Abstract
Nanomedicine era is not far from its realization, but a major concern of targeted delivery still stands tall in its way. Herein we demonstrate the mechanism underlying the anticancer activity of an RNA aptamer (Apt) conjugated to gefitinib-loaded poly (lactic co-glycolic acid) nanoparticles (GNPs). Apt was selected through Cell-SELEX (systemic evolution of ligands by exponential enrichment) process against gefitinib-resistant H1975 lung cancer cells. The selected aptamer exhibited high specificity toward H1975 cells, both qualitatively as well as quantitatively. Software analysis using the MATCH tool predicted Ets1, a proto-oncoprotein, to be the target of the selected aptamer. Interestingly, the localization of identified aptamer varied in descending order of Ets1 expression, wherein maximum localization was observed in H1975 cells than in MDA-MB231, DU-145, H23, H460, A431, A549 and MCF-7 cells, and minimum in L132 cells. Furthermore, Apt-GNP bio-conjugate showed augmented anticancer activity specifically in Ets1-overexpressing cells. In addition, partial depletion of Ets1 in H1975 cells and overexpression of Ets1 in L132 cells reversed the targeting efficacy of the aptamer. Notably, a single intratumoral injection of the Apt-GNP bio-conjugate abrogated the growth of tumor in H1975 xenograft nude mice. Altogether, we present a pioneering platform, involving aptamers, which can be clinically used as a diagnostic marker for metastasis as well as an effective delivery system to escort the pharmaceutical cargo specifically to Ets1-overexpressing highly progressive tumors.
Introduction
Non-small cell lung cancer is the most common type of lung cancer, which is accompanied with a very high reoccurrence rate of 30–60% depending upon the stage of cancer.1 Hyperactive epidermal growth factor receptor (EGFR) signaling, the leading cause of non-small cell lung cancer, leads to unrestrained cellular proliferation and increased survival, resulting in cellular transformation and tumor progression.2 Thus, EGFR emerged as an attractive target for lung cancer therapy. Gefitinib, which is a selective EGFR (ErbB1) tyrosine kinase inhibitor, prevents autophosphorylation of EGFR in various tumor cell lines and xenografts.3 The major hindrance to an effective anticancer activity of gefitinib is the resistance, which arises in the cells after repeated administration of gefitinib. T790M mutation accounts for almost 50% of the cases in which gefitinib resistance arises. T790 is often referred to as the ‘gatekeeper residue'. Substitution of the threonine at this codon with a bulkier residue, such as methionine, is believed to sterically hinder the binding of gefitinib.
To circumvent this problem, we developed a drug delivery platform, specifically against T790M mutant lung cancer cells, involving RNA aptamer and drug-loaded nanoparticles. Ellington and Szostak,4 and Tuerk and Gold5, in 1990, independently described the method of aptamer selection and termed it as systemic evolution of ligands by exponential enrichment (SELEX). This process was designed to select highly specific aptamer sequences against defined targets. Lately, the process of Cell-SELEX has taken over the conventional method of aptamer selection. Cell-SELEX allows the selection of molecular aptamers against cancer cells of interest without any prior knowledge of cell-surface marker proteins, and are thus more flexible and practical to use than other molecular marker-based methods. Aptamers, which can specifically identify the brain tumor-initiating cells,6 liver cancer,7 ovarian cancer8 and prostate cancer cells,9 have been isolated by various research groups.
The novelty of this report lies not in the aptamer selection procedure but in target validation. As stated above, various researchers have reported the selection of cell-specific aptamers, but only a handful studies involve the identification of the aptamer target.10 We used the well-reported Cell-SELEX process for selecting specific aptamer for H1975 T790M mutant lung carcinoma cells (described in Supplementary Figure 1). However, we went a step further and validated the target of aptamer by using bioinformatics approach, which yielded an oncogenic transcription factor Ets1 as the target of our selected aptamer. Our results collectively support the strong candidature of our selected aptamer as a targeting agent for Ets1-overexpressing cells. We provide a pioneering report describing the selection of an RNA aptamer, which can be internalized and retained not only within the cells against which it was selected but also a variety of other metastatic cells that abundantly express the oncogenic transcription factor Ets1.
Results
Selected aptamer exhibits high qualitative and quantitative affinity toward H1975 lung cancer cells
The secondary structure of the resultant sequence obtained after 12 iterative cycles of Cell-SELEX selection was predicted by using Mfold software (Rensselaer Polytechnic Institute, Albany, NY, USA) (Supplementary Figure 2). We used the truncated sequence for our study so as to avoid nonspecific binding (Table 1). Both the target metastatic cancer cells (H1975 cells) and counter-selective noncancer cells (L132 cells) were incubated with Texas Red-labeled aptamer for 60 min. The microscopic images undoubtedly reflect that the localization of aptamer was much higher in H1975 cells as compared with counter-selective L132 cells. Interestingly, in H1975 cells, the aptamer localizes within the nucleus, whereas in L132 cells no significant aptamer internalization was observed (Figure 1a). The higher affinity of aptamers has been attributed to the various three-dimensional structures assumed by them.11 It is worthy to note that the localization of our selected aptamer is not limited to the cell surface or cytosol, but the aptamer localizes efficiently within the nucleus. Therefore, this makes our selected aptamer ideal for targeting drugs within the cell. To determine nonspecific localization, H1975 and L132 cells were treated with Texas Red-labeled initial RNA library. Supplementary Figure 3 shows the representative results for both the cell types treated with Texas Red-labeled library, wherein both the cell types did not exhibit significant localization of the RNA library sequences. Aptamer localization was also studied qualitatively in a panel of other cancer cells, including lung cancer (H23, H460 and A549 cells), skin cancer (A431 cells), breast cancer (MCF-7 and MDA-MB231 cells) and prostate cancer cells (DU-145 cells), and the representative images are shown in Figure 1a. As is evident by the microscopic images, the localization of Texas Red-labeled aptamer was limited in all the cell lines used for the study. H23 lung cancer, MDA-MB231 breast cancer and DU-145 prostate cancer cells show slightly greater accumulation of aptamer, but nuclear localization is not observed in any of these cells. Thus, we inferred from the images that our selected aptamer has maximum ability to internalize and be retained within H1975 lung cancer cells. We further aimed to quantify the internalization of our selected aptamer within the above panel of cells.
Table 1. Sequence of internalized aptamer.
| Aptamer | Size | Sequence |
|---|---|---|
| Apt | 87 | 5′-CAUCGAUGCUAGUCGUAACGAUCCCUGGGGUUAUGAGGUUUCCGGGGAACGAGAACGUUUCUCUCCUCUCCCUAUAGUGAG UCGUAU-3′ |
| Apt-truncated | 25 | 5′-CUGGGGUUAUGAGGUUUCCGGGGAA-3′ |
Figure 1.
(a) Aptamer internalization evaluated in a panel of cells. Cellular internalization of selected RNA aptamers, selected after 12th cycle, was qualitatively analyzed by using the Texas Red-labeled aptamer. Cells were incubated with the Texas Red-labeled aptamer for 60 min under standard conditions. Thereafter, the cells were observed under fluorescent microscope. In all the images, the nucleus is stained as blue (DAPI), and the red color (Texas Red) signifies the presence of aptamer. The extent of aptamer internalization and retention observed in H1975 cells was much more than that observed in any other cells. (b) Quantification of internalized aptamer. Percentage of cells that internalized the selected aptamer was measured by Tali Image-Based Cytometer. The cells were treated with the Texas Red-labeled aptamer for 60 min and then scrapped, washed and loaded onto the Tali Cellular Analysis Slide by pipetting the sample in half-moon-shaped sample loading area. The slide was inserted in the cytometer and the reading was recorded. Three independent experiments were performed for each cell type. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs H1975.
Figure 1b clearly depicts that the highest internalization was observed in H1975 cells (90%). All the other cell types under study showed significantly lower internalization of aptamer. Similarly, to assess any background localization, all the cell types under study were treated with Texas Red-labeled initial RNA library and the amount of internalized RNA sequences was quantified. As observed earlier, cells did not show significant internalization of RNA library (Supplementary Figure 4). This further confirms our above observations and implied that aptamers are specifically binding to and internalizing within H1975 lung cancer cells.
Aptamer conjugated to drug-loaded nanoparticles shows high cytotoxic effect specifically in H1975 cells
To investigate the feasibility of using the selected aptamer for transporting pharmaceutical cargoes within cancer cells, we conjugated them with gefitinib-loaded poly (lactic co-glycolic acid) nanoparticles (GNPs), the synthesis, characterization and mechanism of action of which we have reported earlier.12 Thereafter, we checked if this bio-conjugate (Apt-GNPs) can specifically arrest the growth of H1975 gefitinib-resistant lung cancer cells. Here we sought to evaluate not only the anticancer activity but also the specificity of aptamer-conjugated nanoparticles.
All the cell types under study were treated with comparative dose of gefitinib, GNP, aptamer and Apt-GNP for 48 h, after which the cell viability was assessed. Apt-GNP conjugated formulation showed higher anticancer activity as compared with GNPs and free drug gefitinib, explicitly in H1975 cells (Figure 2a). Interestingly, L132 counter-selective cells along with the other cell types did not show any significant difference between the cell death caused by GNPs and Apt-GNPs, except for H23, MDA-MB231 and DU-145 cells, but the difference was significantly lower as compared with that in H1975 cells. Specific cytotoxic effect in H1975 cells corroborated with our previous results and clearly implies that our selected aptamer specifically identifies the target H1975 cells. We also checked the cytotoxic effect of unconjugated aptamer and GNP on H1975 cells (Supplementary Figure 5). The figure clearly shows that the cytotoxic effect of unconjugated aptamer and GNP was significantly lower than that of the conjugated system. From this data, we infer that both aptamer and GNPs should be conjugated for augmented cytotoxic effect.
Figure 2.
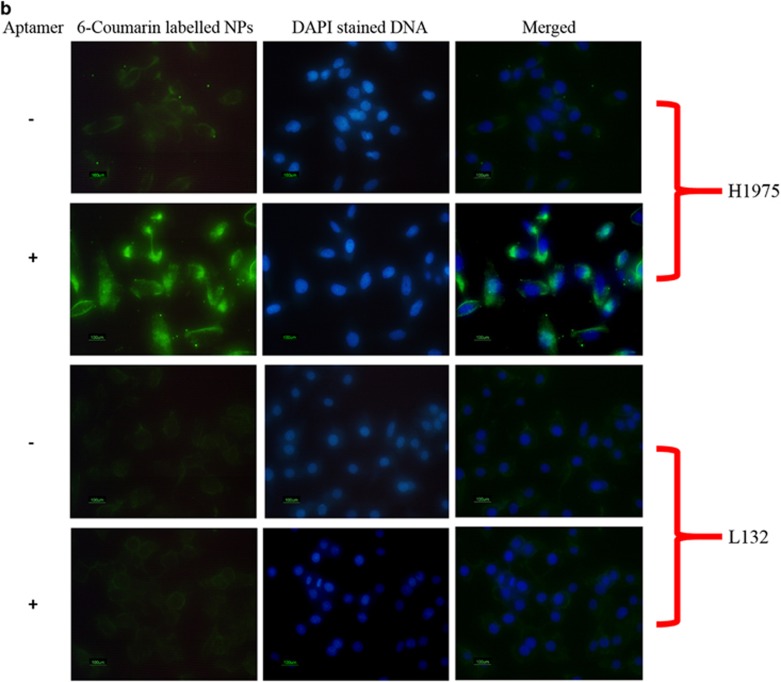
(a) Anticancer activity of gefitinib, GNP, Apt and Apt-GNP. The effect of gefitinib, GNP, Apt and Apt-GNP on the viability of cells was evaluated by using the Tali Image-Based Cytometer. The cells were plated in 100-mm3 dishes and allowed to grow till the cells were 70% confluent. They were then treated with 10 μm gefitinib, GNP and Apt-GNP for 48 h. After treatment, the cells were scrapped and 1 μl of SYTOX dye (Life Technologies) was added per 100 μl of cell suspension and incubated for 1 min in dark. The sample was then loaded on the Tali Cellular Analysis Slide and the slide was inserted in the cytometer. The percentage of viability (%) was then noted. Similarly, untreated cells were also assessed by this method where the viability of cells was 100%. All values are represented as mean±s.e.m.; n=3; ***P<0.001, *P<0.05 vs control and ###P<0.001, #P<0.05 vs GNP. (b) Internalization of Apt-6-coumarin-loaded NP bio-conjugate in H1975 and L132 cells. Both the cells were treated with Apt-6-coumarin-loaded NP bio-conjugate for 60 min and then stained with DAPI. Green stain represents the presence of bio-conjugate, whereas the DAPI stains the nucleus in blue. (c) H1975 cells were treated with 8 μm gefitinib, GNPs, aptamer and apt-GNPs, and then assessed for change in the expression of p300 and H3 acetylation. GNP treatment leads to increased levels of p300 and H3 acetylation. However, Apt-GNP treatment results in even higher expression of p300 and H3 acetylation, thus suggesting higher anticancer activity. Actin and H3 total loading control is also shown. All values are represented as mean±s.e.m.; n=3; **P<0.01, ***P<0.001 vs control.
Thus, from all the above results, we conclude that our selected aptamer specifically identifies H1975 cells and augments the anticancer activity of GNPs probably by increasing the internalization capability and retention of GNPs. For this, we qualitatively assessed the internalization of bio-conjugate by using 6-coumarin-loaded nanoparticles. H1975 and L132 cells were treated with aptamer conjugated to 6-coumarin-loaded nanoparticles and observed under fluorescent microscope (Figure 2b). The fluorescence intensity of 6-coumarin is observed to be much greater in H1975 cells treated with aptamer-conjugated nanoparticles as compared with cells treated with nanoparticle (NPs) alone, which suggests greater internalization and retention of Apt-GNP bio-conjugate as compared with GNPs alone. This further confirms that the increased cytotoxic effect observed in cells treated with the bio-conjugate was due to enhanced localization of NPs within the cells.
We also checked if greater internalization of Apt-GNPs resulted in increased expression of molecular entities altered by GNPs alone. As we have reported earlier, GNPs show higher anticancer activity by augmenting the expression of histone acetyltransferases p300 and CREB-binding protein (CBP). In this study, also we observed a sharp increase in the expression of p300 in GNP-treated cells (Figure 2c). Interestingly, treatment with Apt-GNPs further resulted in an increased expression of p300 in H1975 cells. Similarly, the levels of H3 acetylation were also significantly higher in Apt-GNP-treated cells as compared with untreated, gefitinib-, GNP- or aptamer-treated cells. We propose that our system is utilizing the histone acetyltransferase activator activity of GNPs and selective targeting ability of aptamers to show the increased anticancer activity of gefitinib in H1975 cells.
Ets1 emerged to be the target of the selected aptamer as identified by the MATCH bioinformatic tool
As observed in Figure 1a, the selected aptamer had a tendency to localize within the nucleus, which was very intriguing. Thus, we used the online bioinformatics tool, the MATCH, to check if our selected aptamer showed specificity toward any of the transcription factors present in the cells. The MATCH software (MATCH, Beverly, MA, USA) is used to determine the transcription factor-binding sites within nucleic acid sequences.13 Interestingly, the bioinformatic tool suggested the existence of Ets1 binding site within our selected aptamer.
Ets comprises of a family of transcription factors whose genesis lies in E26, an avian erythroblastosis virus, which carries v-ets oncogene. Ets1 is generally associated with poor prognosis of cancer, which very well explains its high levels in H1975 lung cancer cells. Link between ETS-domain transcription factors and cancer has been well established.14 Direct interaction of aptamer with Ets1 is further confirmed by the fact that ETS-domain proteins bind to sequences, which have a central GGA motif.15 Sequence analysis of our selected aptamer showed that the aptamer indeed has this central moiety, and probably owing to its presence the aptamer was identifying the Ets1 transcription factor or vice versa. Moreover, DNA-binding activity of Ets is also regulated by binding of coregulatory transcription factors. Incidentally, the activity of Ets1 has been closely associated with histone acetyltransferases p300 and CBP. Ets1 recruits CBP and p300, which further accelerates the downstream cascade.16 These all reports can very well explain all our earlier observations.
To confirm that our sequence is binding specifically to H1975 cells owing to the presence of Ets1, we measured the mRNA levels of Ets1 in all the cell types used in the study. The mRNA levels of Ets1 were astronomically higher in H1975 cells as compared with any of the other cell types (Figure 3a). Also, the protein levels of Ets1 were significantly higher in H1975 cells, suggesting active translation of this protein within these cells. Another interesting observation we want to highlight here is that the mRNA and the protein levels of Ets1, although not as high as in H1975 cells, were relatively higher in H23, MDA-MB231 and DU-145 cells as compared with other cell types. This can very well explain our earlier observations of slight increase in aptamer internalization and cell death in these cells.
Figure 3.
Level of Ets1 in a spectrum of cells. (a) The mRNA and protein level of Ets1 was measured and compared in a variety of cells. The RNA from cells was extracted by Trizol method and purified. The RNA was then converted to cDNA and purified. The purified cDNA was then used for quantitative reverse transcription (qRT)–PCR with Ets1 primer (as given in Materials and methods section). The fold change in Ets1 levels was compared with the levels observed in H1975 cells. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs control. The protein level of Ets1 was also assessed in all the cell types by immunoblotting. The protein from the cells was extracted as described in Materials and methods section. Protein estimation was performed by Lowry's method. Equal amounts of protein were then loaded on 10% polyacrylamide gel and transferred on polyvinylidene fluoride membrane. Ets1 was detected by using anti-Ets1 and horseradish peroxidase (HRP)-labeled secondary antibody. The fold change in Ets1 protein levels was calculated with respect to the levels of Ets1 in H1975 cells. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs control. Both the mRNA and protein levels suggest significantly higher levels of Ets1 in H1975 cells as compared with all other cells. (b) Changes in the protein expression of Ets1 and MMP-13 in H1975 cells after the treatment with gefitinib, GNP, aptamer and Apt-GNP. H1975 cells were treated with gefitinib, GNP, aptamer and Apt-GNP for 48 h, and the protein was extracted from these treated cells as described in Materials and methods section. The levels of Ets1 and MMP-13 were checked by immunoblotting. The fold change was calculated with respect to untreated cells. All values are represented as mean±s.e.m.; n=3; ***P<0.001,**P<0.01 vs control. (c) Co-localization of Ets1 and aptamer. H1975 cells treated with Texas Red-labeled aptamer (red) were immunolabeled with Ets1 (green). Blue color stands for DAPI, which stains the DNA.
To further confirm that our selected aptamer specifically identifies Ets1 transcription factor, we investigated whether the treatment of H1975 cells with aptamer-GNP bio-conjugate results in any change in the levels of Ets1 and its downstream molecule matrix metalloproteinases (MMPs). Interestingly, we observed a significant decrease in the protein expression of Ets1 and its downstream molecule MMP-13 in H1975 cells treated with the bio-conjugate (Figure 3b). All our data conclude that aptamer indeed is blocking the Ets1 pathway for cell proliferation and thus showing higher efficacy. Furthermore, as can be seen in Figure 3c, the Texas Red-labeled aptamers co-localize with Ets1 protein in H1975 cells, which strengthens our above observations.
Ets1 depletion in H1975 cells lowers the accumulation of aptamer
To reconfirm that our aptamer was recognizing H1975 cells owing to the presence of high levels of Ets1, we inhibited the expression of Ets1 in H1975 cells by using endoribonuclease-prepared siRNAs (esiRNAs) against it. We checked if the selected aptamer could identify H1975 cells even after silencing Ets1 gene. Complete knockdown of Ets1 resulted in extensive cell death and hence, we could only partially silence it. Figure 4a clearly shows that on increasing the concentration of esiRNA, percentage of knockdown increased.
Figure 4.
Partial knockdown of Ets1 in H1975 cells. (a) H1975 cells were treated with increasing concentrations of esiRNA for 48 h, and then assessed for the percentage of knockdown by both qRT–PCR using Ets1 primers and immunoblotting by using anti-Ets1. The PCR products were run on 2% agarose gel with ethidium bromide and observed under ultraviolet light in a gel docking system. The fold change was calculated with respect to untreated cells. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs control. (b) Aptamer internalized within H1975 cells with and without Ets1 esiRNA. Localization of selected Texas Red-labeled RNA aptamers in a, Ets1 esiRNA untreated cells; and in b, Ets1–esiRNA-treated cells. In all the images, the nucleus is in blue (DAPI) and the aptamer is in red (Texas Red). Significant reduction in aptamer internalization and retention is observed in Ets1 knockdown H1975 cells. (c) Quantification of aptamer internalized within esiRNA (Ets1) untreated and treated cells. H1975 cells treated with Ets1 esiRNA were assessed quantitatively for aptamer internalization by the Tali Image-Based Cytometer. Normal and Ets1 knockdown cells were treated with Texas Red-labeled aptamer for 60 min, and then the cells were scrapped and loaded onto the Tali Cellular Analysis Slide. Three such independent experiments were performed and the percentage of internalization was recorded. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs H1975. (d) Cell viability assay in Ets1–esiRNA-treated H1975 cells. H1975 cells with partial knockdown Ets1 cells were treated with 10 μm gefitinib, GNP and Apt-GNP for 48 h. The cell viability was calculated with H1975 with partially silenced Ets1 as 100% viable cells. All values are represented as mean±s.e.m.; n=3; **P<0.01, ***P<0.001 vs control.
H1975 cells with silenced Ets1 showed lower internalization of Texas Red-labeled aptamer as compared with Ets1-positive cells (Figures 4b and c), both qualitatively and quantitatively. The percentage of H1975 cells internalizing aptamer reduced from 90% to only 40% when the Ets1 was silenced in these cells. This further convinced us that the aptamer is exercising its activity by identifying Ets1 in H1975 cells. Moreover, no significant cell death was observed in Ets1-silenced H1975 cells after treatment with aptamer-conjugated nanoparticles (as compared with GNPs; Figure 4d).
Overexpression of Ets1 in L132 cells renders them aptamer sensitive
To further validate our above observation, L132 cells were transfected with Ets1 plasmid so as to overexpress Ets1 protein in these cells (Figure 5a). Ets1-overexpressing L132 cells showed higher internalization of Texas Red-labeled aptamer (Figures 5b and c). Moreover, higher cell death was observed in Ets1-overexpressing L132 cells treated with Apt-GNP as compared with control L132 cells (Figure 5d). These transfection assays demonstrate, beyond doubt, that the presence or absence of Ets1 in the cells was the deciding factor for the successful anticancer effect of our aptamer-based drug delivery platform.
Figure 5.
Overexpression of Ets1 on L132 cells by transfection with Ets1 plasmid. (a) L132 cells were treated with Ets1 plasmid for 48 h and then assessed for percentage of overexpression by western blotting. All values are represented as mean±s.e.m.; n=3); ***P<0.001 vs control. (b) Aptamer internalized within the L132 cells with and without Ets1 overexpression. Cellular localization of Texas Red-labeled RNAs aptamers in a, Ets1 plasmid-untreated cells; and in b, Ets1 plasmid-treated cells. In all the images, the nucleus is in blue (DAPI) and the aptamer is in red (Texas Red). Significant increase in the localization and retention of aptamer can be observed in Ets1-overexpressed L132 cells. (c) Quantification of aptamer internalization within the Ets1 plasmid untreated and treated cells. L132 cells transfected with Ets1 plasmid were assessed quantitatively for aptamer internalization by the Tali Image-Based Cytometer. Normal and Ets1-overexpressed cells were treated with Texas Red-labeled aptamer for 60 min, and then the cells were scrapped and loaded onto the Tali Cellular Analysis Slide. Three such independent experiments were performed and the percentage of internalization was recorded. All values are represented as mean±s.e.m.; n=3; ***P<0.001 vs H1975. (d) Cell viability assay in Ets1-overexpressed L132 cells. L132 cells with overexpressed Ets1 were treated with 10 μm gefitinib, GNP and Apt-GNP for 48 h. The cell viability was calculated with L132 cells overexpressing Ets1 as 100% viable cells. All values are represented as mean±s.e.m.; n=3; **P<0.01; *P<0.05 vs control and #P<0.05 vs GNP.
Here we would like to highlight an advantage of aptamer-based delivery system over the use of siRNA. One of the limiting factors of siRNA-based therapy is the off-target effect of these antisense oligonucleotides, whereas aptamers have very low likelihood of such off-target effects as suggested by the previous reports.17 Ets family consists of members with oncogenic as well as tumor-suppressive activities. siRNA, even though nonspecifically, inhibits the other members of Ets family, which includes the tumor suppressors Ets2, Elf5 and PDEF (Supplementary Figure 6). Interestingly, aptamers did not decrease the mRNA levels, but instead increase the levels of these tumor-suppressive proteins. Even though the exact mechanism behind this behavior is still unclear, however, the most plausible explanation may involve the compensatory expression of Ets family proteins. We have tried to give a rational explanation for this in Supplementary Text.
Aptamer-NP bio-conjugates exhibit high efficacy in H1975 xenograft model of lung cancer
The fact that Apt-GNP was uniquely able to target H1975 cells in vitro did not ensure their efficacy in vivo. Various issues can come up with the drug delivery platform that involves aptamer, the major concern being its stability. Thus, to ensure that our Apt-GNP system works in the biological milieu, we evaluated the efficacy of our bio-conjugates using xenograft model of lung cancer developed by subcutaneous injection of H1975 cells in the flank of BALB/c nude mice.
As illustrated in Figure 6a, a single intratumoral administration of Apt-GNP was most effective in limiting the growth of tumor as compared with untreated animals. No significant anticancer activity was observed in the animals injected with gefitinib, although GNP-treated animals also showed slight decrease in the growth of tumor. A significant difference was observed in the tumor size of control and Apt-GNP-treated animals after the completion of the study (Figure 6b). Moreover, we also observed a significant difference between the tumor volumes of GNP- and Apt-GNP-treated animals at 20th-day interval. As depicted in Kaplan–Meier survival curve (Figure 6c), all the animals in the Apt-GNP-treated group survived the 21-day period, whereas the animals in the other three groups were killed during the 21-day period when they reached the end point.
Figure 6.
Comparative anticancer activity in H1975 s.c. xenograft nude mice model of lung cancer. (a) Lung cancer was induced in nude mice by implanting H1975 lung cancer cells subcutaneously in the flank of the mice. The tumor was allowed to grow till ~80 mm3. Thereafter, a single intratumoral injection of saline, gefitinib, GNP and Apt-GNP (50 mg/kg) was administered, and the tumor volume was checked over a period of 21 days. Data represents mean±s.e.m. of five mice per group. ***P<0.001 and **P<0.01 vs tumor control at 20th day; ##P<0.01 vs GNP at 20th day. (b) Representative tumors excised out of the mice at the end point for untreated and Apt-GNP-treated mice are shown. Although no significant regression was observed, Apt-GNP were able to limit the growth of the tumor to a great extent. (c) The Kaplan–Meier survival curve shows that 100% of Apt-GNP group survived till 21 days and the animals in other groups were killed during the period of study when they reached the end point (tumor volume 300 mm3). (d) BWL of animals after dosing of animals. BWL was calculated after 21 days of injection. Apt-GNP-treated animals show minimum loss in body weight. (e) Histopathological analysis (hematoxylin and eosin staining) of tumors excised from (a) saline-, (b) gefitinib-, (c) GNP- and (d) Apt-GNP (50 mg/kg)-treated animals **P<0.01 vs gefitinib, #P<0.05 vs GNP. Scale bar=100μm
We assessed the levels of toxicity experienced by the animals of each group by analyzing the effect of gefitinib, GNP and Apt-GNP on body weight loss (BWL; Figure 6d). The saline group demonstrated BWL that paralleled their tumor load. A very high BWL was observed in animals treated with gefitinib and GNP. Significantly lower BWL was observed in Apt-GNP-treated animals. This pattern of BWL can be explained if we consider that in gefitinib- and GNP-treated animals, whole drug does not internalize within the tumor and is thus causing systemic absorption and distribution leading to higher toxicity. Whereas in Apt-GNP-treated animals, the instant binding and internalization of GNPs within the tumor will ensure that whole of the drug is released within the tumor itself.18 After the animals were killed, the tumor was used for histopathological analysis, which clearly indicated the significant fibrotic damage to the tumor tissue of Apt-GNP-treated animals. Some degree of damage can also be observed in the tumor of GNP-treated animals, but saline- and gefitinib-treated animals do not show any such signs of fibrosis (Figure 6e).
Thus, higher efficacy of Apt-GNP as compared with free drug and GNPs, both in vitro and in vivo, suggests the suitability of this platform for the treatment of highly progressive cancers. The aptamer increases the residence time of the NPs within the cancer cells expressing high levels of Ets1 protein. The presence of Ets1 has been detected not only in the nucleus but also in the cytoplasm as well.19, 20 Probably, it is this cytoplasmic Ets1 that helps to retain the aptamer sequence within the cell. Higher accumulation and concentration of the drug not only lead to higher efficacy but also to lower toxicity of it.
Discussion
Various strategies have been designed for intracellular delivery of nanoparticles including modification of surface topology and charge.21 However, a huge limitation of such an approach arises owing to nonspecificity of these nanoparticles, owing to which they are taken up and retained within the cells without any discrimination. Ligand-conjugated nanoparticles have also been studied extensively and have been reported to enhance the cellular uptake of nanoparticles by receptor-mediated endocytosis and thus provide cell-targeting specificity.22 However, again for such an approach, we require well-characterized cancer antigens for the development of ligands against them. Owing to the diverse nature of cancer cells, there always exists a deficit for diagnostic and therapeutic markers.23 The selection of aptamers against purified proteins has an advantage of optimal enrichment during the selection process, but Cell-SELEX process is preferred when the clear marker target is unknown. Herein we selected aptamer against H1975 lung cancer cells that possess T790M mutation in EGFR domain, the leading cause of tyrosine kinase inhibitor resistance. The selection process was performed under normal physiological conditions by Cell-SELEX process. We chose L132 lung cells (noncancer cells) as our counter-selective cells so as to avoid selection of nonspecific aptamers, which may bind to normal lung cells. The selected aptamer exhibited high specificity toward H1975 cells and did not internalize within the normal cells. Thus, they were suitable to carry anticancer drugs and retain them within the cancer cells only.
Various research groups all over the world are engaged in developing effective platform for cancer therapeutics by using aptamers.9, 24, 25 Several reports indicate that aptamer-conjugated drugs show higher effect as compared with unconjugated drugs/nanoparticles,26 although the exact mechanism through which these aptamers augment the anticancer activity has not been evaluated in detail. Our study also shows that the aptamer-conjugated nanoparticles exhibit higher anticancer activity as compared with free drug and NPs alone. It is anticipated that higher accumulation of the drug might be the reason behind it.
Although higher accumulation of the nanoparticles within the tumor does explain higher activity of the conjugates to some extent, it was intriguing that our selected aptamer was showing such high specificity toward H1975 cells and was accumulating specifically within the nucleus. The specific localization of aptamer in the nucleus suggested that it could be interacting either with chromatin or with transcription factors present inside the nucleus. Thus, we checked, through bioinformatic tools, the reason behind aptamer specificity and as a result we obtained Ets1 as our probable target.
A variety of solid tumors including epithelial tumors, sarcomas and astrocytomas possess Ets1.27, 28 High levels of Ets1 in lung and squamous cell carcinoma have been linked with a higher incidence of lymph node metastasis. There is also a growing body of evidence, which suggests that Ets1 has a key role in acquisition of invasive behavior. The genes that respond to Ets1 include metalloproteases MMP-1, MMP-3, MMP-9 and urokinase-type plasminogen activator.29 These proteases are involved in the degradation of extracellular matrix, which is a key event in the process of metastasis. These evidences along with the results of transfection assays (Figures 4 and 5) convinced us that there exists a definitive role of Ets1 behind the augmented anticancer activity of aptamers conjugated to GNPs.
Moreover, our in vivo studies also showed the higher efficacy of Apt-GNP conjugates over free drug. A significant difference was observed in tumor volumes of untreated animals and Apt-GNP-treated ones. However, a statistically significant result in tumor volumes of GNP- and Apt-GNP-treated animals was obtained only at 20th-day interval. The growth of tumor in both these groups paralleled till 2 weeks after which the increase in tumor volume was lowered in Apt-GNP-treated animals. This can be explained on the basis of increased retention of Apt-GNPs within the tumor.
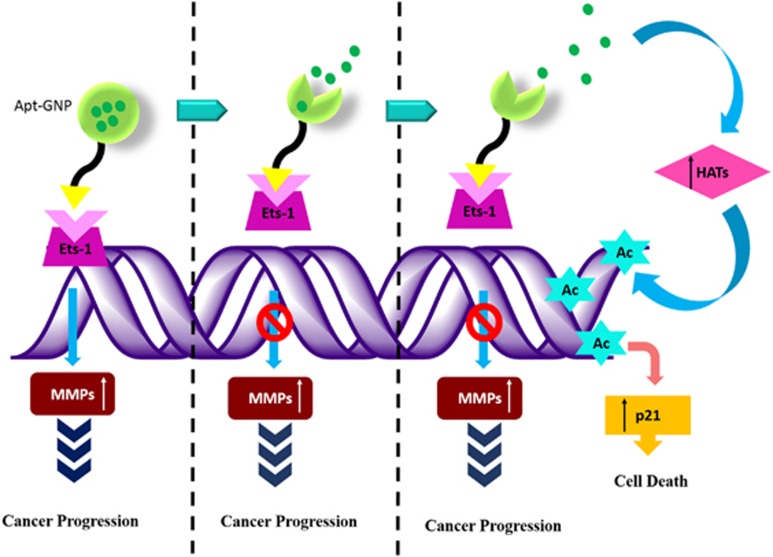
In summary (Figure 7), an aptamer-based drug delivery system was developed, which was although designed to target H1975 T790M mutant lung cancer cells, but our studies show that it works effectively in Ets1 transcription factor overexpressing metastatic cancer cells. Impact of this delivery system generates tremendous significance because of its absolute specificity toward Ets1, which is highly abundant in a vast variety of progressive cancer cells. In addition, the small size of the aptamer ensures lower cost of the targeting vehicle as compared with the antibodies. Our drug delivery system eliminates the probability of chronic accumulation of the targeting system as aptamers are merely composed of nucleic acid, which will degrade once their assigned job is completed. The novelty of our study and advantage of our formulation is not only its increased efficacy but also more importantly, its safety through aptamer conjugation (Apt-GNP). Apart from that, this bio-conjugate system can also be used for diagnostic purposes to identify aggressive tumors. Collectively, this aptamer-based drug delivery platform is an ideal clinical candidate for effective drug delivery as well as for the diagnosis of progressive cancers. Further clinical studies are required to assess the applicability of this delivery system in humans.
Figure 7.
Graphical summary.
Materials and methods
Selection of aptamers by Cell-SELEX
Aptamer selection was performed as described before by Xiao et al.30 with slight modifications as described in the Supplementary Text.
Internalization of selected aptamer
The aptamer sequence obtained after sequencing was tagged with Texas Red and cells were treated with these Texas Red-labeled aptamer for 60 min. After the incubation period was over, the cells were washed extensively with phosphate-buffered saline and were fixed with 2.5% glutaraldehyde for 5 min. Thereafter, the cells were again washed to remove glutaraldehyde and were further treated with 0.1% Tween to increase the permeability of the cells. After 1 min of incubation with 0.1% Tween, the cells were washed and treated with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) solution for 10 s. Thereafter, the cells were observed under fluorescent microscope at × 40 magnification.
Quantification of internalized aptamers
The percentage of cells with internalized aptamer was quantified by using the Tali Image-Based Cytometer (Life Technologies, Carlsbad, CA, USA). The cells were treated with Texas Red-labeled aptamers for 60 min and then washed extensively to remove any unbound aptamers. The cells were then scrapped in phosphate-buffered saline and 25 μl of sample was then loaded on Tali Cellular Analysis Slide (Life Technologies) by pipetting the sample in the half-moon-shaped sample loading area. The slide was then inserted within the slide port of the cytometer and the reading was recorded. Three independent experiments were performed for each cell type.
Conjugation of selected aptamer with GNPs
GNPs in DNase–RNase-free water (10 g/l) were treated with 400 mm 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 100 mm N-Hydroxysuccinimide (NHS) for 15 min at room temperature with mild agitation to give the corresponding NHS-ester. The NHS-activated NPs were washed twice using double distilled water to remove unbound NHS. The activated nanoparticles were then conjugated to 5′-NH2-modified aptamer (Sigma, St. Louis, MO, USA) of 2% weight compared with polymer concentration for 2 h at room temperature with gentle stirring. The resulting aptamer-conjugated poly (lactic co-glycolic acid) nanoparticles were washed three times with DNase–RNase-free water.
Cytotoxicity of aptamer-NP conjugates
In vitro cytotoxicity was determined using the Tali Viability Kit-Dead Cell Green (Invitrogen, Carlsbad, CA, USA). Briefly, cells (5 × 104 cells per well) were treated with 10 μm free drug, GNPs or GNP loaded on aptamer in medium (without fetal bovine serum, unless denoted otherwise; 37 °C; 5% CO2) for 48 h; and were then scrapped and collected by centrifugation. The cytotoxicity assay was performed as per the manufacturer's instructions. Briefly, 1 μl of SYTOX dye was added per 100 μl of cell suspension, vortexed and incubated for 1 min in dark. A volume of 25 μl of sample was then loaded on the Tali Cellular Analysis Slide by pipetting the sample in the half-moon-shaped sample loading area. The slide was then inserted within the slide port of the Tali Image-Based Cytometer. The percentage of viability (%) was then noted. Similarly, untreated cells were also assessed by this method in which the viability of cells was 100%.
Partial knockdown of Ets1 in H1975 cells
H1975 cells were plated in cell culture dishes and allowed to grow in antibiotic-free medium. The cells were allowed to grow till 50% confluence before esiRNA treatment. An amount of 40 nm of esiRNA (Sigma) was used for partial knockdown of Ets1 in cancer cells as described in the Supplementary Text.
Transfection of Ets1 in L132 cells
The full-length cDNA clone of Ets1 was obtained from Origene (Rockville, MD, USA). Transfection of Ets1 in L132 cells was performed as per the manufacturer's instructions and is described in the Supplementary Text.
In vivo anticancer efficacy evaluation
Female nude mice weighing 18–20 g, from National Institute of Nutrition, Hyderabad, India, were obtained (IAEC13/13) and transferred to the pathogen-free environment of the National Toxicology Center in National Institute of Pharmaceutical Education and Research, SAS Nagar, India. The animals were maintained in sterile and clean cages with HEPA filters under standard diet for nude mice with free access to water and at controlled room temperature of 22±2 °C. All the animals were acclimatized for a minimum period of 1 week before the start of experiment and also properly maintained with all necessary attention until the experiment was over.
Injection of gefitinib-resistant cells in nude mice
Xenografts of gefitinib-resistant lung cancer cells were established by injecting 3 × 106 viable cells into the flank/leg region of nude mice to produce tumors. The tumor size was checked twice weekly, and when the tumor was visible it was measured twice weekly with vernier calliper. Thereafter, the animals were treated with gefitinib, GNPs and Apt-GNPs.
Statistical analysis
Results were expressed as the mean value±s.e.m. The comparison of mean values among the various groups was performed by one-way analysis of variance followed by multiple comparisons by Tukey's test. In vivo data was analyzed by using two-way analysis of variance. P-value <0.05 is considered to be significant.
Acknowledgments
The intellectual property rights of the above described delivery system comprising of the aptamer-NP bio-conjugate are protected under Indian Patent Office (Indian Patent application no: 1623/DEL/2014).
Author Contributions
JK designed, performed the experiments and analyzed the data. KT designed, supervised and approved the final version of manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- 1Meert A-P, Martin B, Delmotte P, Berghmans T, Lafitte J-J, Mascaux C et al. The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J 2002; 20: 975–981. [DOI] [PubMed] [Google Scholar]
- 2Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354. [DOI] [PubMed] [Google Scholar]
- 3Arteaga CL, Johnson DH. Tyrosine kinase inhibitors-ZD1839 (Iressa). Curr Opin Oncol 2001; 13: 491–498. [DOI] [PubMed] [Google Scholar]
- 4Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990; 346: 818–822. [DOI] [PubMed] [Google Scholar]
- 5Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990; 249: 505–510. [DOI] [PubMed] [Google Scholar]
- 6Kim Y, Wu Q, Hamerlik P, Hitomi M, Sloan AE, Barnett GH et al. Aptamer identification of brain tumor-initiating cells. Cancer Res 2013; 73: 4923–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y et al. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem 2008; 80: 721–728. [DOI] [PubMed] [Google Scholar]
- 8Van Simaeys D, Löpez-Colön D, Sefah K, Sutphen R, Jimenez E, Tan W. Study of the molecular recognition of aptamers selected through ovarian cancer cell-SELEX. PLoS One 2010; 5: e13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 2002; 62: 4029–4033. [PubMed] [Google Scholar]
- 10Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol 2010; 6: 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010; 9: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Kaur J, Tikoo K. p300/CBP dependent hyperacetylation of histone potentiates anticancer activity of gefitinib nanoparticles. Biochim Biophys Acta 2013; 1833: 1028–1040. [DOI] [PubMed] [Google Scholar]
- 13Kel AE, Gáβling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 2003; 31: 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2: 827–837. [DOI] [PubMed] [Google Scholar]
- 15Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol 1997; 29: 1371–1387. [DOI] [PubMed] [Google Scholar]
- 16Li R, Pei H, Watson DK. Regulation of Ets function by protein-protein interactions. Oncogene 2000; 19: 55. [DOI] [PubMed] [Google Scholar]
- 17Dassie JP, Hernandez LI, Thomas GS, Long ME, Rockey WM, Howell CA et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 2014; 22: 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Farokhzad OC, Jon S, Khademhosseini A, Tran T-NT, LaVan DA, Langer R. Nanoparticle-aptamer bioconjugates a new approach for targeting prostate cancer cells. Cancer Res 2004; 64: 7668–7672. [DOI] [PubMed] [Google Scholar]
- 19Buggy Y, Maguire T, McGreal G, McDermott E, Hill A, O'Higgins N et al. Overexpression of the Ets1 transcription factor in human breast cancer. Br J Cancer 2004; 91: 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Nakayama T, Ito M, Ohtsuru A, Naito S, Sekine I. Expression of the ets-1 proto-oncogene in human colorectal carcinoma. Mod Pathol 2001; 14: 415–422. [DOI] [PubMed] [Google Scholar]
- 21Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov 2010; 9: 615–627. [DOI] [PubMed] [Google Scholar]
- 22Davis ME, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov 2008; 7: 771–782. [DOI] [PubMed] [Google Scholar]
- 23Guo K-T, Ziemer G, Paul A, Wendel HP. CELL-SELEX: novel perspectives of aptamer-based therapeutics. Int J Mol Sci 2008; 9: 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One 2011; 6: e20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA 2006; 103: 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Chu TC, Marks JW, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD et al. Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res 2006; 66: 5989–5992. [DOI] [PubMed] [Google Scholar]
- 27Valter MM, Hügel A, Huang HJS, Cavenee WK, Wiestler OD, Pietsch T et al. Expression of the Ets1 transcription factor in human astrocytomas is associated with Fms-like tyrosine kinase-1 (Flt-1)/vascular endothelial growth factor receptor-1 synthesis and neoangiogenesis. Cancer Res 1999; 59: 5608–5614. [PubMed] [Google Scholar]
- 28Sacchi N, De Klein A, Showalter SD, Bigi G, Papas TS. High expression of ets-1 gene in human thymocytes and immature T leukemic cells. Leukemia 1988; 2: 12–18. [PubMed] [Google Scholar]
- 29Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene 2000; 19: 6533–6549. [DOI] [PubMed] [Google Scholar]
- 30Xiao Z, Levy-Nissenbaum E, Alexis F, Lupták A, Teply BA, Chan JM et al. Engineering of targeted nanoparticles for cancer therapy using internalizing aptamers isolated by cell-uptake selection. ACS Nano 2012; 6: 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.