Abstract
We identified a discrete number of microRNAs differentially expressed in benign or malignant mesothelial tissues. We focused on mir-145 whose levels were significantly downregulated in malignant mesothelial tissues and Malignant pleural mesothelioma (MPM) cell lines as compared to benign tissues (pleura, peritoneum or cysts). We show that promoter hyper-methylation caused very low levels in MPM cell lines. Treatment of MPM cell lines with mir-145 agonists negatively modulated some protumorigenic properties of MPM cells, such as clonogenicity, cell migration and resistance to pemetrexed treatment. The main effector mechanism of the clonogenic death induced by mir-145 was that of accelerated senescence. We found that mir-145 targeted OCT4 via specific binding to its 3’-UTR. Increased intracellular levels of mir-145 decreased the levels of OCT4 and its target gene ZEB1 thereby counteracting the increase of OCT4 induced by pemetrexed treatment which is known to favor the development of chemoresistant cells. In line with this, reintroduction of OCT4 into mimic-145 treated cells counteracted the effects on clonogenicity and replicative senescence. This further supports the relevance of the mir-145-OCT4 interaction for the survival of MPM cells. The potential use of mir-145 expression levels to classify benign vs malignant mesothelial tissues and the differences between pemetrexed-induced senescence and that induced by the re-expression of mir-145 are discussed.
Keywords: Mesothelioma, OCT4, mir-145, mesothelial cysts, chemoresistance, senescence
INTRODUCTION
Malignant pleural mesothelioma (MPM) is a tumor originating from the mesothelial surfaces of the lung and characterized by an overall bad prognosis(1). Chronic inflammation (consequent to asbestos exposure) and genetic predisposition are major players in MPM pathogenesis (2). Additionally, because of the use of asbestos in the ‘70s, the incidence of MPM is expected to peak in the next few years (2). Two main features influence the prognosis of MPM: first, its silent clinical progression, which leads to very late diagnosis and thus strongly limits therapeutic intervention (i.e surgical resection). Second, its extreme resistance to current chemotherapeutic agents which dramatically hampers the disease-free-survival of the treated patients (3, 4). Therefore, identification of diagnostic biomarkers for MPM would be relevant for early diagnosis and patient stratification, in addition to contributing to our understanding of the basic mechanisms of MPM progression.
MicroRNAs (miRs) are short (17–22 nucleotides) noncoding RNAs that regulate gene expression by inhibiting protein translation, through base-pairing mediated interaction with the 3’UTR of target mRNAs(5, 6). Given their ability to simultaneously target multiple genes/pathways, transient or stable modulation of intracellular microRNA levels is a powerful way to transduce adaptive stimuli leading to broad changes in pathway activation. Growing evidence suggest that microRNAs play a major role in oncogenesis, with either tumor suppressor or tumor-promoting modalities(7–9). MicroRNAs are much more stable than mRNAs and this phenomenon allows an assessment of their expression levels retrospectively from frozen or fixed tissues. This feature, together with their rather tissue-specific expression, has prompted the study of microRNAs expression levels as an important diagnostic and prognostic tool for a number of solid and non-solid malignancies (9). Additionally, the study of cancer-associated microRNAs may prove useful in order to elucidate the mechanism(s) behind cell transformation. For example, the identification of low levels of mir-29c* in MPM has provided both a prognostic tool for epithelial MPM and has pointed to aberrant chromatin methylation for the progression of MPM(10). Here we investigated whether specific microRNAs could distinguish malignant mesotheliomas from benign mesothelial lesions (mesothelial cysts, normal pleura and peritoneum). This allowed us to identify a discrete group of microRNAs possibly endowed with diagnostic potential. Among the identified candidates, we further characterized the microRNA-145 (mir-145), whose expression was very low in MPM tissues and cell lines, as compared to benign mesothelial tissues or untransformed mesothelial cells. Mir-145 may represent a potential tumor suppressor, since its expression has been found downregulated in a number of solid tumors, including prostate and bladder tumors, endometrial adenocarcinomas and gliomas (11–14). The homeodomain-containing transcription factor OCT4 is among the multiple targets of mir-145 (13, 15). The OCT4 gene (also named POU5F1) plays an important role in activating self-renewal gene programs in embryonic tissues and stem cells(16). OCT4 is found overexpressed in poorly differentiated and high-grade tumors, with potential to support aberrant proliferation, tumor initiation and chemoresistance (17–21). These properties have been shown to reside in its ability to control Epithelial to Mesenchymal transition (EMT)(22) as shown in mouse models of metastatic colon cancer (23). In fact, RNAi-based downregulation of OCT4 in vitro affects growth and tumorigenicity of ovarian cancer cell lines (24). We found here that mir-145 has the potential to modulate many in vitro pro-tumorigenic features of MPM cells, including growth, clonogenicity and resistance to pemetrexed treatment. Additionally, transfection of mimic-145 into MSTO-211H cells strongly reduced tumor engraftment in vivo. Induction of replicative senescence accounted for the clonogenic death induced by the treatment of MPM cells with mir-145 agonists. Mir-145 directly targeted OCT4 and, indirectly, its EMT-promoting target ZEB1 in MPM cells and the levels of mir-145 and OCT4 inversely correlated in vivo. OCT4 targeting is crucial to the anticancer properties of mir-145. In fact, mimic-145 expression counteracted the increase of OCT4 in pemetrexed resistant MPM cells. Additionally, overexpression of OCT4 at least partially rescued the effects of mimic-145 on the clonogenicity and replicative senescence of the targeted MPM cells. Promoter hyper-methylation may contribute the low levels of mir-145 in MPM cells and malignant MPM tissues. In summary, in this work, we have identified mir-145 downregulation as a characteristic trait of malignant vs benign mesothelial tissues. Additionally, we provide insights into the possible contribution of mir-145-OCT4 interaction to the transformed phenotype of MPM cells.
RESULTS
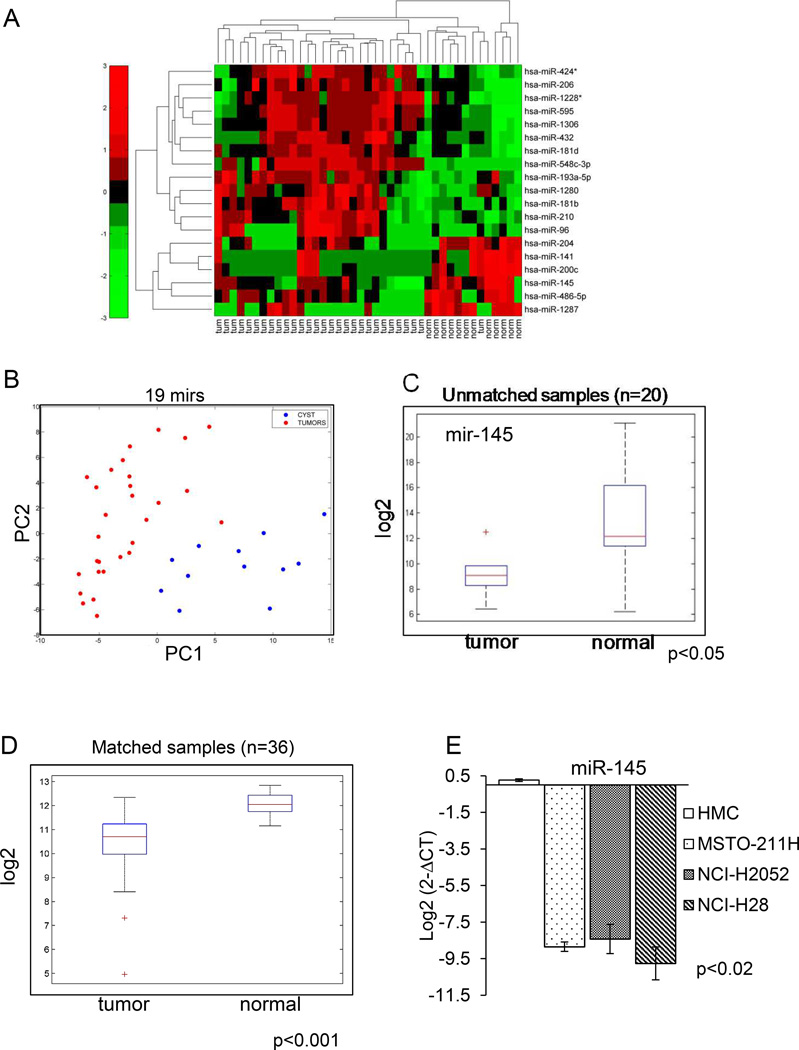
Identification of microRNAs differentially expressed among malignant and benign mesothelial tissues (Fig.1)
Figure 1. MicroRNA expression profile of MPM specimens.
A. Heat map. Levels (Log2) of 19 differentially expressed microRNAs among malignant and benign mesothelium (cysts). B. PCA plot showing the distribution of the training set of samples according to the levels of the 19 mirs as from A. C. Mir-145 expression in benign vs malignant mesothelial tissues. Box plot: Quantitative PCR. Average levels of mir-145 from an independent validation set of samples. RNA collected from fresh biopsies of unmatched mesothelioma (left n=6) and benign pleural tissue (right, n=14) samples, respectively. The “+” indicates an outlier. D. Levels of mir-145 in matched peritoneal vs MPM sample tissues. Box plot: Average levels of mir-145 (intensity values-Log2) from an independent set of samples. RNA collected from fresh biopsies of matched mesothelioma and peritoneal tissue (n=36). The “+” indicate outliers. E. Expression of mir-145 in the indicated cell lines. Q-PCR. Levels of mir-145 in untransformed mesothelial cells (HMC) vs MPM cell lines (Log2). Histogram bars represent mean ± standard deviation of at least three independent replicates. p-values are reported in the lower left side.
We performed an unbiased microRNA screening of MPM specimens (n= 29) and mesothelial benign tissues (mesothelial cysts, n=12). In Table I the main histoclinical features of cysts and MPMs are reported. As expected by the reported incidence distribution of mesothelioma subtypes in Western populations (25) more than 80% of cases were represented by the epithelioid mesothelioma (Table 1). All the analyzed specimens derived from surgically resected, naïve –treated patients. Our analysis revealed striking differences in the microRNA expression profile of benign vs malignant mesothelial tissues and allowed us to identify a discrete number of microRNAs (n=19) which showed statistically significant opposite modulation in benign vs malignant tissues (Fig. 1A, heat map). PCA analysis confirmed the ability of the 19 selected microRNAs to discriminate between the benign and malignant tissues. This was particularly evident on the first component, accounting for 42.24% of the total variance (Fig. 1B). Among the differentially expressed microRNAs, a larger fraction showed higher levels in benign lesions (n= 13) while a restricted number showed strong downregulation in the malignant tissues (n=6)(Fig. 1A). We focused on microRNA-145 (mir-145), because it showed the highest intensity values in its subgroup and because its modulation in our samples suggested the potential to act as a tumor suppressor. Additionally, mir-145 has been recently shown to modulate EMT, a key feature of MPM(26, 27).
TABLE 1.
Features of the MPM specimens analyzed in Fig.1A.
| Histotype | n | % |
|---|---|---|
| Epithelioid | 25 | 86.2 |
| Biphasic | 3 | 10.3 |
| Desmoplastic | 1 | 3.5 |
| T status | ||
| T2 | 9 | 31 |
| T3 | 11 | 37.9 |
| T4 | 5 | 17.3 |
| unknown | 4 | 13.8 |
| Node status | ||
| N0 | 14 | 48.3 |
| N1–3 | 10 | 34.5 |
| unknown | 5 | 17.2 |
Mir-145 is downregulated in MPM specimens and MPM cell lines but not in untransformed mesothelial cells (Fig.1)
We validated the previous results in two ways. Firstly, we assessed the levels of mir-145 by quantitative PCR on unmatched MPM and pleural biopsies obtained from patients (n=20, of which 6 MPM and 14 benign tissue) not belonging to the first set analyzed (Fig. 1C). As shown by the box plot (Fig 1C), we found that mir-145 expression was significantly downregulated in this validation group, thus confirming our initial findings. Secondly, we assessed the mir-145 levels in snap-frozen matched pair samples (n=36) of peritoneal mesothelium vs MPM (Fig. 1D). This, again, revealed that mir-145 levels were significantly lower in the matched tissues (p<0.001)(Fig. 1D). In summary, our analysis revealed a statistically significant downregulation of mir-145 levels in MPM as opposed to the benign tissues, irrespectively whether the latter was derived from cysts, normal mesothelium or peritoneal mesothelium from the same patient (Fig. 1). Mir-145 downregulation was observed in both paraffin-embedded MPM material and from snap-frozen sections. Additionally, the analyzed specimens were derived from three independent institutions (see discussion), thus strengthening further our results. To facilitate a functional characterization of mir-145 in MPM, we tested whether mir-145 downregulation was conserved in representative MPM cell lines as well (Fig. 1E). Quantitative PCR (Q-PCR) analysis showed that mir-145 expression was deeply downregulated in three MPM cell lines (MSTO-211H, NCI-H2052 and NCI-H28) when compared to untransformed mesothelial cells (HMC) (Fig. 1E). Of note, the MPM cell lines were different both in histologic type (epithelioid, sarcomatoid, byphasic) and growth properties (ATCC.org). This suggests that mir-145 downregulation is independent of MPM type and that our in vitro system could recapitulate what observed in the initial microRNA screening of the patient samples (Fig. 1 and table 1). Therefore, we decided to use the mentioned MPM cell lines to characterize the effect of mir-145 downregulation on MPM cells.
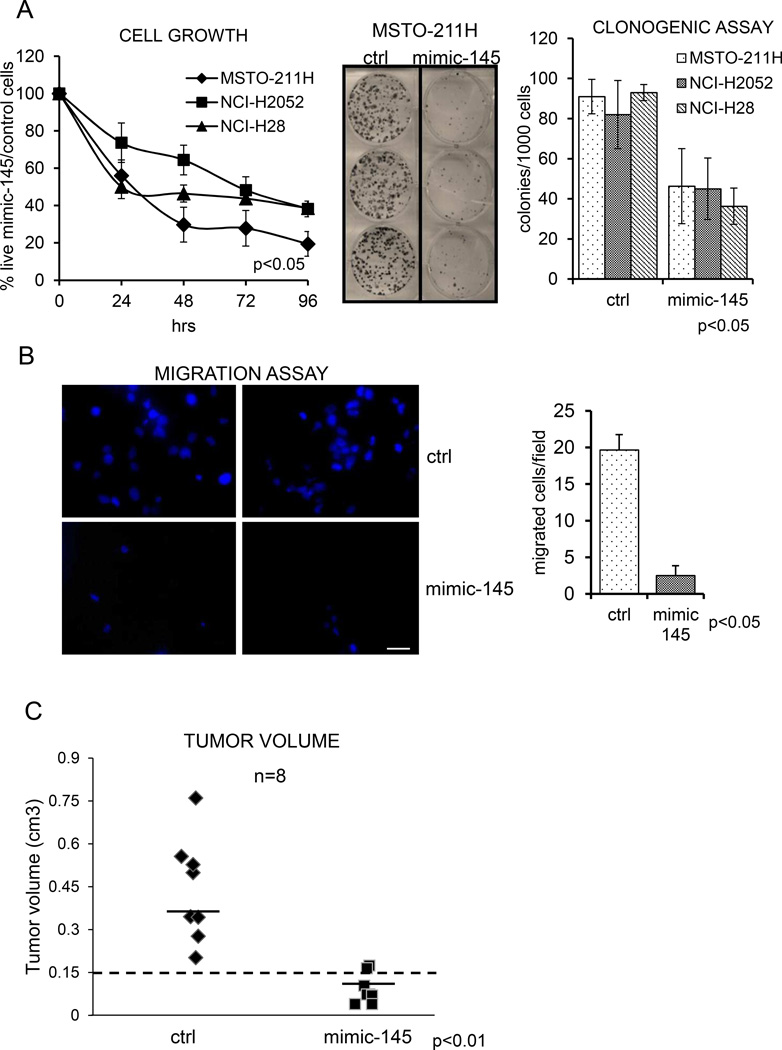
Re-expression of mir-145 affects viability and clonogenicity and migration of the MPM cell lines (Fig.2)
Figure 2. Re-expression of mir-145 affects protumorigenic properties of the MPM cell lines.
A. Left panels: Proliferation of ctrl and mimic-145 transfected MPM cells as assessed by Trypan blue staining. The percentage of live cells (control cells/mimic-145 expressing cells-Y-axis) at the indicated times (xaxis) is reported.. Middle and right panels: Clonogenic assay: representative micrographs of colonies formed by control (ctrl) and mimic-145 transfected MPM cell lines. Histogram bars represent mean ± standard deviation of at least three independent replicates. B Left panel: representative micrographs of transwell migration assays of MSTO-211H cells transfected with ctrl- or mimic-145. DAPI staining was used to quantify the migrated cells on the outer surface of the filter. Scale bar: 100µm. Right panel: histogram shows the percentage of migrated cells at the indicated times. Bars represent mean ± standard deviation of at least two independent replicates. The p-value refers to matched control vs mimic-145 transfected samples. C. mir-145 has anticancer effects in vivo. Briefly, SCID mice were subcutaneously transplantated with MSTO-211H cells (3 × 106 cells/mouse) transfected with either ctrl- or mimic-145 16hrs before injection. No differences in cell viability were observed between the two groups of injected cells at time of injection. C. Graph showing the average tumor volumes measured at day 47 p.i. Horizontal bars represent the average tumor volume of the measurable masses. Dotted line separates engrafted tumors (volume >150mm3 at termination) from non-neoplastic tissue or regressed tumors (volume >150mm3 at termination).
The broad downregulation of mir-145 in the malignant mesothelial tissues (as compared to the benign ones) suggested that mir-145 might act as a tumor suppressor. Therefore, we hypothesized that increased levels of mir-145 could counteract some of the malignant features of the MPM cell lines. To this aim, we transfected control- or mir-145 molecules in MSTO-211H, NCI-H2052 and NCI-H28 MPM cell lines (Fig.2). Transfection of the mir-145 agonist significantly affected the time dependent growth of all three tested MPM cell lines (p<0.05)(Fig. 2A left panel). To detail the observed effect on cell growth, we performed clonogenic assays from the same MPM cell lines (Fig. 2A middle and right panels). The clonogenic assay is useful to assess the long-term in vitro tumorigenic capacity of transformed cells in a way independent of acute toxic effects due to liposome transfection. This revealed a strong inhibition of colony forming ability in the cell lines transfected with mir-145, as compared to control-transfected cells (Fig. 2A middle and right panels, p<0.05).
Finally, we evaluated the effect of mir-145 transfection on the migratory properties of MPM cells. Transwell cell migration assays revealed that the migration of NCI-H2052 and MSTO-211H transfected with mir-145 was strongly reduced, as compared to control-transfected counterparts (Fig 2B, left and right panels, p<0.05). Inhibition of cell migration was also observed by scratch assays as well at early time points (Suppl. Fig. 1, left and right panel). We could not test the NCI-H28 cell line in this experimental context, since the latter cells did not show significant migratory properties at steady state (unpublished observations). Altogether, these observations revealed that forced expression of mir-145 in transformed mesothelial cells affects three protumorigenic features, namely cell growth, clonogenicity and cell migration. Further, to evaluate if the observed effects of mir-145 re-expression would translate in anticancer effects in vivo, we performed xenograft experiments. SCID mice were subcutaneously transplanted with MSTO-211H cells transfected with either ctrl- or mimic-145 16hrs before injection (fig. 2C). No differences in cell viability were observed in the two groups of injected cells at time of injection. We observed a significant inhibition of tumor growth in the mice engrafted with mimic-145 transfected cells. In fact, we observed tumor formation (cut off: volume >150mm3) in 8/8 mice injected with ctrl- transfected cells and in 2/8 mice injected with mimic-45 transfected cells (p<0.05) (fig. 2C). Additionally, the tumors grown from mimic-145 transfected MSTO-211H were significantly smaller than those grown from their control-transfected counterparts (p<0.05) (Fig.2C).
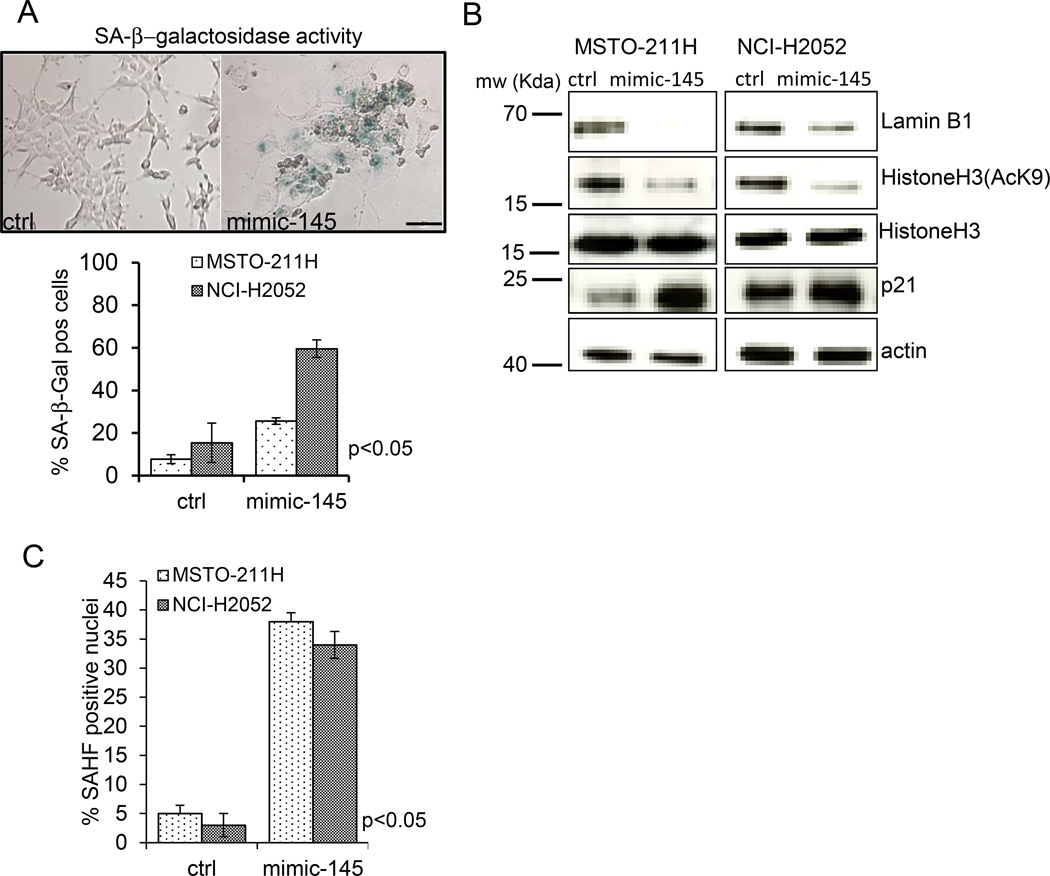
Mir-145 expression triggers senescence of MPM cells (Fig. 3)
Fig. 3. mir-145 induces senescence in MPM cell lines.
A. Upper panel. Representative micrographs of SA-β-gal staining of MSTO-211H cells transfected with control or mimic-145. Scale bar: 100µm. Lower panel. Histogram showing the percentage of SA-β-gal positive cells in ctrl or mimic-145 transfected MSTO-211H and NCI-H2052cells. Histogram bars represent mean ± standard deviation of at least three independent replicates.. B. Western Blot analysis of whole cell lysates obtained from ctrl- and mimic-145-transfected NCI-H2052 cells stained with anti-Lamin B1, anti-histoneH3, anti-acetyl-histoneH3(K9), anti-p21 or anti-actin antibodies (the latter as a loading control). C. Senescence Associated Heterochromatin Foci (SAHF) formation occurs in mimic-145 transfected cells. Histogram showing the percentage of SAHF positive nuclei in ctrl- and mimic-145 treated MSTO-211H and NCI-H2052 cells, respectively. Bars represent mean ± standard deviation of at least two independent replicates. The p-value refers to matched control vs mimic-145 transfected samples.
A closer look at the colonies formed by control- and mimic-145 transfected cells as from Fig. 2A, revealed that the in the mimic-145 expressing cells the colonies were composed of bigger, flat cells and showed reduced cell density, reminiscent of a senescent cell phenotype. Therefore, we investigated whether re-expression of the mir-145 in transformed MPM cell lines could trigger senescence. SA-β–galactosidase staining (at pH 6) of both MSTO-211H and NCI-H2052 cells transfected with ctrl or mimic-145 revealed strong staining in most of the cells expressing mimic-145, with no or little staining observed in the control cells at 5 days after transfection (Fig. 3A upper and lower panel p<0.05). To provide further evidence for the induction of replicative senescence by mir-145 overexpression, we evaluated by Western Blotting the levels of lamin B1, acetylated Histone H3 (AcH3K9) and p21 protein in ctrl- and mimic-145 treated MSTO-211H and NCI-H2052 cells. This revealed a significant reduction of both lamin B1 and AcH3K9 and induction of the p21 proteins in both mesothelioma cell lines transfected with mimic-145 (as compared to ctrl-transfected cells)(Fig. 3B). Lastly, we observed increased formation of senescence-associated heterochromatin foci (SAHF) in the mimic-145 transfected cells (Fig. 3C). SAHF are specialized domains of facultative heterochromatin, known contribute to senescence-associated cell growth arrest by sequestering and silencing proliferation-promoting genes. All the observed changes are strongly suggestive of replicative senescence (28–33). Altogether, these observations support the idea that mir-145 expression induces senescence in MPM cells and that induction of replicative senescence may mediate the anti-clonogenic effects of mimic-145 transfection.
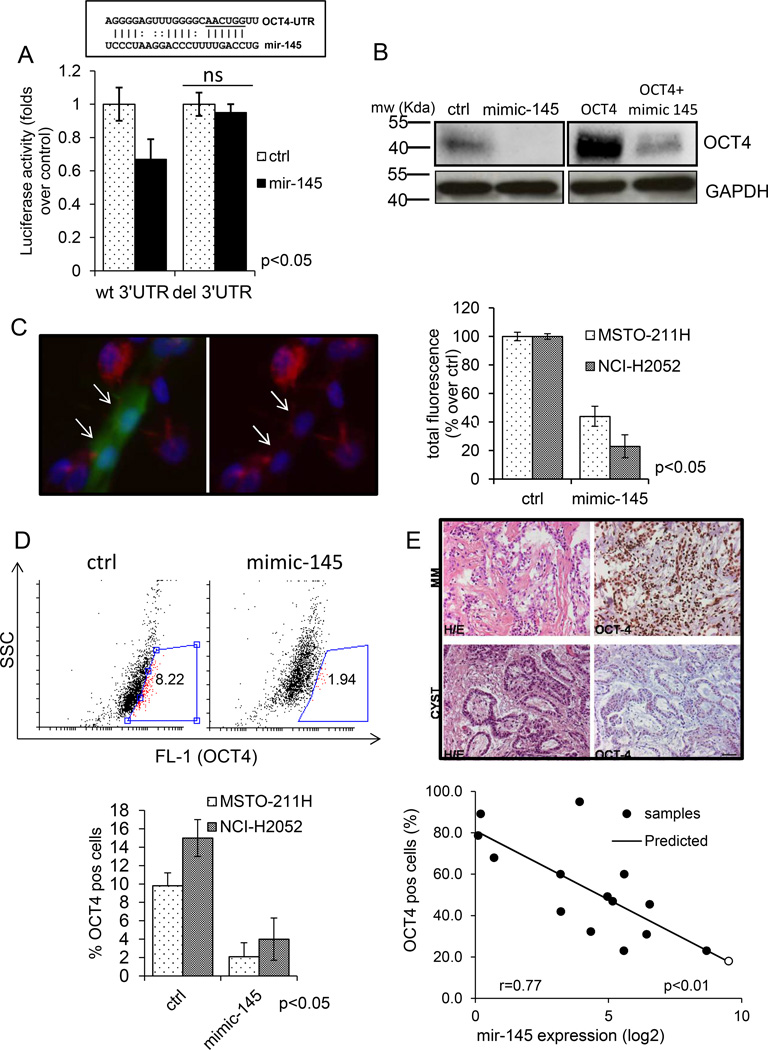
Mir-145 targets OCT4 in MPM cells (Fig. 4)
Figure 4. Mir-145 targets OCT4 in MPM cell lines A. Upper panel.
Schematic representation of the mir-145 target sequence within the OCT4 3’UTR (+138 to +157bp). Lower panel. Transfection of HEK293 cells with mimic-145 inhibited the translation of a luciferase-expressing plasmid containing a wt-3’UTR from OCT4 and not that of a its 3’UTR deleted counterpart. Histogram bars represent mean ± standard deviation of at least two independent replicates Statistics: NS, not significant. B. Representative western blotting analysis of whole cell lysates from control and mimic-145 transfected cells, either mock-transfected (left panels) or transfected with an OCT4 expression vector (retaining the wt 3’UTR region of the gene) (right panels) and stained with anti-OCT-4 antibodies or anti-GAPDH as a loading control. C Left panel: indirect immunofluorescence staining of MSTO-211H cells transfected with GFP (green, arrows) and cotransfected with either control or mimic-145, fixed and stained with OCT4 antibody (red). Nuclei were revealed by DAPI staining (blue). Scale bar: 10µm. Right panel: Semi-quantitative evaluation of the results from MSTO-211H and NCI-H2052 cells transfected as in left panel. Histogram bars represent mean ± standard deviation of at least two independent replicates. The p-value refers to matched control vs mimic-145 transfected samples. D. Upper panel. FACS plot. MSTO-211H cells transfected with control (left) or mimic-145 (right) stained with OCT4 antibodies. Percentage of OCT4 positive cells (gate:red) was calculated after subtracting the background (cells stained with isotype matched antibody). Lower panel. Histogram bars represent mean ± standard deviation of at least two independent replicates of MSTO-211H and NCI-H2052 cells treated as from upper panel. The p-value refers to matched control vs mimic-145 transfected samples. E. The levels of mir-145 and OCT4 inversely correlate in vivo. Upper panel. Representative images of paraffin-embedded sections of mesothelioma (”MM”) or benign mesothelial tissue (“cyst”) stained with Haematoxylin-Eosin (left) or with OCT4 antibody (right). Scale bar, 100µm. Quantitative evaluation of mir-145 levels (x-axis: Log2 expression intensity) and number of OCT4 positive cells (y-axis: percentage) in mesothelioma (black circles) vs benign mesothelial tissues (white circles). Straight line: predicted values. r value from a linear regression model is reported in the text.
The observed biological effects of mir-145 expression on MPM cell lines prompted us to investigate which gene products could mediate its effects. Literature mining and in silico analysis with multiple target finding algorithms revealed several targets of mir-145 with relevance for cancer development (unpublished observations). We focused on OCT4 because of its involvement in modulating aggressive features of solid tumors (19, 20, 23). We and others have previously shown that increased OCT4 levels correlate with the resistance of mesothelioma cells to pemetrexed in vitro (34, 35). In addition, the acquisition of an hypermigratory phenotype is a main feature of transformed cells and requires the reactivation of pathways governing the epithelial to mesenchymal transition (EMT)(36). Initially, we confirmed targeting of the OCT4 3’UTR region by mir-145 by assessing the luciferase activity of HEK293 cells co-transfected with a double luciferase reporter (whose activity was driven by the OCT4 3’ UTR) and a mir-145 expression vector (Fig. 3A). This revealed that transfection of the mir-145 significantly affected the translation of the luciferase mRNA in the targeted cells, as shown by a drop of the enzyme activity (Fig 3A). Additionally, such effect was specific for mir-145, since it was abolished in cells transfected with a mutant 3’UTR construct unable to bind to mir-145 (Fig 3A). Next, we analyzed by western blotting the protein levels of OCT4 in control-vs mimic-145 transfected MST0-211H and NCI-H2052 cells. Our analysis revealed that mir-145 overexpression affected the protein levels of OCT4 (Fig. 3B upper panels). We also tested the effect of mimic-145 on exogenously expressed OCT4 in both MPM cell lines, by transfecting an OCT4 expression vector harboring a wt 3’UTR (37) in mimic-145 treated cells. This confirmed a specific decrease of the exogenously expressed OCT4 protein in the mimic-145 transfected cells (Fig.3B lower panels). Further, we confirmed the downregulation of OCT4 by staining MSTO-211H (and NCI-H2052) cells co-transfected with GFP and mimic-145 with anti-OCT4 antibodies (Fig.3 C). This in fact showed a significant drop of OCT4 staining in the (GFP-positive) mimic-145 transfected cells (Fig. 3C, p<0.05). Downregulation of OCT4 per se is compatible with the biological effects of mimic-145 we observed in vitro, with regard to the inhibition of cell growth, migration and clonogenicity (Fig. 2) In fact OCT4 has been recently shown to control the expression of pivotal EMT promoting genes, including ZEB1, whose expression is relevant for MPM progression (26). Western blotting analysis of lysates derived from control- and mimic-145 transfected cells revealed in fact significantly lower levels of ZEB1 protein in both mimic-145 treated MSTO-211H and NCI-H2052 cells (Suppl. Fig. 2). Lastly, we performed FACS staining of ctrl- and mimic-145 transfected MSTO-211H and NCI-H2052 cells (Fig. 4D). This revealed that mimic-145 transfection strongly reduced the number of OCT4-positive cells (8.94% ± 2.2 vs 1.94 ± 0.8% for ctrl- and mimic-145 transfected cells, respectively) (Fig. 4D, upper and lower panel), therefore mirroring the previous results. In summary, mimic-145 treatment of MPM cell lines affected the levels of endogenous OCT4 as well as the levels of the exogenously expressed protein.
Mir-145 and OCT4 expression levels inversely correlate in vivo (Fig. 4E)
Since the forced expression of mir-145 triggers OCT4 downregulation, we predict that lower mir-145 levels may correlate with increased expression of OCT4 in vivo. To verify this, we performed immunohistochemistry staining for OCT4 of paraffin embedded sections derived from the same samples used for our initial microRNA screening (Fig. 1A), for which the levels for mir-145 were previously determined (Fig 1A). This showed that, as compared to the reference benign tissues (cysts), the levels of OCT4 were higher in all the MPM samples ranging from 21–89% positive cells vs 18% in the cyst (Fig. 4-E.). Additionally we found a significant correlation between the extent of mir-145 downregulation and the number of OCT4 positive cells (Fig. 4E-linear regression analysis: r: 0.77; r2: 0.59; p value <0.01), thus enforcing the relevance of mir-145 targeting of OCT4.
OCT4 antagonizes the miR-145 -mediated suppression of MPM protumorigenic features (Fig.5)
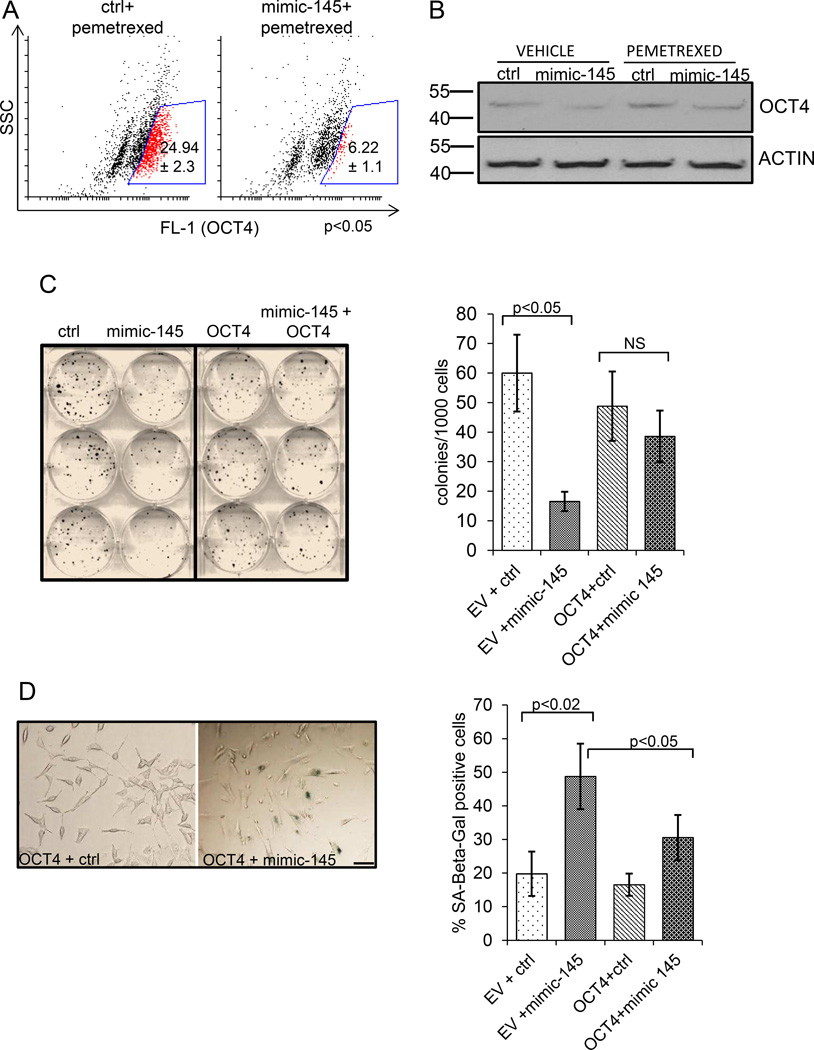
Figure 5. Antagonistic effects of mir-145 and OCT4 on the tumorigenicity of MPM cells.
A. mimic-145 transfection counteracts the OCT4 increase in pemetrexed-treated MPM cells. FACS staining of MSTO-211H cells transfected with control (left) or mimic-145 (right) (24hrs), treated with pemetrexed (72 hrs) and stained with OCT4 antibodies. Percentage of OCT4 positive cells (gate:red) was calculated after subtracting the background (cells stained with isotype matched antibody). Reported values represent the mean ± standard deviation of at least two independent replicates.. B. Representative western blot analysis of whole cell lysates from vehicle or pemetrexed-treated control- and mimic-145 transfected cells, either and stained with anti-OCT-4 antibodies or anti-ACTIN as a loading control. C. Overexpression of OCT4 rescues the effects of mimic-145. Left panel. Clonogenic assay. Representative micrograph of MSTO-211H cells transfected with either ctrl or mimic-145 and subsequently transfected with empty vector (EV) or OCT4 expression vector (retaining the wt 3’UTR sequence).Cells were stained with Crystal Violet 7 days later. Right panel. Quantitative analysis of clonogenic assays. Histogram bars represent mean ± standard deviation of at least three independent replicates. Statistics: NS, not significant. D. Left panel. SA-β-gal staining of MSTO-211H transfected with empty (EV) or OCT4-expressing vectors and subsequently (16hrs later) transfected with ctrl- or mimic-145. Scale bar: 20µm Right panel. Histograms showing the percentage of SA-β-gal positive cells as from left panel. Histogram bars represent mean ± standard deviation of at least three independent replicates.
To correlate the anti-cancer effects of mimic-145 with its activity toward the OCT4 gene product, we hypothesized that mimic-145 may antagonize the increase of OCT4 induced by pemetrexed treatment. In fact, we and others have shown that emergence of chemoresistant clones within pemetrexed and cisplatin-treated MPM cell lines is linked to the upregulation of EMT and stemness-related genes, including OCT4 (34, 35). Therefore, we evaluated the ability of mimic-145 to affect the levels of OCT4 in pemetrexed treated cells. FACS staining of ctrl- and mimic-145 transfected MSTO-211H cells treated with pemetrexed revealed in fact a significant increase of the number of OCT4 positive cells in the ctrl-transfected ones (Fig. 5A, left panel) (as compared to vehicle treated cells-fig. 4B). This effect was strongly reduced in their mimic-145 transfected counterparts (Fig. 5A, right panel) (24.94 ± 2.3 vs 6.22 ±1.1%, respectively).Similar results were obtained when using NCI-H2052 cells (data not shown). Additionally, we performed western blotting analysis with anti-OCT4 antibodies of lysates derived from the same cells as in fig. 5A: this revealed a significant decrease of OCT4 induction in MSTO-211H cells transfected with mimic-145 and treated with pemetrexed as compared to the similarly treated, ctrl-transfected cells(Fig. 5B). Based on these results, we predicted that overexpression of OCT4 could counteract the tumor-suppressive properties of the mimic-145. To this aim, we transfected control – or mimic-145 treated MSTO-211H and NCI-H2052 cells with an OCT4 expressing vector (or with an empty vector) and we evaluated the properties of the transfected cells (Fig.5B–C). We found that exogenous expression of OCT4 (and not that of the empty vector-EV) significantly attenuated the effect of mimic-145 on the clonogenicity of the transfected cells. In fact, mimic-145 +OCT4 transfected cells formed significantly more colonies than their control counterparts (Fig. 5C, p<0.05). Further, we evaluated the effect of OCT4 overexpression on the extent of cell senescence induced by the expression of mimic-145 (Fig. 5D). This revealed a significant reduction of the number of SA-β-Galactosidase positive cells in the OCT4 + mir-145 expressing cells as opposed to the EV+mir-145 transfected cells (Fig. 5D, p<0.05). In summary, these observations further strengthen the physical and functional relevance of the mir-145-OCT4 interaction. Our observations are also reminiscent of previous work showing the potential for OCT4 overexpression to bypass the senescence of human primary breast cells (38).
Promoter methylation may contribute to mir-145 silencing in MPM cell lines (Fig. 6)
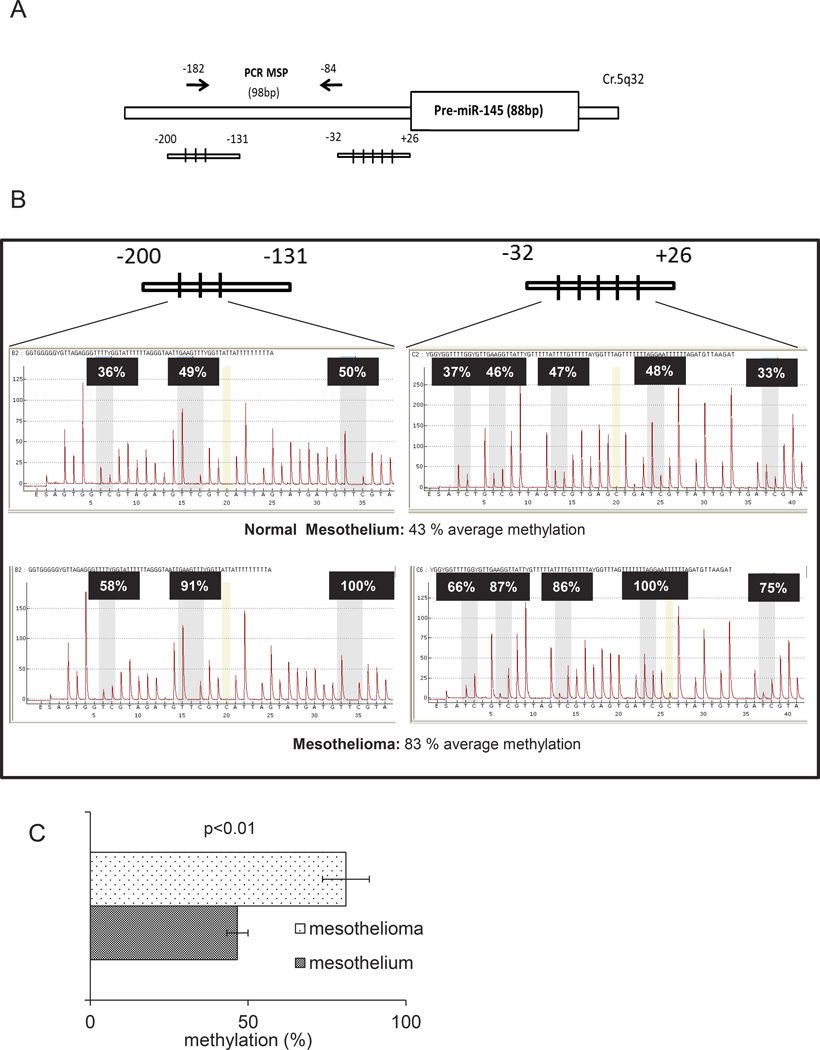
Figure 6. DNA methylation affects the levels of mir-145 in MPM samples.
A. Schematic representation of the miR-145 promoter regions containing the CpG islands analyzed in 6B. B. Representative results of pyrosequencing assays from normal mesothelium (upper panel) and malignant mesothelium (lower panel). C. Histogram showing the average levels of methylation of the mir-145 regulatory regions in normal mesothelial tissues (n=5) and mesothelioma samples (n=3). Histogram bars represent mean ± standard deviation of at least two independent replicates.
We have found the levels of mir-145 downregulated in neoplastic mesothelial tissues and MPM cell lines, as opposed to benign tissues and untransformed mesothelial cells, respectively. We next investigated how the levels of mir-145 could be downregulated in tumor samples. Therefore we investigated the methylation status of all the DNA CpG islands represented within the regulatory regions of the mir-145 promoter (Fig. 6A). We performed pyrosequencing assays on normal and malignant mesothelial tissues, respectively (Fig. 6B, upper and lower panels). This showed a general decrease of DNA methylation in all CpG islands analyzed (within the mir-145 promoter), in the mesothelia as compared to the mesothelioma tissues (average methylation: 46% ± 7% vs 83% ± 3%, for mesothelium vs mesothelioma, respectively, p<0.01) (Fig. 6C). The difference in methylation was even more evident for the CpG islands located closer to the transcription start site (Fig. 6B upper and lower panels). Further, we analyzed the methylation of the same CpG islands in MSTO-211H and NCI-H2052 cells treated with either vehicle or 5-Azacytidine (5-AZA-10uM, 96hrs), a DNA demethylating agent currently tested in clinical trials for myelodisplastic syndromes (MDS)25. 5’AZA treatment is expected to counteract the transcriptional repression due to promoter DNA hyper-methylation. This revealed a dramatic change in the methylation status of the CpG islands in the cells treated with 5’AZA (Suppl. Fig.3A and 3B) which readily matched the increase of mir-145 levels as assessed by semiquantitative PCR (Suppl. Fig. 3C). These observations support the idea that increased DNA methylation may be responsible for the low levels of mir-145 in both mesotheliomas and mesothelioma cell lines.
DISCUSSION
We investigated whether specific microRNAs could distinguish a large group of mesotheliomas from benign mesothelial lesions cysts, pleura or peritoneum. To reduce a possible “single-institution bias” in the sample collection, the specimens used for both the screening and the validation have been independently derived from different institutions, all reference centers for mesothelioma therapy, and analyzed with two different platforms. The use of mesothelial cysts as normal control tissues is unprecedented. Mesothelial cysts are relatively rare structures composed of normal pleural or peritoneal mesothelium which develop as liquid-filled translucent fibrous walls lined by a single layer of mesothelial cells(39). Therefore, mesothelial cysts may represent a valuable alternative to the use of immortalized cell lines or non-mesothelial tissues as a control. In the experimental system here used, we found that, with regard to mir-145 expression, all the sources of benign mesothelial tissue (cysts, pleural mesothelium, peritoneum) behaved similarly. Our initial screening has identified a number of microRNAs (n=19) whose levels of expression may segregate benign from malignant mesothelial tissues. Notably, some of the microRNAs identified in this study have been found by others and already shown to possess the power to discriminate between lung adenocarcinoma and mesothelioma tissues (mir-141, mir-200c, mir-193)(40). This represents an indirect biological validation of our findings. On the other hand, some other microRNAs (mir-145, mir-204, mir-486-5p, mir-1287) have not been described yet for mesothelioma, to the best of our knowledge. We have focused on mir-145 for its potential to act pleiotropically as a tumor suppressor via targeting multiple relevant cancer genes (12, 41–48). We provide evidence here that hypermethylation of mir-145 regulatory region is responsible for the low levels of this microRNA in both malignant mesothelial tissues and mesothelioma cell lines. We found that overexpression of mir-145 by mimic compounds affected the levels of OCT4 and, indirectly, of its EMT-associated gene target, such as ZEB1. Both OCT4 and ZEB1 are intimately connected to the maintenance of stemness, epithelial to mesenchymal transition and cancer chemoresistance (18–20, 24). Mir-145 downregulation conveys clear protumorigenic advantages to mesothelioma cells. Indeed, proliferation, clonal growth and migration of MPM cells are readily reduced by the transfection of “mimic” compounds and in vivo tumorigenicity is reduced as well. All this is unprecedented for MPM and shows how microRNA modulation is complex and how a change in the levels of a single microRNA may affect multiple pathways, impinging on key tumor features. EMT and chemoresistance are in fact major determinants of mesothelioma development and progression (26, 35). Mir-145 has multiple targets which are relevant to cancer progression. Although it is likely that multiple mechanisms underlie the anticancer effects of mir-145 re-expression in MPM cell lines, our observations suggest that OCT4 downregulation can be a main one. In fact, add-back of OCT4 into mimic-145 treated MPM cells at least partially restores the clonogenicity of the cells, while reducing significantly the number of senescent cells. These observations agree with the data from Beltran et al., showing that lentiviral expression of OCT4 in human mammary epithelial cells (HMECs) can “bypass” the spontaneous senescence occurring in those primary cultures after few passages(38). Conversely, the pro-senescence effects of mir-145 re-expression in MPM cells are reminiscent of the data from Bonifacio et al., (49) showing mir-145 upregulation in senescent human fibroblasts. Induction of chemoresistant cell populations is a side effect of pemetrexed and cisplatin treatment which limits their efficacy in vivo. This is fuelled by cytokines and growth factors secreted by senescent MPM cells in response to chemotherapy treatment, according to a Senescence Associated Secretory Phenotype (SASP) model(35, 50–52). The resulting chemoresistant cells are endowed with very high levels of EMT and stemness genes, including OCT4 and ZEB1(35). Indeed, we show here that pemetrexed treatment induces OCT4 and increases the number of OCT4 positive cells and the emergence of the latter cells may represent a mechanism of tumor progression(35). Mimic-145 treatment may, at least partially, counteract this phenomenon given its ability to downregulate OCT4 expression in pemetrexed treated samples.
One unanswered question of this study is whether mir-145 may act as a part of a larger group of microRNAs. In fact, we found that the distribution of the expression of mir-145 in our collected samples was strikingly similar to that of other 5 microRNAs (Fig.1A). Therefore, it is possible that all or some of the mentioned microRNAs may act as a functional cluster by targeting converging pathways. This is supported by the fact that at least some of the microRNAs identified in this study are known to target common EMT-promoting factors (for example, mir-141 and 200c target ZEB1/2)(53–55). In this regard, it would be interesting in future work to investigate the relationship between mir-145 and the other microRNAs sharing a similar distribution, to unravel possible functional interactions. With regard to this, it is interesting that targeting of the ZEB1-EMT axis is a frequent predicted function of the mentioned microRNAs (56). This reflects the prominent role of these EMT players in MPM development(26, 27).
Materials and Methods
Sample collection
41 unmatched specimens (29 MPM and 12 mesothelial cysts) were collected from two different institutions in Italy (Regina Elena Cancer Institute, Rome and General Hospital S. Vincenzo, Taormina). All the specimens were from formalin fixed and paraffin embedded tissues (FFPE) and initial diagnosis confirmed by expert histopatologists. 20 fresh biopsies (14 normal pleura vs 6 MPM) were obtained from Regina Elena Cancer Institute. An additional 36 snap-frozen samples (matched peritonea vs MPM) were used as a validation set of the results obtained with the initial screening and derived from the New York University Medical Center, New York, United States. The patients were not treated with any previous radio or radio-chemotherapy, before to undergoing surgery. Independent Ethical Committees from all the Institutions have approved the use of the samples.
RNA extraction, labeling and microarray hybridization
RNA from FFPE samples and cell lines was extracted using the miRneasy® FFPE kit (QIAGEN) and TRIZOL, respectively, following the manufacturer’s instructions. TRIZOL was used for the extraction of snap-frozen tissue specimens as well. Total RNA (200 ng) coming from tissues was labeled and hybridized to Human miRNA Microarray V2 Rel.16 (Agilent). Scanning and image analysis were performed using the Agilent DNA Microarray Scanner (P/N G2565BA) equipped with extended dynamic range (XDR) software, according to the Agilent miRNA Microarray System with miRNA Complete Labeling and Hyb Kit Protocol manual. Feature Extraction Software (Version 10.5) was used for data extraction from raw microarray image files using the miRNA_105_Dec08 FE protocol. For the matched peritonea vs MPM samples, samples were extracted with TRIZOL and 5ug of Total RNA was processed as described elsewhere (57).
microRNA expression profiling and data analysis
Analysis of expression of 887 human microRNAs were performed using in house built routine by Matlab (The Mathworks Inc.). Signals were pre-processed with quantile normalization and log2 transformed. Samples with duplicates available were tested for quality control. Unpaired T-test and Wilcoxon test were used to identify differentially expressed features between mesothelioma samples and cysts. Deregulated miRNAs with pvalue less than 0,001 and FDR (False Discovery Rate) less than 0,05 were selected. Unsupervised hierarchical clustering and Principal Component Analysis were used to separate different subgroups of samples basing on the identified signature. Validation of the signature was obtained by RT-PCR.
Target Prediction
Diana mirPath (http://diana.cslab.ece.ntua.gr/pathways) using Targetscan 5 and Pictar was used for the in silico putative target prediction of the selected microRNAs.
Cell lines and culture conditions
Human cell lines HEK293, MSTO-211H, NCI-H2052 and NCI-H28 were obtained from ATCC (Rockville; MD, USA). HMC Human Primary Mesothelial cells were obtained from TEBU-BIO (France). All cell lines were cultured as monolayers at 37 C and 5% CO2 in DMEM/F12+GLUTAMAX (Invitrogen-GIBCO, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen-GIBCO), 5 ug/ml Insulin (SIGMA).
Cell transfection
Pre-miRNA Precursor-Negative Control (Ambion) (Ctrl), Pre-miRNA145 (Ambion)(mimic145) at final concentration of 5nM were transfected into the indicated MPM cell lines using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. OCT4 expression vectors were from Warren et al. (37), Addgene plasmid 26816. Lipofectamine 2000 (Invitrogen) was used to transfect the DNA vectors, according to the manufacturer’s instructions.
Luciferase target assay
HEK-293T cells were transiently co-transfected in 6-well dishes using Lipofectamine 2000 (Invitrogen) with 2µg of OCT4-3’-UTR-(wt and mutant)-Luciferase vectors (a kind gift of Dr. Kenneth Kosik, UCSF Santa Barbara, CA, United States) 200 ng of the transfection control Renilla vector (phRLTK, Promega), and 500ng of mir-145 expression vector pEP-mir-145 (Cell Biolabs, San Diego, CA USA). Cells were harvested 48 hours post transfection and luciferase activities were analyzed by the dual-luciferase reporter assay system (Promega, Madison, WI).
Reagents
Pemetrexed (Eli Lilly and Company, Indianapolis, IN, USA) and 5-aza-2′-deoxycytidine (Sigma, St Louis, MO) were dissolved according to the manufacturer’s instructions.
Cell viability assay
Manual counting and Trypan Blue dye exclusion assay were used to assess cell growth and viability were indicated.
5-Aza-2′-deoxycytidine treatment
Cells were plated in six-well plates on Day 0, and treated with a final concentration of 10 µM 5aza-2′-deoxycytidine (Sigma, St Louis, MO) on alternative days beginning from Day 1, until Day 3. The treated cells were harvested on Day 4.
DNA isolation, sodium bisulfite conversion and pyrosequencing Analysis
Genomic DNA was isolated using the QIAamp DNA microKit (Qiagen, Hilden, Germany). Sodium bisulfite modification of 450 ng DNA was performed using the DNA Methylation kit (Diatech Pharmacogenetics) according to the manufacturer’s protocol. 5 out 12 microliters of modified DNA was subjected to PCR amplification of the specific promoter region of pri-miR-145 using the Corbett Life Science Rotor-Gene™ 6000. The primer sequences are listed in Supplementary Table 1. The resultant PCR products were analyzed by gel electrophoresis to confirm the size of the product and rule out the formation of primer dimers. The specific PCR products were then subjected to quantitative pyrosequencing analysis using a PyroMark Q96 ID (Qiagen) according to the manufacturer’s protocol. The genomic location of the bisulfite pyrosequencing assays and the number of CpG sites investigated in each assay are shown in Figure 6. The pyrosequencing analysis was performed with PyroMarker CpG software 1.0.11 (Qiagen).
In vitro migration assay
Wound healing assay. Cells seeded in 6-well tissue culture and grown to 95% confluence wounded with a sterile 10-µL pipet tip to remove cells by one perpendicular line. PBS1× washing was used to remove loosely attached cells. Digital micrographs were taken after scratching at the indicated times. Transwell migration assay. Migration assay was performed using a 24-well Boyden chamber with a non-coated 8-mm pore size filter in the insert chamber (Falcon). Cells (mimic 145- and control-transfected MSTO-211H) (5×104) were suspended in 0.5 ml DMEM/F12 media without containing FCS and seeded into the insert chamber. Cells were allowed to migrate for 48 h into the bottom chamber containing 0,7 ml of DMEM/F12 media containing 5% FCS in a humidified incubator at 37°C in 5% CO2. Migrated cells which had attached to the outside of the filter were visualized by staining with DAPI and counted.
Clonogenic assays
MPM cell lines were grown to 70% confluence and pulse treated with the indicated drugs or transfected as indicated. 16hrs later, cells were detached and seeded at 500–1500 cells/well into 6-well dishes (COSTAR) in drug-free media. Fresh media (25%) was added every three days. Colonies were stained with crystal violet and colonies (>50 cells) counted after 10– 14 days.
cDNA synthesis and Q-PCR
Total RNA was extracted using the TRIZOL Reagents (GIBCO). Real time-PCR of miRNA expression was performed using TaqMan MiRNA® Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. RNU6B and RNU49 were used as endogenous controls to standardize miRNA expression. For measurement of the precursor miRNA expression, PCR was perfomed using 2× red Mix DNA Polymerase MasterMix (RBC Bioscence) using the primers published by Zhang et al.49. PCR was performed at 95°C for 5 min, 32 cycles at 95°C for 30 s, 60°C for 40 s and 72°C for 40 s followed by a final extension at 72°C for 10 min. The PCR products were loaded on 2 % agarose gel for analysis.
Lysate preparation and immunoblotting analyses
Cells were lysed in buffer with 50 mM Tris-HCl pH 8, with 1% NP-40 (Igepal AC-630) 150mM NaCl, 5mM EDTA. Extracts were centrifuged at 14000 ×rpm for 15min to remove cell debris. Protein concentrations were determined by colorimetric assay (Bio-Rad). Western blotting was performed using the following primary antibodies: mouse monoclonal anti-GAPDH (Sigma Aldrich A228), TUBULIN (Sigma Aldrich T6074), OCT4 C-10 (Santa Cruz Biotechnology, inc. sc-5279, OCT4A (C30A3) Rabbit mAb Cell Signaling #2840), ZEB1 (Cell Signaling D80D3), p21 (Cell Signaling 12D1), laminB1 (ABCAM 16048), histone H3 Acetyl K9 (ABCAM 61231), Histone H3 (ABCAM 10799). Secondary antibodies used were goat anti-mouse, conjugated to horseradish peroxidase (Amersham Biosciences, Piscataway, NJ, USA). Immunostained bands were detected by chemiluminescent method (Pierce Rockford, IL, USA).
SA-β-gal staining
SA-β-galactosidase staining was performed on adherent cells according to manufacturer’s instructions (Cell Signaling, Danvers, MA, USA).
Immunofluorescence microscopy
Cells (5×103 cells per chamber) were seeded into 8-chambers culture slides (BD Falcon, Franklin Lakes, NJ). The next day, cells were rinsed with ice-cold PBS and fixed with 4% paraformaldehyde for 10 min at room temperature followed by permeabilization with 0,1% Triton X-100. The cells were subjected to immunofluorescence staining with Oct-3/4 Prediluted Monoclonal Antibody (PM 313 AA, BIOCARE Medical, Concord, CA 94520 USA) for 2 h at room temperature. The cells were then washed with cold PBS three times for 3 min each, and counterstained with DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride). Fluorescent staining of Senescence Associated Heterochromatin Foci (SAHF). To visualize SAHF, DAPI was used to stain for DNA after fixation and permeabilization of the cells. The cells were examined by fluorescence microscopy (Zeiss Axiophot). Fluorescence intensities from images of six randomly selected microscopic fields of cells were semi-quantitatively analyzed by densitometry (ImageJ software, NIH Image). The Corrected Total Cell Fluorescence (CTCF) was calculated by the formula: CTCF = Integrated Density – (Area of selected cell * Mean fluorescence of background readings).
Immunohistochemistry
OCT4 was analyzed by immunohistochemistry using the anti-OCT4 (Santa Cruz Biotecnology, inc. sc-5279) in a series of 10 mesothelioma patients subjected to surgery at the Regina Elena Cancer Institute (Rome, Italy) between 2005 and 2008. Formalin-fixed, paraffin-embedded 5µm sections were stained with Haematoxylin and Eosin or stained with anti-Oct-4 antibody (SC5279 LOTTJ1612, Santa Cruz, 1:50, BENCHMARK ULTRA VENTANA, Roche, Tucson, USA). Seven randomly chosen fields from each sample were scored.
Flow Cytometry
Cells were detached by PBS/EDTA 2mM and resuspended for ab staining at 1×106 cells/100uL in PBS/BSA1%. The following antibodies were used: anti-OCT4 rabbit monoclonal (INVITROGEN), IgG2a isotype matched negative control (Invitrogen), AlexaFluor-488 conjugated anti rabbit antibody. Data were acquired using a FACS Calibur instrument and analysis was performed by using FlowingSoftware 2.0 (Cell Imaging Core, University of Turku, Finland).
Xenograft transplantation
Suspensions of 3 × 106 MSTO-211H cells were injected subcutaneously in PBS 1×/Matrigel (BD Biosciences) into 5-week old male SCID mice (Charles River, Milan). Body weight and clinical signs of the mice were determined every 3 d. Tumor volume was calculated by using the formula: V = π/6 × length × width (by manual caliper). All tumorigenicity assays were performed according to the guidelines set by the internal Ethical Committee.
Statistical analysis
Data were analyzed using Student’s t test with correction for data sets with unequal variances. EXCEL software was used to graph and calculate linear regression of the fig. 4E.
Supplementary Material
REFERENCES
- 1.Kaufman AJ, Pass HI. Current concepts in malignant pleural mesothelioma. Expert review of anticancer therapy. 2008 Feb;8(2):293–303. doi: 10.1586/14737140.8.2.293. [DOI] [PubMed] [Google Scholar]
- 2.Carbone M, Ly BH, Dodson RF, Pagano I, Morris PT, Dogan UA, et al. Malignant mesothelioma: facts, myths, and hypotheses. Journal of cellular physiology. 2012 Jan;227(1):44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazarika M, White RM, Jr, Booth BP, Wang YC, Ham DY, Liang CY, et al. Pemetrexed in malignant pleural mesothelioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005 Feb 1;11(3):982–992. [PubMed] [Google Scholar]
- 4.Mujoomdar AA, Tilleman TR, Richards WG, Bueno R, Sugarbaker DJ. Prevalence of in vitro chemotherapeutic drug resistance in primary malignant pleural mesothelioma: result in a cohort of 203 resection specimens. The Journal of thoracic and cardiovascular surgery. 2010 Aug;140(2):352–355. doi: 10.1016/j.jtcvs.2009.11.072. [DOI] [PubMed] [Google Scholar]
- 5.Tang G, Tang X, Mendu V, Tang X, Jia X, Chen QJ, et al. The art of microRNA: various strategies leading to gene silencing via an ancient pathway. Biochimica et biophysica acta. 2008 Nov;1779(11):655–662. doi: 10.1016/j.bbagrm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Ji X. The mechanism of RNase III action: how dicer dices. Current topics in microbiology and immunology. 2008;320:99–116. doi: 10.1007/978-3-540-75157-1_5. [DOI] [PubMed] [Google Scholar]
- 7.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008 Sep 1;7(17):2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Garzon R, Cimmino A, Fabbri M, Croce CM. MicroRNAs and leukemias: how strong is the connection? Leukemia research. 2006 Jun;30(6):653–655. doi: 10.1016/j.leukres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006 Nov;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Pass HI, Goparaju C, Ivanov S, Donington J, Carbone M, Hoshen M, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer research. 2010 Mar 1;70(5):1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. International journal of cancer Journal international du cancer. 2013 Jun 1;132(11):2479–2491. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- 12.Adammek M, Greve B, Kassens N, Schneider C, Bruggemann K, Schuring AN, et al. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertility and sterility. 2013 Jan 8; doi: 10.1016/j.fertnstert.2012.11.055. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, Liu S, Xin H, Jiang J, Younglai E, Sun S, et al. Up-regulation of microRNA-145 promotes differentiation by repressing OCT4 in human endometrial adenocarcinoma cells. Cancer. 2011 Sep 1;117(17):3989–3998. doi: 10.1002/cncr.25944. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. International journal of cancer Journal international du cancer. 2010 Mar 1;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 15.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009 May 15;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 16.Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem cells and development. 2009 Sep;18(7):1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samardzija C, Quinn M, Findlay JK, Ahmed N. Attributes of Oct4 in stem cell biology: perspectives on cancer stem cells of the ovary. Journal of ovarian research. 2012;5(1):37. doi: 10.1186/1757-2215-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ, Gimotty PA, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012 Nov 22;31(47):4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai LL, Yu CC, Chang YC, Yu CH, Chou MY. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2011 Sep;40(8):621–628. doi: 10.1111/j.1600-0714.2011.01015.x. [DOI] [PubMed] [Google Scholar]
- 20.Linn DE, Yang X, Sun F, Xie Y, Chen H, Jiang R, et al. A Role for OCT4 in Tumor Initiation of Drug-Resistant Prostate Cancer Cells. Genes & cancer. 2010 Sep;1(9):908–916. doi: 10.1177/1947601910388271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, et al. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009 May;57(7):724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- 22.Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer research. 2010 Dec 15;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 23.Dai X, Ge J, Wang X, Qian X, Zhang C, Li X. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncology reports. 2013 Jan;29(1):155–160. doi: 10.3892/or.2012.2086. [DOI] [PubMed] [Google Scholar]
- 24.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a subpopulation of stem cell-like cells in ovarian cancer. Oncogene. 2010 Apr 8;29(14):2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 25.Rudd RM. Malignant mesothelioma. British medical bulletin. 2010;93:105–123. doi: 10.1093/bmb/ldp047. [DOI] [PubMed] [Google Scholar]
- 26.Casarsa C, Bassani N, Ambrogi F, Zabucchi G, Boracchi P, Biganzoli E, et al. Epithelial-to-mesenchymal transition, cell polarity and stemness-associated features in malignant pleural mesothelioma. Cancer letters. 2011 Mar 28;302(2):136–143. doi: 10.1016/j.canlet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Schramm A, Opitz I, Thies S, Seifert B, Moch H, Weder W, et al. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. European journal of cardio-thoracic surgery : official journal of the European Association for Cardio-thoracic Surgery. 2010 Mar;37(3):566–572. doi: 10.1016/j.ejcts.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Molecular biology of the cell. 2012 Jun;23(11):2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006 Aug 11;126(3):503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 30.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003 Jun 13;113(6):703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 31.Romanov VS, Pospelov VA, Pospelova TV. Cyclin-dependent kinase inhibitor p21(Waf1): contemporary view on its role in senescence and oncogenesis. Biochemistry Biokhimiia. 2012 Jun;77(6):575–584. doi: 10.1134/S000629791206003X. [DOI] [PubMed] [Google Scholar]
- 32.Wajapeyee N, Deibler SK, Green MR. Genome-wide RNAi screening to identify regulators of oncogene-induced cellular senescence. Methods Mol Biol. 2013;965:373–382. doi: 10.1007/978-1-62703-239-1_25. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg WC, Denning MF. P21Waf1 control of epithelial cell cycle and cell fate. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 2002;13(6):453–464. doi: 10.1177/154411130201300603. [DOI] [PubMed] [Google Scholar]
- 34.Kai K, D'Costa S, Yoon BI, Brody AR, Sills RC, Kim Y. Characterization of side population cells in human malignant mesothelioma cell lines. Lung Cancer. 2010 Nov;70(2):146–151. doi: 10.1016/j.lungcan.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 35.Canino C, Mori F, Cambria A, Diamantini A, Germoni S, Alessandrini G, et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 2012 Jun 28;31(26):3148–3163. doi: 10.1038/onc.2011.485. [DOI] [PubMed] [Google Scholar]
- 36.Yilmaz M, Christofori GEMT. the cytoskeleton, and cancer cell invasion. Cancer metastasis reviews. 2009 Jun;28(1–2):15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 37.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010 Nov 5;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP, et al. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast cancer research : BCR. 2011;13(5):R94. doi: 10.1186/bcr3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granville L, Laga AC, Allen TC, Dishop M, Roggli VL, Churg A, et al. Review and update of uncommon primary pleural tumors: a practical approach to diagnosis. Archives of pathology & laboratory medicine. 2005 Nov;129(11):1428–1443. doi: 10.5858/2005-129-1428-RAUOUP. [DOI] [PubMed] [Google Scholar]
- 40.Jean D, Daubriac J, Le Pimpec-Barthes F, Galateau-Salle F, Jaurand MC. Molecular changes in mesothelioma with an impact on prognosis and treatment. Archives of pathology & laboratory medicine. 2012 Mar;136(3):277–293. doi: 10.5858/arpa.2011-0215-RA. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013 Jan 3;32(1):61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Chopp M, Zheng X, Katakowski M, Buller B, Jiang F. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncology reports. 2013 Jan;29(1):67–72. doi: 10.3892/or.2012.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, et al. MicroRNA-145 Is Downregulated in Glial Tumors and Regulates Glioma Cell Migration by Targeting Connective Tissue Growth Factor. PloS one. 2013;8(2):e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speranza MC, Frattini V, Pisati F, Kapetis D, Porrati P, Eoli M, et al. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget. 2012 Jul;3(7):723–734. doi: 10.18632/oncotarget.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinge CM, Radde BN, Imbert-Fernandez Y, Teng Y, Ivanova MM, Abner SM, et al. Targeting the intracellular MUC1 C-terminal domain inhibits proliferation and estrogen receptor transcriptional activity in lung adenocarcinoma cells. Molecular cancer therapeutics. 2011 Nov;10(11):2062–2071. doi: 10.1158/1535-7163.MCT-11-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Guo H, Qian G, Ge S, Ji H, Hu X, et al. MiR-145, a new regulator of the DNA fragmentation factor-45 (DFF45)-mediated apoptotic network. Molecular cancer. 2010;9:211. doi: 10.1186/1476-4598-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, et al. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. Journal of experimental & clinical cancer research : CR. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, et al. miR-145 inhibits breast cancer cell growth through RTKN. International journal of oncology. 2009 May;34(5):1461–1466. [PubMed] [Google Scholar]
- 49.Bonifacio LN, Jarstfer MB. MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PloS one. 2010;5(9) doi: 10.1371/journal.pone.0012519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu D, Reyes-Mugica M, Rebbaa A. Role of the beta catenin destruction complex in mediating chemotherapy-induced senescence-associated secretory phenotype. PloS one. 2012;7(12):e52188. doi: 10.1371/journal.pone.0052188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cahu J, Bustany S, Sola B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell death & disease. 2012;3:e446. doi: 10.1038/cddis.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodier F. Detection of the senescence-associated secretory phenotype (SASP) Methods Mol Biol. 2013;965:165–173. doi: 10.1007/978-1-62703-239-1_10. [DOI] [PubMed] [Google Scholar]
- 53.Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, et al. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Annals of surgical oncology. 2012 Jul;19(Suppl 3):S656–S664. doi: 10.1245/s10434-012-2217-6. [DOI] [PubMed] [Google Scholar]
- 54.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Molecular biology of the cell. 2011 May 15;22(10):1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008 Oct;7(20):3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 56.Ceppi P, Peter ME. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2013 Mar 4; doi: 10.1038/onc.2013.55. [DOI] [PubMed] [Google Scholar]
- 57.Nakanishi H, Taccioli C, Palatini J, Fernandez-Cymering C, Cui R, Kim T, et al. Loss of miR-125b-1 contributes to head and neck cancer development by dysregulating TACSTD2 and MAPK pathway. Oncogene. 2013 Feb 18; doi: 10.1038/onc.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.