Abstract
Mutations in Wilms' tumor 1 (WT1) cause a wide spectrum of renal manifestations, eventually leading to end-stage kidney failure. Insufficient understanding of WT1's molecular functions in kidney development has hampered efficient therapeutic applications for WT1-associated diseases. Recently, the generation and characterization of mouse models and application of multiple state-of-the-art approaches have significantly expanded our understanding of the molecular mechanisms of how WT1 mutations lead to kidney failure. Here, we discuss the WT1 binding consensus and illustrate the major roles of WT1 in different cell populations in kidney biology. WT1 controls metanephric mesenchyme (MM) self-renewal and proliferation mainly by regulating FGF and BMP-pSMAD signaling pathways as well as Sall1 and Pax2, encoding key transcription factors; WT1 drives MM differentiation and mesenchyme–epithelial transition by targeting Fgf8 and Wnt4; WT1 defines podocyte identity by activation of other podocyte-specific transcription factors, including Mafb, Lmx1b, FoxC2, and Tcf21. These factors potentially cooperate with WT1 regulating the expression of components and regulators of the cytoskeleton for establishing podocyte polarity, slit diaphragm structure, and focal adhesion to the glomerular basement membrane. Understanding of WT1's function in kidney biology including WT1-regulated pathways will give insights that will eventually help therapeutic applications.
Keywords: metanephric mesenchyme, podocyte, super-enhancers, Wilms' tumor 1
Kidney failure is a worldwide public health problem, with increasing incidence and prevalence, high costs, and poor outcomes.1 Some of those renal manifestations are caused by Wilms' tumor 1 (WT1) mutations, including Wilms' tumor, the Denys-Drash syndrome (DDS), the Frasier syndrome (FS), and the steroid-resistant nephrotic syndrome. To understand WT1 function in kidney development and homeostasis, multiple mouse models have been generated and characterized. Particularly, the generation of Wt1 conditional knockout mice crossed to inducible and cell-specific Cre mice allowed to dissect Wt1 function with regard to specific cell populations and the time of development. To decipher the molecular mechanisms of Wt1 mutations leading to kidney failure, multiple state-of-the-art experimental approaches have been applied, such as chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) using embryonic and adult kidneys. In this review, we present and summarize our current understanding of WT1's function in kidney biology including WT1-controlled regulatory networks. We also discuss how this knowledge might be translated into therapeutic applications.
WT1 AND ITS EXPRESSION PATTERN IN THE MAMMALIAN KIDNEY
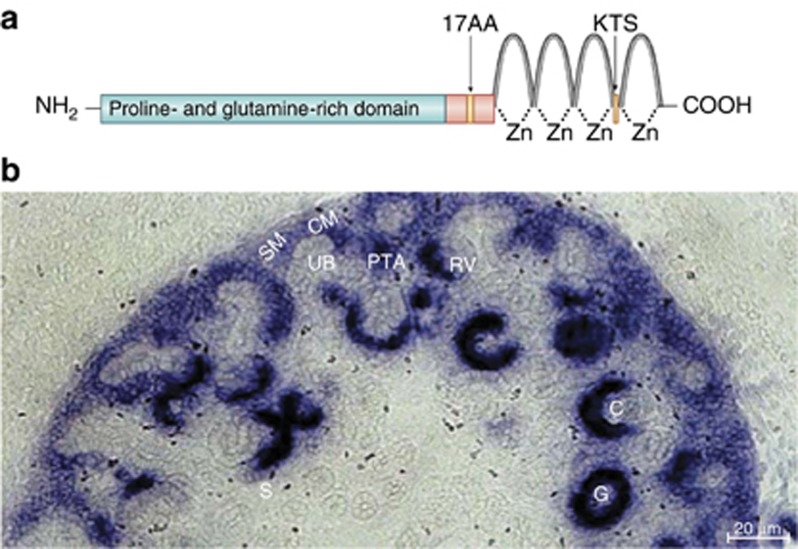
The human WT1 gene spans 50 kb of genomic DNA and comprises 10 exons, which generate a 3 kb mRNA. It encodes a protein with an amino-terminal proline- and glutamine-rich protein interaction domain and four carboxy-terminal Krüppel-type (Cys2-His2) zinc fingers, major characteristics of a typical transcription factor. Expression of WT1 is complex, with at least 36 isoforms derived from alternative splicing, usage of alternative translation start sites, and RNA editing. There are two major alternative splice sites resulting in four isoforms: proteins with or without the central 17 amino acids from alternative splicing of exon 5 and proteins including or excluding the three amino acids lysine, threonine, and serine (KTS) between the third and fourth zinc finger domain (Figure 1a). The corresponding isoforms are termed WT1+KTS and WT1−KTS and are expressed in a constant ratio of about 2:1 in humans.2 WT1 mutations in human patients are commonly associated with dysfunction of the urogenital system, indicating the essential roles of WT1 for kidney and gonad development. In mammals, the definitive kidney, the metanephros, forms at the end of the posterior intermediate mesoderm through distinct consecutive steps.3 In brief, first, a specialized region called metanephric mesenchyme (MM) is formed in the posterior intermediate mesoderm; second, the MM induces an outgrowth of the nephric duct called ureteric bud (UB); third, the UB invades the MM and induces adjacent MM cells to condense around the UB tip, forming the cap mesenchyme; the remaining MM cells are called stromal mesenchyme; fourth, the cap mesenchyme cells induce UB to branch, whereas a subset of the cap mesenchyme (pre-tubular aggregate) on the ventral side of the UB tips undergoes mesenchymal–epithelial transition to form a renal vesicle. This fuses with the ureteric stalk, generating a comma-shaped body and subsequently an S-shaped body with a proximal-distal cleft. The most proximal cleft is infiltrated by endothelial cells and forms the glomerular tuft. Finally, reciprocal epithelial–mesenchymal interactions induce UB to branch repeatedly in a highly reproducible manner, and new nephrons form from the mesenchyme adjacent to each new UB tip. The ureteric branches form the collecting duct tree that connects the nephrons to the ureter and drain the urine into the bladder. The earliest WT1 signal is detected in intermediate mesoderm at E9.0, and then the signal increases in cap mesenchyme after UB induction. Higher expression is also detected in pre-tubular aggregates, proximal parts of renal vesicle, and comma- and S-shaped body. The highest expression is in capillary loops and finally restricted to podocytes of the glomerulus, where expression is maintained throughout life (Figure 1b).
Figure 1.
Wilms' tumor 1 (WT1) protein structure and Wt1 expression pattern in developing murine kidney. (a) Structure of WT1 protein showing alternative splice regions consisting of 17 amino acids encoded by exon 5 and amino acids lysine, threonine, and serine (KTS) encoded by the 3′-end of exon 9 and the different functional domains. (b) The expression of Wt1 in developing kidney (E15.5) detected by in-situ hybridization (picture provided by Dagmar Kruspe). Wt1 is expressed in cap mesenchyme (CM) surrounding the ureteric bud (UB) and at very low level in stromal mesenchyme (SM). Wt1 expression is increased in pre-tubular aggregates (PTAs) that begin to undergo mesenchymal to epithelial transition to form the renal vesicle (RV). Wt1 is expressed in the proximal part of renal vesicles and the S-shaped bodies (S), which give rise to podocyte precursors in the capillary loop (C). The highest expression of Wt1 is found in podocytes within glomeruli (G). Scale bar=20 μm.
WT1 ALTERATIONS AND RENAL ANOMALIES
WT1 was initially identified using a positional cloning approach, as it is located in a genomic region, deletion of which results in the WAGR syndrome (Wilms' tumor, aniridia, genitourinary anomalies, and mental retardation).4, 5 Since then many WT1 mutations were identified, including missense, nonsense, splice site mutations, as well as deletions.6 WT1 mutations have been found to be associated with a large spectrum of phenotypes at the clinical level, including Wilms' tumor, the DDS, the FS, and the isolated steroid-resistant nephrotic syndrome, all of which progress to end-stage kidney disease. Overall, there is a solid genotype–phenotype association of WT1 diseases.6, 7, 8 Approximately 20% of WTs, the most commonly found kidney tumor in children, are caused by WT1 mutations. A high number of bilateral tumors are associated with truncation mutations, particularly in the 5′ half of the WT1 gene. Mutations within exon 8 or 9 of WT1 usually lead to the DDS, characterized by diffuse mesangial sclerosis, urogenital abnormalities, and a high risk of developing Wilms' tumor. Disruption of the KTS splicing, caused by splice site mutations, results in the loss of the WT1+KTS isoform and leads to the FS, characterized by pseudohermaphroditism and focal segmental glomerulosclerosis. In addition, there are idiopathic steroid-resistant glomerulopathies caused by mutations in WT1. Wilms' tumors or nephroblastoma often contain metanephric blastema, stromal and epithelial derivatives. It is therefore thought to arise by a failure of kidney progenitor cells to differentiate. On the other hand, glomerulopathies as seen in DDS, FS, and steroid-resistant nephrotic syndrome are considered to result from the loss of WT1 function during podocyte development and maintenance. In the following, we will discuss WT1 as a transcription factor and discriminate between its functions in kidney progenitor cells and in podocytes.
SEQUENCE AND LOCALIZATION OF WT1 BINDING SITES
WT1 has been suggested to recognize and bind the EGR1-like GC-rich DNA sequence.9 However, a physiologically relevant WT1 binding sequence cannot only be deduced from in vitro experiments. Recently, chromatin immunoprecipitation (ChIP) followed by microarray hybridization or high-throughput sequencing (ChIP-seq) experiments provided insight into the genomic DNA binding characteristics of WT1 during kidney development and homeostasis.10, 11, 12, 13 The WT1 binding motif derived from ChIP followed by microarray hybridization shows a high similarity to the EGR1 binding motif, whereas the motif based on ChIP-seq experiments demonstrates a slight variation in the form of an exchange from C to A. Considering all the motifs identified from ChIP-seq experiments, WT1 recognizes the sequence of Gg/tGGGAGg/t.11, 12, 13 Interestingly, in many gene regulatory regions, we found two similar motifs containing GGGAGg/t close to each other, suggesting that WT1 can form and bind as a homodimer.12 This could explain the dominant manner of manifestation of diseases caused by WT1 mutations.
In the past, numerous potential target genes of WT1 were identified by characterization of the promoter regions via various approaches, such as reporter assays, electrophoretic mobility shift assay, and co-transfection assays (Supplementary Table S1 online). Examination of genomic WT1 binding sites in kidneys revealed that they were significantly enriched in the promoter region near the transcription start sites. Meanwhile, combining all the WT1 peaks from ChIP-seq results shows around 45% of WT1 binding sites localized more than 50 kb away from transcription start sites, indicating that WT1 regulates gene expression via both proximal and distal regulatory elements, thus highly extending the WT1 potential target gene list.10, 11, 12, 13 In addition, the differences of WT1 binding loci between developing and adult kidneys as well as the different gene expression patterns between Wt1 mutant MM and podocytes indicate that the gene sets regulated by WT1 significantly depend on the specific cellular context.
WT1 IS INVOLVED IN SELF-RENEWAL AND DIFFERENTIATION OF MM
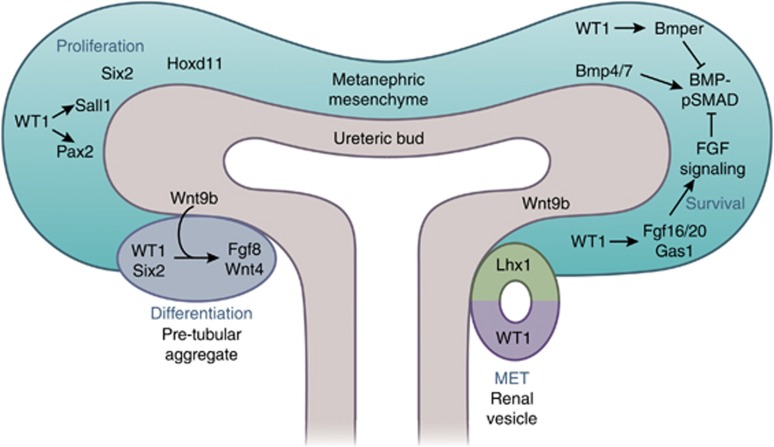
In view of the important function of WT1 in kidneys, several animal models (e.g., mouse, zebrafish, and Xenopus) have been applied. Particularly, generation and characterization of different mouse models have greatly elucidated the WT1 function.14 Wt1 knockout mice have no kidneys with failure of UB outgrowth and MM degeneration by apoptosis at E12. In Wt1−/− MM cells, a lack of FGF and enhanced BMP-pSMAD signaling was observed. Mechanistically, WT1 directly regulates FGF20/16 expression and activates FGF signaling for MM survival.11 After the MM responds to FGF signals, WT1-dependent GAS1 can promote MM proliferation by activation of intracellular FGF-stimulated AKT signaling.15 Also, WT1 directly activates the expression of BMP modulator Bmper to repress pSMAD signaling for permission of MM survival.11 In addition, mutation of WT1 in Wilms' tumor is frequently accompanied by canonical Wnt activation.16 WT1 is indicated to inhibit Wnt signaling in the MM by activating CXXC5 expression, encoding a Wnt/β-catenin inhibitor.10 The interaction of BMP, FGF, and Wnt signaling has been implicated for the survival of MM.17 Interestingly, WT1 can affect all key signaling pathways in MM by transcriptional regulation of respective modulators. As shown for Wt1, inactivation of other transcription factors encoding genes like Sall1/4, Six1/2, Eya1, Pax2, and Hoxd11 also results in kidney agenesis.17 Among them, WT1 directly regulates Sall1 and Pax2 in MM (Figure 2).10
Figure 2.
Wilms' tumor 1 (WT1) contributes to metanephric mesenchyme self-renewal and its differentiation. WT1, Sall1, Pax2, Six2, and Hoxd11 are individually required for survival and self-renewal of metanephric mesenchyme (MM). Among them, WT1 is upstream of Sall1 and Pax2 transcription factors. BMP4 induces the apoptosis of MM; however, BMP7 promotes the MM differentiation. In balance of the BMP-pSMAD signaling in MM, WT1 directly regulates Bmper, a BMP-pSMAD inhibitor, and activates the FGF signaling pathway via regulating Fgf20/Fgf16 and Gas1 expression to antagonize BMP-pSMAD signaling for the survival and self-renewal of MM. High expression level of Wnt9b beneath the ureteric tip increases the Wnt singling activity nearing the MM; there, the β-catenin-LEF/TCF complex works together with Six2 and WT1 activating the critical differentiation factors, Fgf8 and Wnt4, to form pre-tubular aggregate (PTA). Signals produced by PTA promote the mesenchyme–epithelial transition (MET) to form renal vesicles. The proximal part of the renal vesicle expressing Wt1 will develop into the epithelial cells of the glomerulus and the distal part of the renal vesicle expressing Lhx1 will develop into the epithelial cells of the nephron tubule.
In addition to self-renewal and expansion of nephron progenitors, signals for initiation of nephrogenesis are required in order for kidney development to occur. One such signal is Wnt9b, which is expressed at a high level beneath the ureteric tip and induces the adjacent nephron progenitor cells to differentiate and to form pre-tubular aggregates. WT1 expression level is increased in the pre-tubular aggregate domain (Figure 1b). Fgf8 and Wnt4 are expressed in pre-tubular aggregates and are required for mesenchymal–epithelial transition to develop the renal vesicle.17 On the basis of direct evidence from ChIP experiments, β-catenin and LEF/TCF complexes together with Six2 regulate Fgf8 and Wnt4 transcription in pre-tubular aggregate.18 Interestingly, WT1 also binds to the cis-regulatory regions of Fgf8 and Wnt4 and activates their expression.11, 19 Somatic deletion of Wt1 at E13.5 disrupts MM epithelialization and results in the lack of nephrons at birth.20 Taken together, WT1 controls MM differentiation and mesenchymal–epithelial transition mainly by regulation of Fgf8 and Wnt4, probably cooperating with β-catenin, LEF/TCF, and Six2 complexes (Figure 2).
WT1 REGULATES PODOCYTE MATURATION AND HOMEOSTASIS
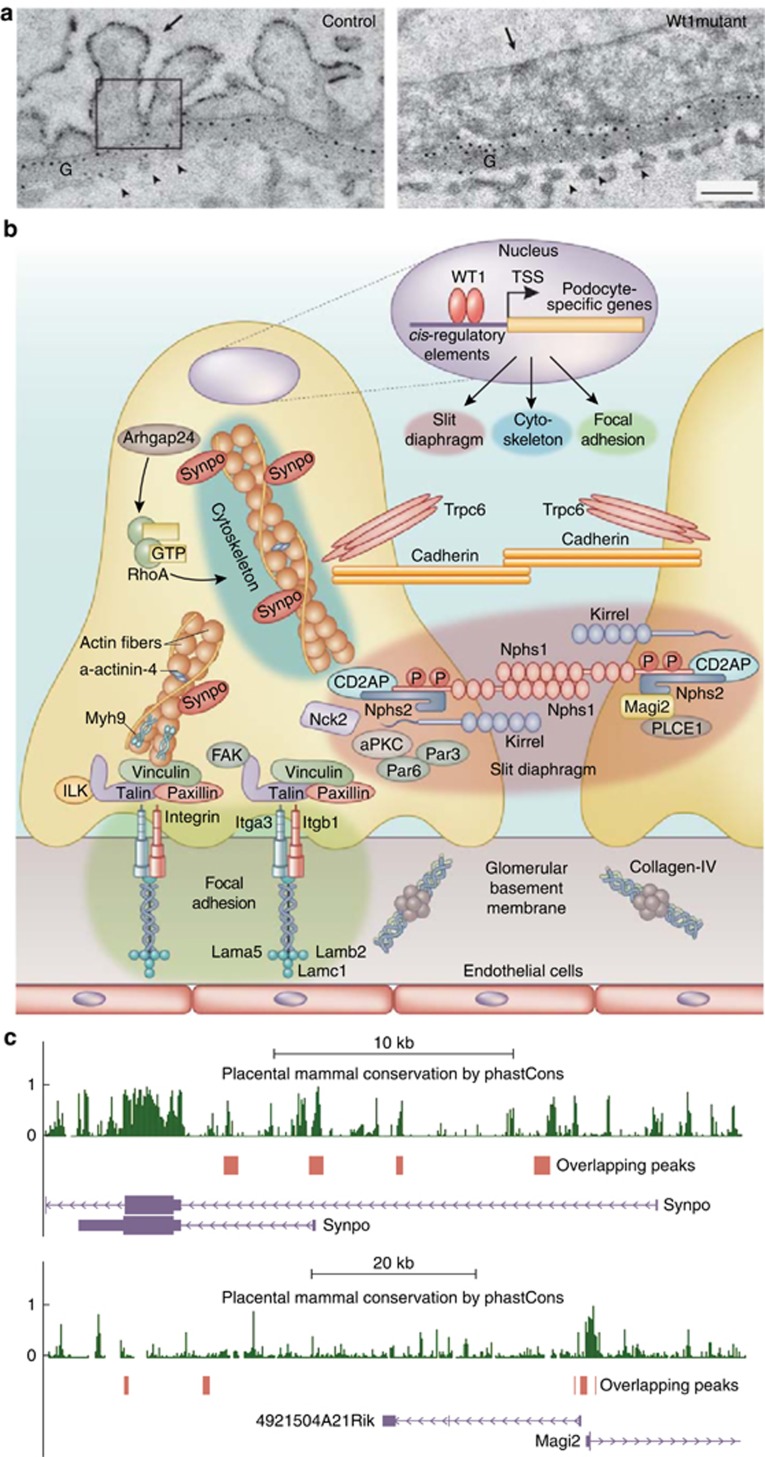
After MM mesenchymal–epithelial transition, Wt1 expression continues to increase along with the development of podocyte progenitors and is later restricted to adult podocytes. Mature podocytes consist of four morphologically distinct segments: a cell body, major processes, secondary processes, and foot processes (FPs). Between interdigitating FPs of neighboring podocytes, a unique structure, the slit diaphragm (SD), is formed. FPs are functionally defined by three membrane domains: the apical membrane domain, the SD, and the basal membrane domain that is attached to the glomerular basement membrane. The integrity of podocytes and their interaction with the glomerular basement membrane is crucial for maintenance of the intact glomerular filtration barrier. Alteration of the intercellular junctions and cytoskeletal structure of podocytes or their detachment from the membrane results in the development of albuminuria.21 Investigations in humans, mouse, and fish have revealed a large number of gene products involved in podocyte development and maintenance (Supplementary Table S2 online). WT1 mutations leading to disruption of podocyte development or maintenance are associated with glomerulopathies in DDS, FS, and steroid-resistant nephrotic syndrome. Podocyte-specific Wt1 knockout mice display defects in podocyte differentiation and lead to kidney failure and death within 24 h after birth.12 Inducible adult podocyte Wt1 knockout mice show FP effacement, proteinuria, and glomerulosclerosis, thus confirming the essential roles of Wt1 in podocyte maturation and maintenance (Figure 3a).22, 23 Recent ChIP-Seq and transcriptome analyses have shed light on the WT1-controlled transcription network in adult podocytes (Figure 3b).12, 13
Figure 3.
WT1 defines the podocyte-specific morphology. (a) Ultrastructural kidney analysis of Wt1 mutant and control adult mice treated with doxycycline for 6 days. Podocytes (arrow) and endothelial cells (arrowheads) were abnormally distant from the GBM (G), which was itself irregularly shaped in the mutant tissue. Black dots indicate negative charges by polyethylenimine staining. No obvious charge differences between the GBMs of mutant and control glomeruli are detectable. Scale bar=0.5 μm. This figure is adapted with permission from Dong et al.12 (b) Schematic WT1-controlled networks for establishing podocyte-specific morphology. WT1 regulates the slit diaphragm complex components, including Nephrin (Nphs1), Podocin (Nphs2), Membrane-associated guanylate kinase, WW and PDZ domain–containing protein 2 (Magi2), CD2-associated protein (CD2AP), NCK adaptor protein 2 (NCK2), Kin of IRRE-like protein 1/2/3 (Kirrel) and phospholipase C, and epsilon 1 (Plce1). WT1 controls focal adhesion by regulating Integrin α3 (Itga3), Integrin β1 (Itgb1), Laminin α5 (Lama5), and Laminin β2 (Lamb2). WT1 also controls the cytoskeleton components and its regulators to establish the podocyte polarity—namely, Synaptopodin (Synpo), α-actinin-4, myosin, heavy polypeptide 9, non-muscle (Myh9), Rho GTPase-activating protein 24 (Arhgap24), and Atypical protein kinase C (aPKC). (c) Multiple Wt1 binding sites to cis-regulatory regions of Synpo and Magi2 as viewed in the UCSC browser within the indicated genomic intervals. Conservation denotes placental mammal basewise conservation by phastCons score. The peaks indicate the overlapping WT1 binding sites generated from Kreidberg and colleagues' and our ChIP-seq data.12, 13
WT1 controls the expression of SD component encoding genes
The SD is a specialized protein complex structure of podocytes, which serves as a blood filtration unit and constitutes a fundamental component of the glomerular filtration barrier. The SD, as a specialized intercellular junction connecting neighboring FPs, contains elements of tight, adherens, and gap junctions. The SD is also considered as a complex signaling hub for regulating the plasticity of podocyte FPs and the loss of SD integrity leading to proteinuria.24 Interestingly, WT1 binds to the proximal promoters of many genes encoding proteins that localize to the SD structure of podocytes, such as Nphs1, Nphs2, Magi2, Ptpro, Kirrel, Plce1, Cldn5, and Nck2.12, 13 This supports the hypothesis that sharing transcription factor binding sites conserved in distance and orientation can help control the expression of genes that act together in the same biological context.25 Notably, the expression of SD protein encoding genes is controlled differentially by WT1 variants. Magi2 expression depends on the WT1+KTS isoform, whereas Nphs1 and Nphs2 expressions are regulated by the WT1−KTS isoform.26
WT1 controls the polarity of podocytes and cytoskeleton arrangement
Apart from the SD structure, a complex cytoskeleton underlies the delicate architecture of podocytes to build and maintain their apicobasal and planer cell polarity. Interruption of cytoskeleton structure and loss of polarity are among characteristic features of injured podocytes.27 For example, loss of the actin cytoskeletal proteins, ACTN4, SYNPO, and MYH9, results in autosomal recessive nephrotic syndromes. In addition, mutation of actin regulatory protein, ARHGAP24, a RhoA-activated Rac1 GTPase-activating protein, also causes early or adult onset nephrotic syndrome. Podocyte-specific knockout of the polarity protein aPKC, a central component of Par3-Par6-aPKC complex, in mice also results in the nephrotic syndrome (Supplementary Table S2 online). WT1 binds the cis-regulatory elements of those genes in ChIP-Seq experiments, indicating that WT1 controls the polarity of podocytes and cytoskeleton arrangement.12, 13
WT1 controls the cell-matrix adhesion of podocytes
Tight adherence of podocyte to the glomerular basement membrane is another prerequisite for maintaining the integrity of the glomerular filtration barrier due to its exposure to permanent transcapillary filtration pressure. The major cell-matrix adhesion receptor in podocytes is integrin α3β1 connecting to laminin 521 in the glomerular basement membrane.28 Mutations in ITGA3 and LAMB2 in human patients or podocyte-specific knockout of Itgb1 and Lama5 in mice result in FP effacement and proteinuria (Supplementary Table S2 online). All the cis-regulatory regions of these genes have WT1 binding signals, suggesting that WT1 controls the cell-matrix adhesion of podocytes.12, 13
WT1 and the concept of super-enhancers
By taking a closer look at the WT1 binding loci in the podocyte-specific genes, it is apparent that, in many cases, several WT1 binding sites localize to intragenic or intergenic regions in the gene's cis-regulatory domains, such as in Synaptopodin and Magi2 (Figure 3c). Correlation analysis with expression data from FACS-sorted podocytes suggests that genes harboring multiple WT1 binding sites usually have a higher expression level in podocytes than genes having no or only one Wt1 binding sequence. The analysis of these enhancer peaks shows a significant enrichment for MAFB, Homeobox, Tcf-family, and Fox-class binding sites, indicating that these factors may cooperate with WT1 to regulate podocyte-specific gene expression.13 WT1 binds to its own promoter, suggesting an auto-regulatory loop.11, 12, 13 WT1 also regulates the expression of Mafb, Lmx1b, Foxc2, and Tcf21, which are podocyte-specific transcription factors. Mutation of each of them causes the loss of kidney function (Supplementary Table 2 online). These findings are reminiscent of the concept of super-enhancers, which consist of clusters of enhancers that are densely occupied by master regulators and Mediator for enhancing the cell-specific gene expression pattern.29 In light of this paradigm, the recent data suggest that WT1 works together with Mafb, Lmx1b, FoxC2, and Tcf21 to define podocyte identity as a master regulator by activating the podocyte-specific super-enhancers, together with Mediator.
CONCLUSION AND PERSPECTIVE
WT1 mutations result in a large spectrum of phenotypes, either caused by dysfunction of kidney progenitors or podocytes. Significant advances have recently been made in the understanding of WT1 function in kidney biology and kidney pathophysiology associated with WT1 mutations, providing promising therapeutic applications. Recently, application of in utero delivery of a pseudotyped rAAV 2/9 vector provides a possible avenue for the targeting of gene therapies of WT1 mutation–related kidney diseases.30 In addition, understanding the molecular mechanism of kidney development has fostered the establishment of protocols for the generation of mature kidney cell types and the three-dimensional nephron-like structures from pluripotent cells.31 Transplantation of the de novo nephrons generated from stem cells under a mouse kidney capsule could integrate the already existing blood stream into the glomerulus, thus enhancing potential treatment of kidney diseases.31 Although these therapeutic directions point toward a translational relevance, in addition, developing pharmacological agents can help combat kidney disorders. A drug screening platform can be established from reconstituted kidneys in vitro, derived from pluripotent stem cells from WT1 mutant patients. Simultaneously, a better understanding of the complex WT1-dependent molecular mechanisms involved in podocyte biology coupled with directed differentiation protocols will assist in the generation of large quantities of pure podocytes, thus, paving the way for novel clinical applications.
Acknowledgments
We thank Abinaya Nathan and Thomas Bates for critically reading and improving this manuscript. The cartoons were drawn with help from Abinaya Nathan. We are grateful to other members of our laboratory for their helpful discussions and contributions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG; EN280/8-1).
All the authors declared no competing interests.
Footnotes
SUPPLEMENTARY MATERIAL
Table S1. Wt1 target genes.
Table S2. List of genetic mutations or deletion or transgenic animal models that involved in podocyte function.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
Supplementary Material
References
- 1Eckardt KU, Coresh J, Devuyst O et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet 2013; 382: 158–169. [DOI] [PubMed] [Google Scholar]
- 2Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet 2006; 2: R196–R201. [DOI] [PubMed] [Google Scholar]
- 3Little MH, McMahon AP. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb Perspect Biol 2012; 4 a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Gessler M, Poustka A, Cavenee W et al. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature 1990; 343: 774–778. [DOI] [PubMed] [Google Scholar]
- 5Call KM, Glaser T, Ito CY et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell 1990; 60: 509–520. [DOI] [PubMed] [Google Scholar]
- 6Lipska BS, Ranchin B, Iatropoulos P et al. Genotype-phenotype associations in WT1 glomerulopathy. Kidney international 2014; 85: 1169–1178. [DOI] [PubMed] [Google Scholar]
- 7Chernin G, Vega-Warner V, Schoeb DS et al. Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol 2010; 5: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Royer-Pokora B, Beier M, Henzler M et al. Twenty-four new cases of WT1 germline mutations and review of the literature: genotype/phenotype correlations for Wilms tumor development. Am J Med Genet Part A 2004; 127A: 249–257. [DOI] [PubMed] [Google Scholar]
- 9Hashimoto H, Olanrewaju YO, Zheng Y et al. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 2014; 28: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Hartwig S, Ho J, Pandey P et al. Genomic characterization of Wilms' tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development 2010; 137: 1189–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Motamedi FJ, Badro DA, Clarkson M et al. WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat Commun 2014; 5: 4444. [DOI] [PubMed] [Google Scholar]
- 12Dong L, Pietsch S, Tan Z et al. Integration of cistromic and transcriptomic analyses identifies Nphs2, Mafb, and Magi2 as Wilms' Tumor 1 target genes in podocyte differentiation and maintenance. J Am Soc Nephrol 2015. [DOI] [PMC free article] [PubMed]
- 13Kann M, Ettou S, Jung YL et al. Genome-wide analysis of Wilms' Tumor 1-controlled gene expression in podocytes reveals key regulatory mechanisms. J Am Soc Nephrol 2015. [DOI] [PMC free article] [PubMed]
- 14Ozdemir DD, Hohenstein P. Wt1 in the kidney–a tale in mouse models. Pediat Nephrol 2014; 29: 687–693. [DOI] [PubMed] [Google Scholar]
- 15Kann M, Bae E, Lenz MO et al. WT1 targets Gas1 to maintain nephron progenitor cells by modulating FGF signals. Development 2015; 142: 1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Maiti S, Alam R, Amos CI et al. Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res 2000; 60: 6288–6292. [PubMed] [Google Scholar]
- 17Kopan R, Chen S, Little M. Nephron progenitor cells: shifting the balance of self-renewal and differentiation. Curr Top Dev Biol 2014; 107: 293–331. [DOI] [PubMed] [Google Scholar]
- 18Park JS, Ma W, O'Brien LL et al. Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev Cell 2012; 23: 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Essafi A, Webb A, Berry RL et al. A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev Cell 2011; 21: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Hu Q, Gao F, Tian W et al. Wt1 ablation and Igf2 upregulation in mice result in Wilms tumors with elevated ERK1/2 phosphorylation. J Clin Invest 2011; 121: 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Brinkkoetter PT, Ising C, Benzing T. The role of the podocyte in albumin filtration. Nat Rev Nephrol 2013; 9: 328–336. [DOI] [PubMed] [Google Scholar]
- 22Gebeshuber CA, Kornauth C, Dong L et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med 2013; 19: 481–487. [DOI] [PubMed] [Google Scholar]
- 23Chau YY, Brownstein D, Mjoseng H et al. Acute multiple organ failure in adult mice deleted for the developmental regulator Wt1. PLoS Genet 2011; 7: e1002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Grahammer F, Schell C, Huber TB. The podocyte slit diaphragm–from a thin grey line to a complex signalling hub. Nat Rev Nephrol 2013; 9: 587–598. [DOI] [PubMed] [Google Scholar]
- 25Cohen CD, Klingenhoff A, Boucherot A et al. Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci USA 2006; 103: 5682–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Lefebvre J, Clarkson M, Massa F et al. Alternatively spliced isoforms of WT1 control podocyte specific gene expression. Kidney Int 2015. [DOI] [PubMed]
- 27Welsh GI, Saleem MA. The podocyte cytoskeleton–key to a functioning glomerulus in health and disease. Nat Rev Nephrol 2012; 8: 14–21. [DOI] [PubMed] [Google Scholar]
- 28Sachs N, Sonnenberg A. Cell-matrix adhesion of podocytes in physiology and disease. Nat Rev Nephrol 2013; 9: 200–210. [DOI] [PubMed] [Google Scholar]
- 29Whyte WA, Orlando DA, Hnisz D et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Picconi JL, Muff-Luett M, Wu D et al. Kidney-specific expression of GFP by in-utero delivery of pseudotyped adeno-associated virus 9. Mol Ther Methods Clin Dev 2014; 1: 14014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Humphreys BD. Kidney structures differentiated from stem cells. Nat Cell Biol 2014; 16: 19–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.