolide, lincosamide and streptogramin antibiotic (MLS) resistant, ablates recognition by TLR13 (termed immune escape) 5, 8. On the other hand, bacterial transfer (t) RNA is recognized via TLR7, and Gram‐negative bacterial RNA activates human (h) TLR8 9, 10, 11. Whether Gram‐positive bacterial RNA drives a strong immune activity immediately upon acute infection also in humans as it had been observed in mice is largely unknown. Previous studies searching for a human pattern recognition receptor (PRR) equivalent to mTLR13 have concluded that mainly TLR2 is operative in response to Gram‐positive bacteria 5, 12.
Here, we demonstrate that not only Sa19, but also variants of it including mitochondrial (mt) 16S rRNA‐borne derivatives strongly stimulate human immune cells via TLR8 exclusively. Ligand recognition of TLR8 was found to be promiscuous, in that its ligand consensus motif was broader than “UGG” 13, since “UAA” and “UGA” motifs were also activating. Human TLR8 thus senses both bacterial pathogen‐derived rRNAs and self‐mitoribosomal (mtr) RNA wherein the latter descends from bacteria according to the endosymbiotic theory. Specifically, we observed that TLR8‐mediated RNA sensing is critically involved in recognition of viable Escherichia coli and S. aureus. For example, the inhibition of lysosomal function, which affects bacteria‐driven TLR8 and also TLR7 activation, allowed antibiotics to inhibit consequent inflammation. Our results identify TLR8 as major bacteria and mitochondria sensor and implicate a novel therapeutic interference in acute inflammation pathology.
Results and Discussion
Specific bacterial and mitochondrial RNAs activate human immune cells dependent on MyD88 and Unc93B1 but largely independent on TLR7
In quest for a human equivalent of TLR13, we analyzed whether the 23S rRNA segment Sa19—with 19 bases of appropriate length 14—or heat‐inactivated S. aureus (hiSa) stimulate PBMCs. Both compounds were active. Thus, blockade of TLR2 with a neutralizing and human/mouse cross‐reactive monoclonal antibody (mAb, named T2.5) was ineffective unless the hiSa solution was incubated with single‐stranded (ss) RNA‐specific RNase A (Fig 1A) 5, 15. We thus concluded that both Sa19 “like” ssRNA segments as well as TLR2 ligands are candidates of S. aureus‐derived immune stimulatory PAMPs.
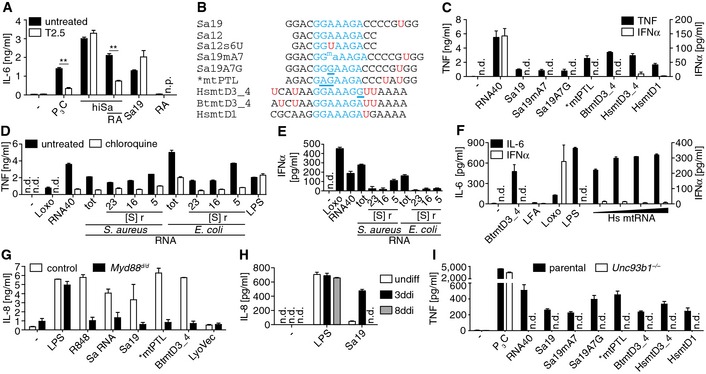
Figure 1. Specific bacterial and mitochondrial ribosomal RNA segments and fractions induce proinflammatory but not type I IFN cytokine production through an endosomal TLR .
-
ACytokine release by PBMCs challenged with heat‐inactivated S. aureus (hiSa), and/or RNase A (RA)‐treated solution, or Sa19 (TLR13 activating ORN) upon TLR2 blockade (T2.5; n.p., not performed; **P ≤ 0.01; n = 3; unpaired t‐test).
-
BAlignment of Sa19 and mitochondrial 16S rRNA Sa19 “like” segment (mt, mitochondrial; PTL, peptidyl transferase loop; D, domain; _, transition region) sequence with common core motif (blue) and uracils (red U); *conserved in human (Hs), cattle (Bt), mouse and rat; G/A underlined, mutated core motif; ma, 6N methylation of adenosine 7.
-
CCytokine release of PBMCs transfected with ORNs including TLR7/8 ligand RNA40 (n = 3).
-
D, EActivity of PBMCs upon bacterial RNA challenge, pretreatment with chloroquine in (D) only (S, Svedberg; r, ribosomal; tot, total; n = 3).
-
FActivity of PBMCs upon mitochondrial (mt) RNA challenge (Hs, human; triangle, increasing doses; LFA, Lipofectamine 2000; n = 3).
-
GActivity of PBMCs challenged with ORNs and total S. aureus (Sa) RNA (Myd88 d/d, mutant MyD88 expression not impairing LPS‐driven IL‐8 production; n = 2).
-
HResponsiveness of undifferentiated (undiff) and 3‐ or 8‐day PMA‐differentiated (ddi) THP‐1 cells to Sa19 challenge (n = 3).
-
IActivity of parental and Unc93b1 −/−‐3ddiTHP‐1 cells challenged with ORNs (n = 3).
Also, self‐RNA–antimicrobial peptide complexes stimulate human dendritic cells (DCs) via TLRs and the endosymbiotic theory implies that self‐mitochondria originate from bacteria 16, 17. Since 23S bacterial rRNA comprises Sa19, we analyzed the sequence of its 16S mtrRNA human, cattle, mouse and rat orthologs and designed specific 19‐mer Sa19 “like” oligoribonucleotides (ORNs, Figs 1B and EV1A and B). The 16S mtrRNA Sa19 orthologous segment termed *mtPTL, like Sa19 located within domain V of the large rRNA is conserved yet mutated toward “GAGAAGA”, while integer Sa19 “GGAAAGA” motifs are contained in other regions of 16S mtrRNAs 18. We also used two Sa19 adenosine (A) 7 variants that lack murine immune cell stimulatory capacity (Figs 1B and EV1C and D) 5, 7. None of the Sa19 “like” RNA segments applied activated mTLR13, and only *mtPTL carrying a TLR7‐prone “UAU” motif activated the murine immune system at all and through TLR7 (Fig EV1C–H) 14, 19. Total mtRNA was refractory (Fig EV1D), as if *mtPTL was rendered inactive in the context of total mtRNA. This finding was reminiscent of TLR13 “muteness” toward the E. coli Sa19 within total RNA 5.
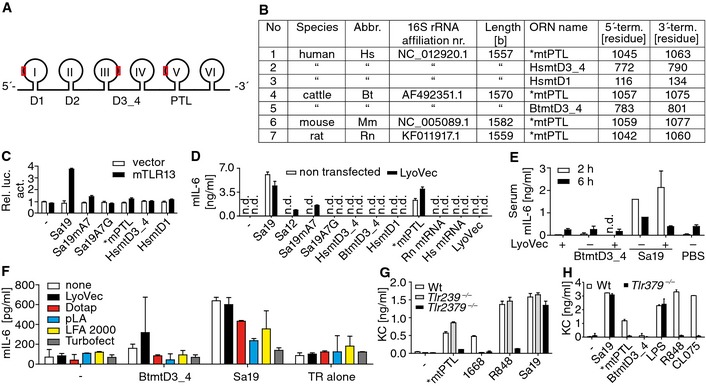
Figure EV1. Scheme of bacterial and mitochondrial (mt) largest rRNAs, Sa19 “like” segments (ORNs) within the latter, and mostly non‐responsiveness of the murine immune system to ORNs and mtRNA .
-
ASchematic of both, bacterial 23S and mitochondrial 16S rRNAs (I to VI, domains; red rectangles, Sa19 like segments within 16S mtrRNA; D, domain; PTL, peptidyl transferase loop within V).
-
BSa19 “like” 16S rRNA segment information (abbr., abbreviation; term., terminal; *interspecies sequence conservation).
-
CLuciferase activity (Rel. luc. act.) of transfected and challenged HEK293 cells (n = 2).
-
DCytokine release by bone marrow‐derived macrophages (BM; LyoVec, transfection reagent; Hs, human; Rn, rat; n = 2).
-
ESerum cytokine concentrations of mice challenged i.v. with phosphorothioate‐stabilized ORNs (n = 3).
-
FCytokine release of splenocytes transfected with ORNs with various transfectants (pLA, poly‐L‐arginine; LFA, Lipofectamine 2000; TR, transfection reagent only; n = 2).
-
G, HCytokine release of challenged wild‐type (Wt) and k.o. BMs (n = 3).
We next analyzed human PBMCs for their responsiveness to different bacteria and self‐derived RNAs. While all Sa19 “like” ORNs applied including TLR13‐silent, point‐mutated, and methylated ones induced robust TNF production in PBMCs, surprisingly none triggered substantial type I interferon (IFN) release (Fig 1C). This result indicated a lack of TLR7 involvement because TLR7 ligands such as RNA40 and bacterial tRNA induce PBMC type I interferon (IFN) production by activating plasmacytoid DC TLR7 14, 19. Next, we analyzed bacterial rRNA activities (Fig EV2A) 5, 9, 10. 23S, 16S, and 5S rRNA of both Gram‐positive and Gram‐negative bacteria induced substantial TNF release from PBMCs, provided their lysosomes were functional (Fig 1D). While total bacterial RNA encompassing tRNA triggered substantial IFNα release, neither bacterial rRNAs, nor total mtRNA hardly induced IFNα release, while the latter RNA elicited strong IL‐6 production (Figs 1E and F, and EV2B).
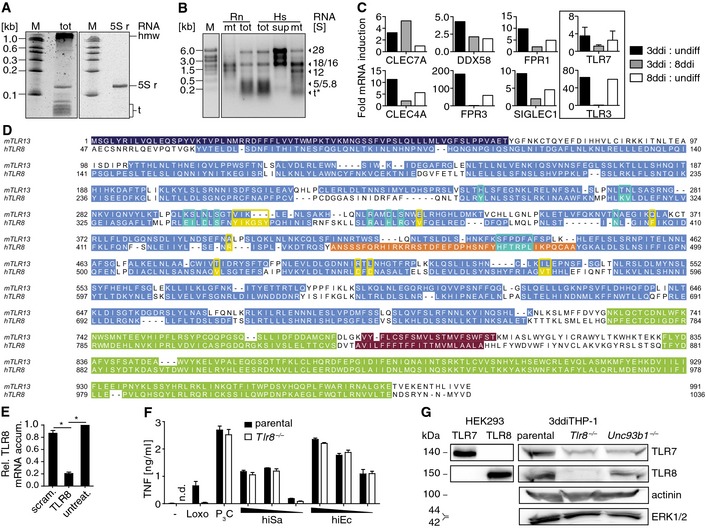
Figure EV2. RNA subpopulation isolation, candidate mRNAs, mTLR13‐hTLR8 alignment, TLR8 mRNA knockdown, and responsiveness as well as TLR7/8 expression of differentiated k.o. THP‐1 cells.
-
A, BElectrophoresis gels of total S. aureus RNA with tRNA and 5S rRNA (left panel) or isolated 5S rRNA (right panel; PAGE) and total or purified mammalian mtRNA (agarose), respectively (kb, kilobases; M, RNA size marker; tot, total; S, Svedberg; r, ribosomal; hmw, high molecular weight; t, transfer; mt, mitochondrial RNA; sup, supernatant at initial mitochondria separation step; Rn, rat; Hs, human; *S not applicable).
-
CReceptor mRNA amounts increased ≥ 2‐fold in 3‐day differentiated (ddi) as compared to undifferentiated (undiff) and 8ddiTHP‐1 cells, except for the data marked by a frame (depicted for comparison; dual TLR7 probe set; CLEC7A, C‐type lectin domain family 7A; FPR3, formyl peptide receptor 3; DDX58, DEAD box polypeptide; CLEC4A, C‐type lectin domain; FPR1, formyl peptide receptor; SIGLEC1, sialic acid binding Ig‐like lectin; n = 1), according to the comparative gene array‐based transcriptome profiling.
-
DAlignment of murine (m) TLR13 and human (h) TLR8 sequences (46 residue signal and leucine‐rich repeat N‐terminal domain, LRRNT, of the latter is not depicted for clarity; dark blue, signal peptide and LRRNT; light blue, LRRs; violet, transmembrane domains; green, LRR C‐terminal and cytoplasmic Toll‐IL1 receptor resistance gene domains; orange, z‐loop; yellow and turquoise, residues known to contribute to hTLR8 ligand recognition site one and two, respectively).
-
ERelative (Rel.) mRNA accumulation (accum.) in 3ddiTHP‐1 cells upon transfection of siRNAs (scram., scramble control; untreat., untreated; *P ≤ 0.01; unpaired t‐test; n = 3).
-
FActivity of 3ddiTHP‐1 cells challenged with different amounts of heat‐inactivated (hi) S. aureus and E. coli (Sa and Ec, respectively; –, unchallenged; n.d., not detected; Loxo, loxoribine; P3C, Pam3 CSK 4; corresponding to Fig 3C; n = 3).
-
GImmunoblot analysis of R848 overnight‐primed 3ddiTHP‐1 cell lysates (genotypes indicated, TLR7+ or TLR8+ HEK293 cell lysates for positive and cytoplasmic protein detection as loading controls; n = 3).
PBMCs of a human individual expressing a nonfunctional Glu53Δ MyD88 mutant 20 failed to respond to S. aureus RNA and to Sa19 “like” ORNs (Fig 1G). Given that 3‐day differentiated (3ddi) THP‐1 cells turned out to be responsive toward Sa19 (Fig 1H), we applied THP‐1 cells lacking expression of Unc93B1 (a chaperon translocating TLRs from the endoplasmic reticulum to endosomes) 21, 22. Like undifferentiated and 8ddiTHP‐1 cells, Unc93b1 −/−‐3ddiTHP‐1 counterparts failed to respond toward Sa19 “like” ORNs (Fig 1I).
Upregulation or overexpression of TLR8 confers uridine‐dependent specific bacterial and mitochondrial RNA responsiveness while TLR8 knockout abrogates it
Unc93b1 −/−‐3ddiTHP‐1 cells were also insensitive to large bacterial rRNAs (Fig 2A) 12. However, they responded well to hiSa and E. coli (Ec) via TLR2 (Fig 2B and C). Next, we comparatively profiled transcriptomes of undifferentiated, 3ddi‐, and 8ddiTHP‐1 cells. Selection of non‐interleukin receptors that were at least twofold upregulated on the mRNA level in 3ddiTHP‐1 cells led to seven candidate molecules out of which we considered mTLR13‐like TLR8 as most promising candidate (Figs 2D and EV2C). In contrast to mTLR13, hTLR8 carries a z‐loop (Fig EV2D) providing a functionally important cleavage site that contributes to ligand binding, and is deleted in mTLR8 13. In order to substantiate our selection of hTLR8, we analyzed its “gain and loss of” function. All Sa19 “like” ORNs activated hTLR8 overexpressing HEK293 cells, and BtmtD3_4 triggered also mRAW264.7 cell activity through hTLR8 (Fig 2E and F). Furthermore, knockdown of TLR8 mRNA abrogated responsiveness to BtmtD3_4, and to R848 (Figs 2G and EV2E).
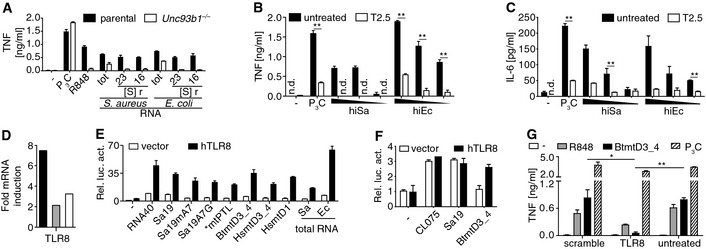
Figure 2. Transcriptome as well as gain‐ and loss‐of‐function analyses implicate TLR8 as bacteria and mitochondria ribosomal RNA sensor.
-
ACytokine release of 3‐day differentiated (ddi) THP‐1 cells toward stimulation with TLR ligands and bacterial RNAs (S, Svedberg; r, ribosomal; tot, total; n = 3).
-
B, CResponsiveness of Unc93b1 −/−‐3ddiTHP‐1 cells treated with T2.5 and challenged with heat‐inactivated (hi) bacteria (triangle, decreasing doses; n = 3).
-
DRatios of constitutive mRNA amounts in 3‐ versus 8ddi (gray bar) and of each of both versus undifferentiated THP‐1 cells (black or white bar, respectively) according to a transcriptome profiling result (n = 1).
-
E, FNF‐κB‐driven relative luciferase activity (Rel. luc. act.) of hTLR8‐overexpressing HEK293 and murine RAW264.7 cells, respectively, upon challenges (vector, empty plasmid; n = 3).
-
GTransfection of TLR8 siRNA impairs 3ddiTHP‐1 cell responsiveness (n = 3).
Moreover, responsiveness of Tlr8 −/−‐3ddiTHP‐1 cells to specific ORNs and bacterial rRNAs was strongly impaired, while total bacteria RNA recognition was widely operative (Fig 3A and B) 22. Otherwise, their sensitivity for hiSa and hiEc was normal, but largely TLR2 driven (Figs 3C and EV2F). Sensing of imidazoquinoline R848 and the guanosine (G) nucleotide analogue loxoribine by TLR7 was weak in Tlr8 −/− cells yet abrogated by TLR7 mRNA expression knockdown (Figs EV2F and G, and EV3A) 19, 23. Besides loxoribine and RNA40, only total bacterial RNA and to a low degree also S. aureus 5S rRNA triggered IFNα release by PBMCs, which emphasized TLR8 specificity and lack of TLR7 specificity of the ORNs, bacterial rRNAs, as well as mtRNA (Fig 1C, E, and F) 14. Of note, mtRNA‐driven activation of 3ddiTHP‐1 cells was absent from Tlr8 −/− and Unc93b1 −/−‐3ddiTHP‐1 cells (Fig 3D).
Figure 3. TLR8 recognizes UR/URR motif containing RNA .
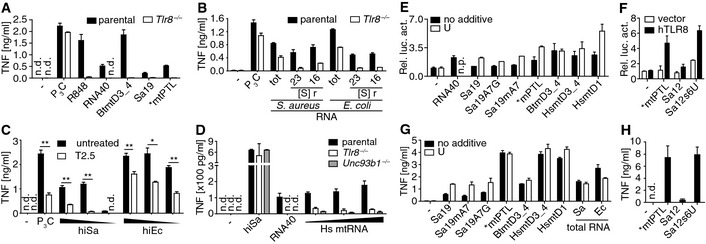
-
A, BCytokine release of 3‐day differentiated (ddi) THP‐1 cells challenged with (A) ORNs or (B) bacterial RNA fractions (tot, total; S, Svedberg; r, ribosomal).
-
CResponsiveness of Tlr8 −/−‐3ddiTHP‐1 cells challenged with heat‐inactivated (hi) bacteria (triangle, decreasing doses; *P ≤ 0.05; **P ≤ 0.01; unpaired t‐test) upon TLR2 blockade (T2.5).
-
DResponsiveness of 3ddiTHP‐1 cells upon challenge with human (Hs) mitochondrial (mt) RNA (triangle, increasing doses).
-
E–HNF‐κB‐driven relative luciferase activity (Rel. luc. act.) of hTLR8+ HEK293 cells or cytokine release by PBMCs all transfected with the RNAs indicated and additional uridine (U) if indicated (n.p., not performed; vector, empty plasmid).
Figure EV3. TLR7 mRNA knockdown in k.o. THP‐1 cells and consequent responsiveness, Sa12 derivative sensitivity, bacterial infection‐driven type I IFN or IL‐6 production and inhibition of PBMCs or whole blood, respectively, and further species PBMC responsiveness.
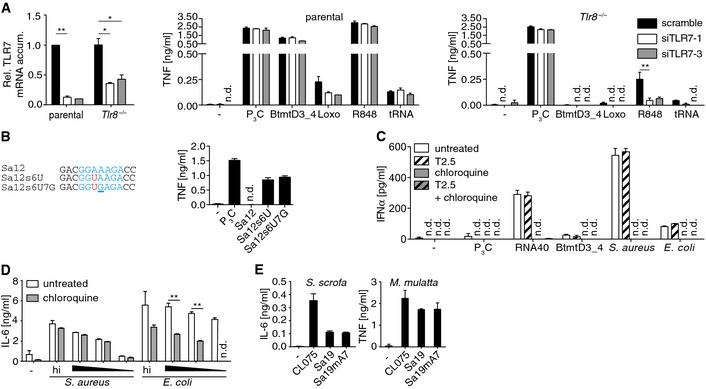
- Knockdown upon transfection of control (scramble) or two different TLR7 mRNA‐specific siRNAs (siTLR7‐1/3) in and cytokine release of respective 3‐day differentiated THP‐1 cells upon challenge (Rel., relative; accum., accumulation; Loxo, loxoribine; R848, small molecule, 50 μg/ml; tRNA, 200 ng/well of 96‐well plate E. coli transfer RNA; n = 3).
- Sequence alignments of Sa12 and Sa12s6U with another Sa12 variant carrying in addition, as compared to the latter, ORN and, reminiscent of the respective motif in HsmtD1, a UGA motif (blue, core sequence; red, A6U; underlined, A7G). Diagram depicts PBMC activities upon challenges with Sa12 and derivatives (n = 3).
- PBMC type I IFN production upon pretreatments (T2.5, TLR2‐neutralizing mAb; chloroquine, lysososmal function inhibitor) and challenge with TLR ligands or infection (5 × 104 cfu/ml; n = 2; corresponding to Fig 4B).
- Cytokine release of whole blood culture upon lysosome inhibition and challenge with heat‐inactivated (hi) and viable bacteria (triangle, decreasing dose of infection; n = 3; corresponding to Fig 4C).
- Cytokine release of Sus (S.) scrofa and Macaca (M.) mulatta PBMCs upon challenge with ORN variants (n = 3).
TLR7 and most likely also TLR8 preferentially bind U/G rich viral‐, si‐, and self‐RNA 14, 19, 23, 24, 25. Structural data imply that the RNA specificity of TLR8 might be guided by the U and G content of ssRNA 13. We thus transfected Sa19 “like” ORNs differing in their U content (Fig 1B) and admixed U nucleosides into TLR8+ cells. The stimulatory power of ORNs that contained sparse U (such as Sa19 and Sa19 point mutants) substantially became enhanced by co‐application of U nucleoside. Notably, ORN HsmtD1 containing like Sa19 just one U strongly activated cells. U‐less Sa12 containing a CGG motif failed to activate TLR8, while its mutant Sa12s6U carrying an A6U mutation resulting in containment of a UAA motif, included in duplicate in BtmtD3_4, while HsmtD1 carries a UGA motif, strongly activated cells (Fig 3E–H). Our data imply UR/URR rather than mere UG/UGG as RNA ligand consensus motif (Figs 3F and H, and EV3B) 13.
Bacterial infection‐driven cell activation is TLR8 dependent and inhibited by chloroquine in whole blood
Next, we explored the impact of hTLR8 on protective interventions during infection. To test this, THP‐1 cells pretreated with TLR2 neutralizing mAb were seeded with viable S. aureus, or with E. coli. In Tlr8 −/−‐3ddiTHP‐1 cells, inhibition of activation by TLR2 blockade was substantial (Fig 4A). Furthermore, a combination of chloroquine—a broadly established lysosomal function inhibitor—and T2.5 pretreatment significantly impacted PBMCs. Specifically, a contribution of TLR2 to both, TNF and IFNα production upon Gram‐positive bacteria‐driven activation, was barely detectable since chloroquine alone was efficient, which was true also for Gram‐negative bacterial infection in respect to IFNα but not TNF release (Figs 4B and EV3C). The failure of the control stimulus (and TLR8 ligand) BtmtD3_4 to induce substantial IFNα production while triggering that of TNF to similar degrees as compared to both infections implicated involvement of further endosomal pattern recognition in bacterial infections such as of tRNA through TLR7 (Figs 4B and EV3C). Also within whole blood, chloroquine inhibited TNF release more strongly upon infection with Gram‐positive bacteria as compared to infection with Gram‐negative bacteria, while its impact on IL‐6 production (strongly induced by lipopeptide/TLR2) was the opposite (Figs 4C and EV3D). Altogether, these data imply TLR8 as one major bacteria‐ and self‐mitochondria sensor and hint at an anti‐inflammatory potential of TLR8 blockade in sepsis. Whether this potential extends toward other clinical syndromes, such as trauma‐induced sterile inflammation, or autoimmunity, remains to be evaluated.
Figure 4. Lysosomal function inhibition affecting TLR8 activity is anti‐inflammatory upon both, Gram‐positive and Gram‐negative bacterial infection ex vivo .
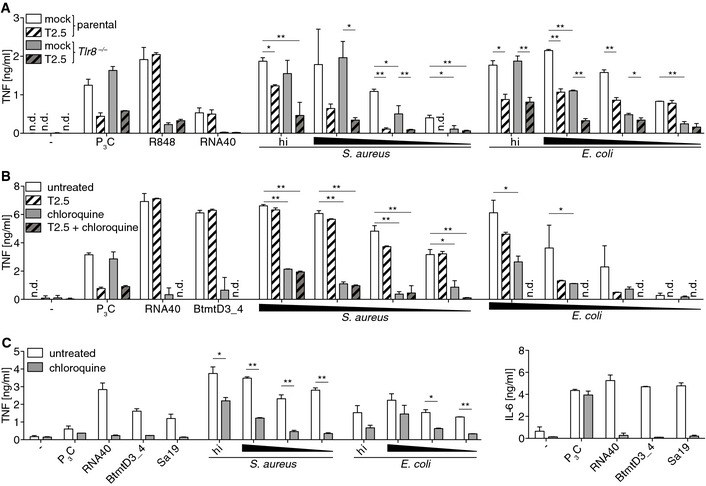
- Cytokine release of 3‐day differentiated THP‐1 cells of the indicated genotypes upon TLR2 blockade (T2.5) and challenge with TLR ligands, heat‐inactivated (hi) or viable bacteria.
- PBMC activity upon endosome function inhibition (chloroquine) and TLR2 blockade (T2.5) followed by TLR ligand challenge or bacterial infection.
- Activity of whole blood culture upon endosome function inhibition (chloroquine) and challenge with TLR ligands, heat‐inactivated (hi), or viable bacteria.
Perspective and concluding remarks
Upon submission of this study, four reports on Gram‐positive bacterial RNA sensing by hTLR8 or on mTLR13 structure have been published and largely summarized 26, 27, 28, 29, 30. Thus, S. aureus as well as Streptococcus pyogenes and Streptococcus agalactiae and Listeria monocytogenes total RNAs activate hTLR8 but not TLR7 26, 28. Total RNAs of further Gram‐positive and probiotic bacteria, as well as Enterococcus faecalis (EC‐12)‐derived 23S and 16S rRNA drive TLR8‐dependent, yet TLR7‐independent IL‐12 production 27. Our results extended these and earlier findings by implicating 5S beyond 23S and 16S rRNA of S. aureus and also Gram‐negative E. coli as well as mtrRNA as immune stimulatory P‐/DAMPs that activate hTLR8 with their UR/URR motif segments 11, 13.
Function restricting TLR8 mutation in mice might underlie their TLR13 expression and thus the A7 methylation mediated Sa19 camouflage, which is inoperative in other tetrapodes including human, macaque, and hog (Figs 1C, EV3E and EV4). Evolvement of TLR8 as a bacteria sensor might have resulted from perpetuating confrontation with either soil bacteria producing an MLS antibiotic, or bacteria expressing resistance conferring methyltransferase only, or bacteria carrying a respective 23S rRNA mutation upon fish–tetrapod evolutionary transition 31. Since immune escape of such bacteria could have handicapped host survival, promiscuous ssRNA sensing via TLR8 is of advantage. The price to be paid, however, is reactivity toward endogenous ssRNA with which the immune system of hTLR8 transgenic mice can hardly cope 32, 33.
Figure EV4. Phylogenetic tree illustrating TLR13 and/or TLR8 expression by Coelomata species including human and mouse.
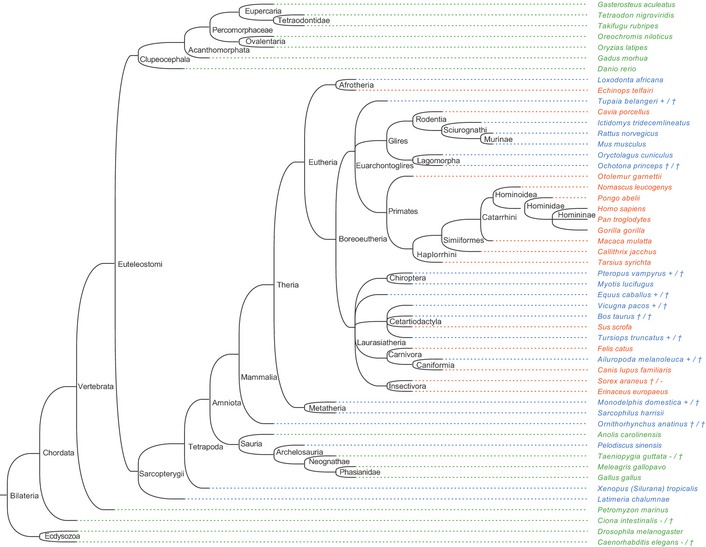
Species‐specific expression of either TLR13 exclusively (green species name), both TLR13 and TLR8 (blue), or TLR8 exclusively (orange). Additional symbols behind the species names indicate if expression of at least one of the two TLRs is uncertain and specific information on gene appearance and protein sequence annotation as found. Separated by a slash, the first mark specifies TLR8 expression, while the second one specifies TLR13 expression (+, expression certain: annotation as protein alone or additional gene annotation in genome sequence; †, expression of functional protein due to potential pseudogene carriage not obligatory: no protein annotation while gene is being annotated in genome sequence; –, expression unlikely: no protein annotation and not annotated in genome or the latter not analyzed due to unavailability of assembled genome).
Materials and Methods
Materials
RNase A, phorbol 12‐myristate 13‐acetate (PMA), U nucleosides, chloroquine, and gentamicin were applied at 20 μg/ml, 200 nM, 5 mM, 5 or 20 μg/ml (PBMCs or whole blood, respectively), and 10 μg/ml respectively (all Sigma). Cells were pre‐incubated for 30 min for TLR2 blockade with 20 μg/ml T2.5 3 (Hycult Biotech). Pam3CSK4 was applied at 0.1 μg/ml (EMC Microcollections). LPS (0111:B4), CL075, RNA40, loxoribine, and R848 were applied to cells at 0.1 or 1 μg/ml (human or murine cells, respectively), 2.5 μg/ml, 0.5 μg/ml, 600 μM, and 10 or 5 μg/ml (human or murine cells, respectively), respectively (all Invivogen). ODN 1668 was applied at a concentration of 10 μM (MWG).
Bacterial challenges
HiSa and hiEc were applied at concentrations of 1 × 108, 107, and 106 cfu/ml or a concentration of 1 × 107 cfu/ml only. The hiSa solution was incubated with RNase A for 1 h at 37°C wherein the 37°C incubation alone did not abrogate the stimulation capacity 5. THP‐1 cells were infected with 1 × 108, 107, and 106 cfu/ml of S. aureus and 1 × 107, 106, and 105 cfu/ml of E. coli, PBMCs with 1 × 106, 5 × 105, 1 × 105, and 1 × 104 cfu/ml of S. aureus and 5 × 104, 1 × 104, 5 × 103, and 1 × 103 cfu/ml of E. coli, and whole blood with 1 × 107, 106, and 105 cfu/ml of S. aureus and 1 × 106, 105, and 104 cfu/ml of E. coli. After 1 h, gentamicin was added to stop bacterial growth 3.
Cell culture
BM or splenocytes from mice and PBMCs generated from humans of wt genotype if not indicated otherwise as well as RAW264.7 (murine macrophagoid), HepG2 (human hepatocytoid), THP‐1 (human monocytoid), and HEK293 (human fibroblastoid) cell line cells were grown under regular cell culture conditions 3. THP‐1 cells were differentiated with PMA (1 × 107 cells/96‐well plate) for either 24 h followed by 3 days culture in PMA‐free medium (3ddi), or 72 h followed by 5 days culture in PMA‐free medium (8ddi). Whole blood was drawn into monovettes (Braun, 8 ml) prefilled with Bivalirudin (5.3 mg) and aliquoted in 200 μl portions 34. Blood was drawn and PBMCs generated from buffy coats (provided by P. Horn) upon approval by the local ethics committee from healthy donors (14‐5804‐B0, University Hospital Essen) and from a patient expressing a nonfunctional Glu53Δ/157_159ΔGAG MyD88 mutant, approved through ethics vote of the local committee (282/11, University of Freiburg, Germany).
Bacteria
S. aureus (DSMZ 20231) and a clinical isolate clone of E. coli 3 were used for infections and challenges as heat‐inactivated bacteria in vitro or in vivo, or as sources of RNA preparations used for cell challenges by their transfection. Bacteria were inoculated with 16‐h‐grown preparatory cultures (agitation, 37°C, E. coli in LB, S. aureus in BHI medium). In the logarithmic growth phase, bacteria were pelleted and collected in PBS for RNA preparation, heat inactivation 100°C, 15 min), or infection. Bacterial samples (taken immediately prior to inactivation) were titrated by plating.
Cytokine measurement and immunoblot analysis
Enzyme‐linked immunosorbent assay (ELISA, human IL‐8, IL‐6, and TNF, used also for macaque samples, mouse IL‐6, TNF, and KC all R&D, capture and detection antisera αswine IL‐6, Bethyl Laboratories, recombinant swine IL‐6, Kingfisher Biotech Inc.) and luminex or ELISAs (eBioscience) for human IFNα were performed according to the manufacturer's instructions. Supernatants from murine BMs and splenocytes or THP‐1 cells (human) were sampled for ELISA 16 h after challenge start, while those of human PBMCs and whole blood were sampled after a 24‐h time period.
Mitochondria and subsequent mtRNA preparation
Liver was harvested from Wistar rats. Tissue as well as harvested cultured HepG2 cells was homogenized in ice‐cold isolation buffer (2 mM Hepes, 220 mM mannitol, and 70 mM sucrose, pH 7.4). Homogenized suspension was spun at 494 g (Sorvall ST16 cell centrifuge) for 3 min at 4°C. Supernatant was transferred to a fresh tube and spun. This washing step was repeated twice. Clear supernatant was spun at 26,000 g for 10 min at 4°C. Pellet was resuspended in ice‐cold isolation buffer, and the centrifugation step was repeated twice which resulted in a pure mitochondria preparation. Pellets were resuspended in TRI reagent (Sigma) for RNA isolation (as described before 5). A total of 0.25, 0.5, 1, or 2 μg per 200 μl mtRNA was applied to PBMCs and 0.5, 1, or 2 μg per 200 μl to THP‐1 cells.
Transfection of ORNs, and bacterial and mtRNA preparations
RNA40 was transfected principally using LyoVec (Invivogen) which was also used for ORNs, bacterial and mitochondrial RNAs challenge of BMs. RAW264.7 cells were transfected with Sa19 by LyoVec and with BtmtD3_4 by pLA (Sigma). HEK293 cells were transfected with ORNs by Lipofectamine 2000 (Life Technologies). All bacterial and mitochondrial RNA preparations were transfected with Lipofectamine 2000 in THP‐1 cells and PBMCs except for BtmtD3_4 and *mtPTL for the transfection of which pLA was used. ORNs (IBA) were transfected at a concentration of 100 pmol per 200 μl. Bacterial RNAs were transfected as follows: 1 μg (total RNA), 600 ng (23S and 16S rRNA), and 400 ng (5S rRNA) bacterial RNAs per 200 μl to challenge cells.
TLR8 mRNA knockdown
Forty‐eight hours upon differentiation, THP‐1 cells were transfected with siRNA (Qiagen) toward human TLR8 (ID SI02642458) as well as scrambled variant (ID 1027280, negative control). Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) was used according to the manufacturer's instructions. Twenty‐four hours post‐transfection cells were challenged with stimulants in fresh medium. Supernatants were analyzed by ELISA. RNA was isolated and analyzed via RT–PCR as described before 5. The primers used for PCR were (MWG eurofins) as follows: 18S rRNA, (sense) 5′‐GTAACCCGTTGAACCCCATT‐3′, (antisense) 5′‐CCATCCAATCGGTAGTAGCG‐3′; human Tlr8, (sense) 5′‐GAGCATCAACCAAAGCAAGA‐3′, (antisense) 5′‐TGCCGTAGCCTCAAATACTG‐3′.
TLR8 knockout
Tlr8 −/−‐THP‐1 cells were generated using the CRISPR/Cas9 technology as previously described 22. In brief, cells were transfected with two plasmids encoding a CMV‐mCherry‐Cas9 expression cassette and a U6 promoter‐driven sgRNA, respectively, by electroporation. The CRISPR target site used for TLR8 was AGGTCAGCATTGACGACTGAAGG. THP‐1 cells expressing mCherry‐Cas9 upon electroporation were enriched by FACS sorting and cloned by limiting dilution. Two weeks later, single‐cell clones were picked and expanded. Of these clones, genomic DNA was extracted and subjected to a first‐level PCR using the following primer pair: 5′‐ACACTCTTTCCCTACACGACGCTCTTCCGATCTCTGTAGTCGACGATTGCTGCACT‐3′ and 5′‐TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCATACAAGTATGGGGCAGCGCT‐3′ (TLR8 gene‐specific sequences in bold, Illumina adapter sequences underlined). In second‐level PCRs, barcode primers unique for individual clones were used and PCR products were subjected to deep sequencing using the MiSeq (Illumina) benchtop sequencing system. Raw sequencing data were analyzed by applying the online tool OutKnocker.org and cell clones harboring out‐of‐frame mutations within the TLR8 gene were identified.
Gene array
For gene expression analysis of undifferentiated versus 3ddi versus 8ddiTHP‐1 cells, RNA was prepared using TRI reagent (as described before 5) from cells analyzed in parallel for their Sa19 responsiveness (Fig 1H). Total RNA concentration and purity were measured with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies/Thermo Scientific). RNA integrity was assessed using an Agilent Bioanalyzer nano chip. Biotinylated cRNA was prepared according to the Affymetrix ExpressKit protocol starting from 200 ng total RNA. Following fragmentation, 10 μg of cRNA was hybridized for 16 h at 45°C on GeneChip HG‐U133Plus_2. GeneChips were washed and stained in the Affymetrix Fluidics Station 450 using the Affymetrix hybridization, wash, and staining Kit. Arrays were scanned in a GeneChip 3000 scanner with G7 update. The data were analyzed with MAS5 using Affymetrix default analysis settings and global scaling to a target intensity of 1,000 as normalization method. The microarray data from this publication have been submitted to the NCBI GEO database http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67264.
Reporter gene assay
Murine TLR13 and human TLR8 expression plasmids were transfected together with luciferase reporter plasmids into HEK293 cells for analysis of NF‐κB‐driven firefly and constitutive renilla luciferase activities upon calcium phosphate precipitation, as previously described 5. RAW264.7 cells were transfected with the same plasmids using TurboFect (Life Technologies) following the manufacturer's instructions. 16 h after challenge, cell lysate was analyzed for luciferase activity (96‐well plate Berthold luminometer).
Statistics
Results were analyzed for statistical significance using the Student's t‐test for unpaired samples. Each illustrated column represents mean ± SD (error bars) of triplicate data points except for whole blood experiments results of which are represented by duplicate data points.
Further Methods are described in the Appendix Supplementary Methods.
Author contributions
AK, MO, and CC designed and performed most experiments, analyzed data, interpreted results, and participated in manuscript writing; DB and SR performed protein sequence alignment and in silico expression analyses; JK and PH performed comparative analysis of MyD88‐defective human blood; AMS, JS, and HH prepared PBMCs and performed analyses; SS and JB designed experiments; HW designed experiments and wrote the manuscript; VH generated Tlr8 −/−‐THP‐1 cells; and CJK overlooked the project, designed most experiments, analyzed and interpreted results, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank S. Schimanski, T. Scholtysik, B. Bathke, and A. Kibler for technical work, P. Horn for buffy coats, L. Klein‐Hitpass for gene array analyses, O. Witzke for pig blood, G. Iliakis and V. Mladenova (all University Hospital Essen, Germany) for access to an ultracentrifuge, C. Distler for macaque blood and J. Rassow (both Ruhr University Bochum, Germany) for guidance toward rat liver mitochondria isolation, and German Research Foundation (DFG) for funding of TRR60 and GK1949 projects (to C. K.).
EMBO Reports (2015) 16: 1656–1663
References
- 1. Takeuchi O, Akira S (2010) Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- 2. Hotchkiss RS, Sherwood ER (2015) Immunology. Getting sepsis therapy right. Science 347: 1201–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spiller S, Elson G, Ferstl R, Dreher S, Mueller T, Freudenberg M, Daubeuf B, Wagner H, Kirschning CJ (2008) TLR4‐induced IFN‐gamma production increases TLR2 sensitivity and drives Gram‐negative sepsis in mice. J Exp Med 205: 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deshmukh SD, Kremer B, Freudenberg M, Bauer S, Golenbock DT, Henneke P (2011) Macrophages recognize streptococci through bacterial single‐stranded RNA. EMBO Rep 12: 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S et al (2012) TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance‐forming modification. Science 337: 1111–1115 [DOI] [PubMed] [Google Scholar]
- 6. Hidmark A, von Saint Paul A, Dalpke AH (2012) Cutting edge: TLR13 is a receptor for bacterial RNA. J Immunol 189: 2717–2721 [DOI] [PubMed] [Google Scholar]
- 7. Li XD, Chen ZJ (2012) Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife 1: e00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wardyn SE, Kauffman LK, Smith TC (2012) Methicillin‐resistant Staphylococcus aureus in central Iowa wildlife. J Wildl Dis 48: 1069–1073 [DOI] [PubMed] [Google Scholar]
- 9. Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M et al (2012) Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 209: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jockel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, Ehm G, Schnare M, Nain M, Kaufmann A et al (2012) The 2′‐O‐methylation status of a single guanosine controls transfer RNA‐mediated Toll‐like receptor 7 activation or inhibition. J Exp Med 209: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cervantes JL, La Vake CJ, Weinerman B, Luu S, O'Connell C, Verardi PH, Salazar JC (2013) Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J Leukoc Biol 94: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fieber C, Janos M, Koestler T, Gratz N, Li XD, Castiglia V, Aberle M, Sauert M, Wegner M, Alexopoulou L et al (2015) Innate immune response to Streptococcus pyogenes depends on the combined activation of TLR13 and TLR2. PLoS ONE 10: e0119727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, Miyake K, Shimizu T (2015) Toll‐like receptor 8 senses degradation products of single‐stranded RNA. Nat Struct Mol Biol 22: 109–115 [DOI] [PubMed] [Google Scholar]
- 14. Hornung V, Guenthner‐Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A et al (2005) Sequence‐specific potent induction of IFN‐alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11: 263–270 [DOI] [PubMed] [Google Scholar]
- 15. Meng G, Rutz M, Schiemann M, Metzger J, Grabiec A, Schwandner R, Luppa PB, Ebel F, Busch DH, Bauer S et al (2004) Antagonistic antibody prevents toll‐like receptor 2‐driven lethal shock‐like syndromes. J Clin Invest 113: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, Homey B, Barrat FJ, Zal T, Gilliet M (2009) Self‐RNA‐antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med 206: 1983–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams TA, Foster PG, Cox CJ, Embley TM (2013) An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504: 231–236 [DOI] [PubMed] [Google Scholar]
- 18. Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515: 283–286 [DOI] [PubMed] [Google Scholar]
- 19. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S (2004) Species‐specific recognition of single‐stranded RNA via toll‐like receptor 7 and 8. Science 303: 1526–1529 [DOI] [PubMed] [Google Scholar]
- 20. Alsina L, Israelsson E, Altman MC, Dang KK, Ghandil P, Israel L, von Bernuth H, Baldwin N, Qin H, Jin Z et al (2014) A narrow repertoire of transcriptional modules responsive to pyogenic bacteria is impaired in patients carrying loss‐of‐function mutations in MYD88 or IRAK4. Nat Immunol 15: 1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pelka K, Phulphagar K, Zimmermann J, Stahl R, Schmid‐Burgk JL, Schmidt T, Spille JH, Labzin LI, Agrawal S, Kandimalla ER et al (2014) Cutting edge: the UNC93B1 tyrosine‐based motif regulates trafficking and TLR responses via separate mechanisms. J Immunol 193: 3257–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmid‐Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V (2014) OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res 24: 1719–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diebold SS, Kaisho T, Hemmi H, Akira S, Reise Sousa C (2004) Innate antiviral responses by means of TLR7‐mediated recognition of single‐stranded RNA. Science 303: 1529–1531 [DOI] [PubMed] [Google Scholar]
- 24. Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL (2005) Nucleic acids of mammalian origin can act as endogenous ligands for Toll‐like receptors and may promote systemic lupus erythematosus. J Exp Med 202: 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA (2004) Recognition of single‐stranded RNA viruses by Toll‐like receptor 7. Proc Natl Acad Sci USA 101: 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, Flo TH, Mollnes TE, Espevik T, Stenvik J (2015) TLR8 senses Staphylococcus aureus RNA in human primary monocytes and macrophages and induces IFN‐beta production via a TAK1‐IKKbeta‐IRF5 signaling pathway. J Immunol 195: 1100–1111 [DOI] [PubMed] [Google Scholar]
- 27. Nishibayashi R, Inoue R, Harada Y, Watanabe T, Makioka Y, Ushida K (2015) RNA of Enterococcus faecalis strain EC‐12 is a major component inducing interleukin‐12 production from human monocytic cells. PLoS ONE 10: e0129806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eigenbrod T, Pelka K, Latz E, Kreikemeyer B, Dalpke AH (2015) TLR8 senses bacterial RNA in human monocytes and plays a nonredundant role for recognition of Streptococcus pyogenes . J Immunol 195: 1092–1099 [DOI] [PubMed] [Google Scholar]
- 29. Eigenbrod T, Dalpke AH (2015) Bacterial RNA: an underestimated stimulus for innate immune responses. J Immunol 195: 411–418 [DOI] [PubMed] [Google Scholar]
- 30. Song W, Wang J, Han Z, Zhang Y, Zhang H, Wang W, Chang J, Xia B, Fan S, Zhang D et al (2015) Structural basis for specific recognition of single‐stranded RNA by Toll‐like receptor 13. Nat Struct Mol Biol 22: 782–787 [DOI] [PubMed] [Google Scholar]
- 31. Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G (2014) Bacterial phylogeny structures soil resistomes across habitats. Nature 509: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guiducci C, Gong M, Cepika AM, Xu Z, Tripodo C, Bennett L, Crain C, Quartier P, Cush JJ, Pascual V et al (2013) RNA recognition by human TLR8 can lead to autoimmune inflammation. J Exp Med 210: 2903–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Snyder JM, Treuting PM, Nagy L, Yam C, Yi J, Brasfield A, Nguyen LP, Hajjar AM (2014) Humanized TLR7/8 expression drives proliferative multisystemic histiocytosis in C57BL/6 mice. PLoS ONE 9: e107257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coch C, Luck C, Schwickart A, Putschli B, Renn M, Holler T, Barchet W, Hartmann G, Schlee M (2013) A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS ONE 8: e71057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


