Abstract
Amphotericin B (AmB) is the archetype for small molecules that form ion channels in living systems, and has recently been shown to replace a missing protein ion transporter and thereby restore physiology in yeast. Molecular modeling studies predict that AmB self-assembles in lipid membranes with the polyol region lining a channel interior that funnels to its narrowest region at the C3-hydroxyl group. This model predicts that modification of this functional group would alter conductance of the AmB ion channel. To test this hypothesis, the C3-hydroxyl group was synthetically deleted and the resulting derivative, C3deoxyAmB (C3deOAmB), was characterized using multidimensional NMR experiments and single ion channel electrophysiology recordings. C3deOAmB possesses the same macrocycle conformation as AmB and retains the capacity to form transmembrane ion channels, yet the conductance of the C3deOAmB channels is threefold lower than that of AmB channels. Thus, the C3-hydroxyl group plays an important role in AmB ion channel conductance, and synthetic modifications at this position may provide an opportunity for further tuning of channel functions.
The antifungal polyene macrolide natural product amphotericin B (AmB) is the archetypical small molecule capable of forming ion channels in living systems,1 and we recently reported that this small molecule can functionally substitute for a missing protein ion transporter and thereby restore physiology in yeast.2 To maximally harness this remarkable functional capacity, it is necessary to understand the molecular underpinnings that govern AmB ion channel formation, conductance, gating, and selectivity. However, despite more than half a century of research, the structure of the AmB ion channel remains unknown.
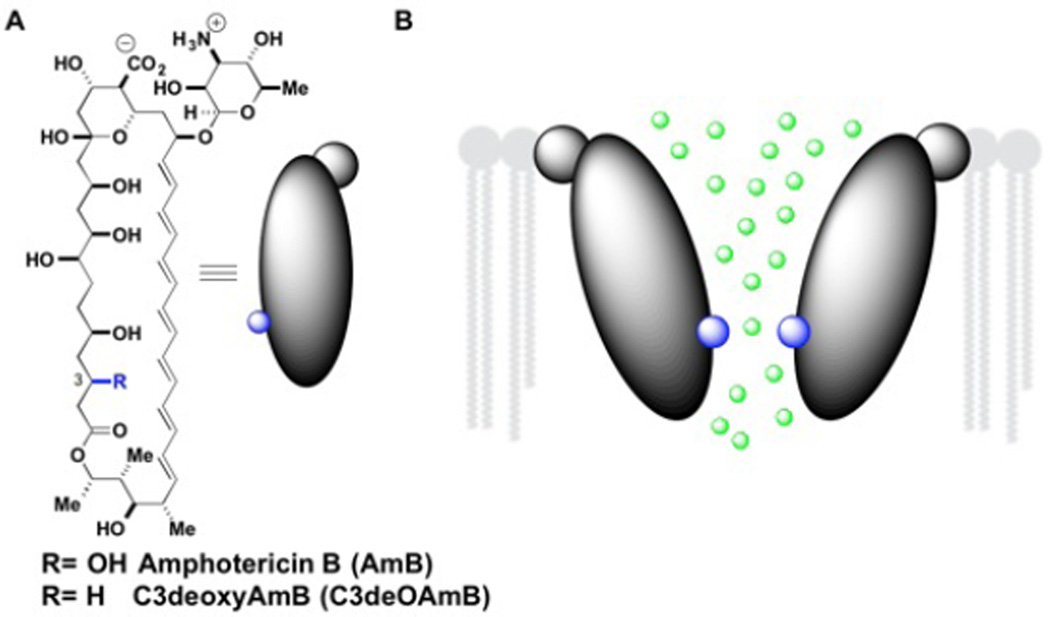
Modeling studies predict that AmB self-assembles into multimeric structures in which the polyol region lines a water-filled channel interior.3 Furthermore, the AmB channel is predicted to have a wide entrance near the C15 alcohol funneling to its narrowest region near the C3 alcohol (Figure 1).4 This model therefore predicts that modifications at the C3 position would permit channel formation but alter ion conductance.
Figure 1.
A.) Amphotericin B and C3deOAmB structures. B.) AmB ion channel model funneling to narrowest region at C3-OH (highlighted in blue)
No derivatives with modifications at the polyol region of AmB have been studied using electrophysiological recordings, which are critical for characterizing differences in single ion channel conductances. This is likely in large part a consequence of the synthetic difficulties in obtaining such site-specifically modified AmB derivatives. Synthetic modification of the C13 hemiketal,5 and genetic manipulations of the producing organism6 and related organisms7 have yielded polyol modified derivatives. However, the impacts of these modifications on single ion channel conductance have not been reported.
To test the hypothesis that the C3-hydroxyl group plays an important role in ion channel conductance, we targeted its chemoselective deletion.8 As one of 10 distinct hydroxyl groups appended to AmB, this represented a substantial synthetic challenge. We recognized, however, that the unique β-positioning of the C3 alcohol relative to the C1 carbonyl may provide an opportunity for selective elimination to form an alpha, beta-unsaturated macrolactone followed by chemoselective conjugate reduction.
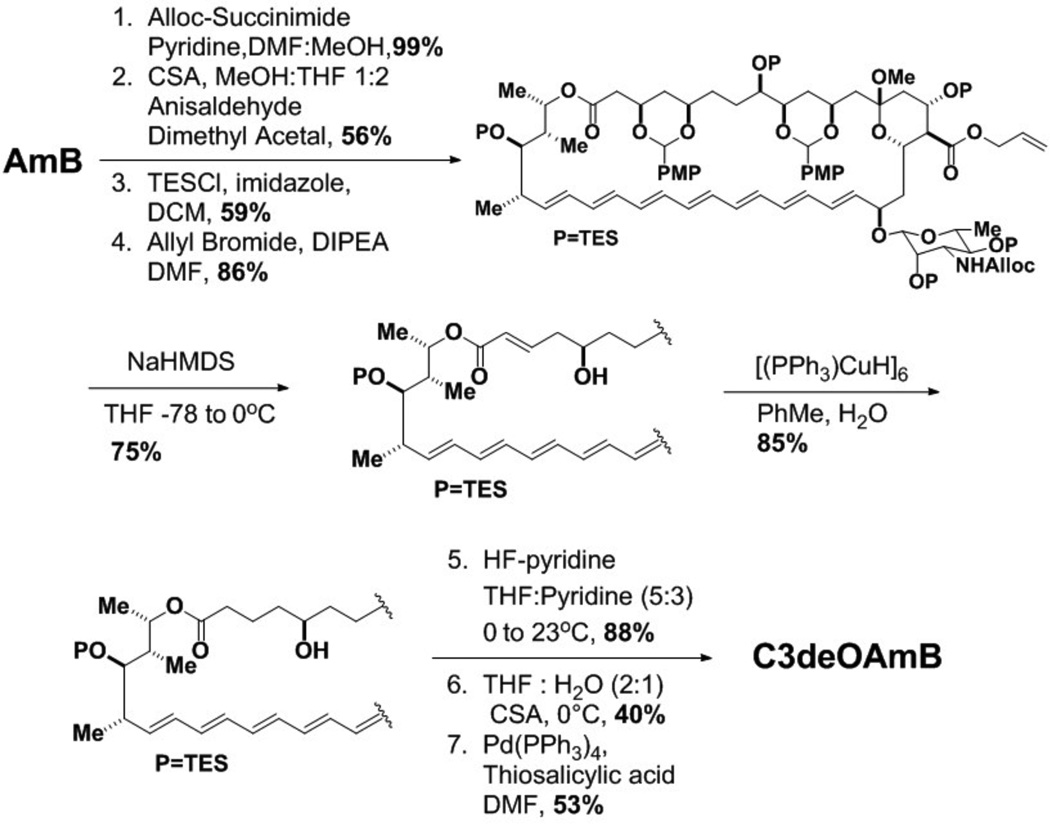
This plan was ultimately reduced to an efficient 9-step synthesis as shown in Scheme 1. Starting with the natural product, a series of functional group protections delivered intermediate 1 (Scheme 1 and Supporting Information). Gratifyingly, exposure of 1 to NaHMDS at low temperatures chemoselectively eliminated the C3 p-methoxyphenylacetal, presumably via an E1cB type mechanism, yielding intermediate enone 2. Subsequent site-selective Stryker reduction9 of carbonyl-conjugated C3,C4 double bond provided deoxygenated intermediate 3. A final series of deprotections yielded the targeted single-atom modified variant, C3deOAmB.
Scheme 1.
Synthesis of C3deOAmB
It was unclear at the outset whether this functional group deletion would cause changes in macrocycle conformation, which in turn would complicate the analysis of voltage clamp electrophysiological recordings of the corresponding ion channels. Specifically, in the crystal structure of a derivative of AmB, the C3 hydroxyl group is involved in a hydrogen bonding network that includes both the C1 carbonyl and C5 hydroxyl group.10 Disruption of such a hydrogen bonding network might result in a change in macrocycle shape. To test this, we independently determined the ground state conformations of both AmB and C3deOAmB using stochastic conformation generation methods constrained by extensive NOESY and phase sensitive COSY NMR data processed using amplitude-constrained multiplet evaluation.11 Optimization allowed us to perform these experiments without the use of solubilizing protective groups.
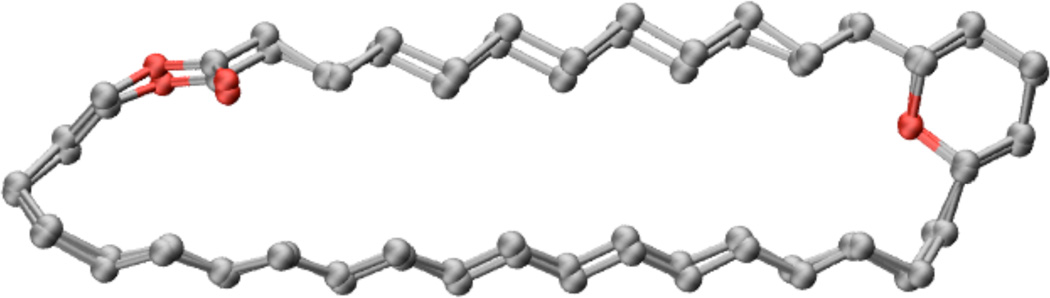
We determined the lowest energy conformations of the AmB and C3deOAmB structures using the Low Mode MD conformational search method12 with NOESY-based internuclear distance restraints and phase-sensitive COSY-based dihedral restraints.11 As shown in Figure 2, the structure-based alignment of the lowest energy conformation of the C3deOAmB skeleton with the lowest energy conformer of AmB exhibits a heavy atom RMSD of 0.33 ± 0.03 Å, which is comparable to the precision of each structure. Thus, within the uncertainty of this NMR-based structure determination, the ground state conformations of these two compounds are identical.
Figure 2.
Overlay of the lowest energy conformations of AmB and C3deOAmB. The heavy atom RMSD is 0.33 ± 0.03 Å.
Further supporting this conclusion, both the 13C and 1H chemical shifts show excellent agreement between AmB and C3deOAmB, with the exception of the modified C3 site and its immediate neighbors, for which the chemical shifts change due to well understood electronegativity effects (Figure S6–S9).13
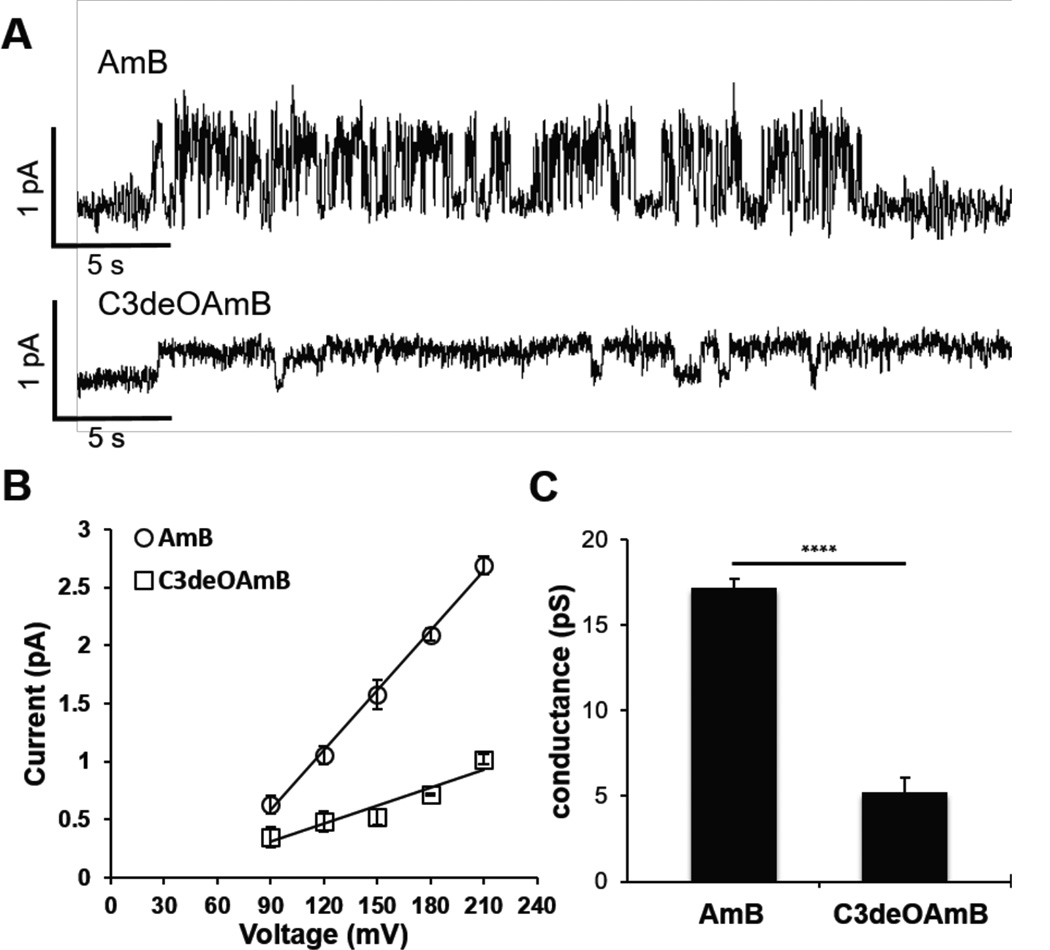
Finally, we tested the key hypothesis that removal of the C3 hydroxyl group would impact ion channel conductance. Specifically, HPLC-purified AmB and C3deOAmB were compared head-to-head in voltage-clamp single ion channel recording studies. Planar lipid bilayers were formed from a 70:30 ratio of DPhPC lipids to ergosterol at pH = 7.0. AmB or C3deOAmB were then added to both sides of the membrane as a solution in DMSO and channel activity was investigated. Shown in Figure 3A are representative single ion channel traces of both AmB (top) and C3deOAmB (bottom) (Figure S1–S2). The conductance of each molecule was determined by measuring the current of the single channels formed from each molecule at a range of voltages (Figure 3B). AmB has a single ion channel conductance of 17.2 ± 0.5 pS. Under identical conditions C3deOAmB also forms single ion channels, however, the conductance significantly decreases to 5.2 ± 0.9 pS, a three-fold reduction (Figure 3C). Preliminary experiments in liposomes also support differences in ion selectivity between AmB and C3deOAmB channels (see Supporting Information).
Figure 3.
A.) Single ion channel activity of AmB (top) and C-3deOAmB (bottom) in planar lipids bilayers. DPhPC Lipids with 30% Ergosterol. 1 M KCl pH = 7.0, 5 mM HEPES +150 mV. B.) Current vs voltage response of AmB and C3deOAmB from 90 to 210 mV in 30 mV increments. 2 M KCl, pH = 7.0, 5 mM HEPES.14 AmB r2 = 0.9969. C3deOAmB r2 = 0.9174. C.) Bar plot illustrating the differences in ion channel conductance between AmB and C3deOAmB. **** p < 0.001. graphs depict means ± SD.
Thus, deletion of a single atom at the C3 position of AmB is synthetically accessible, does not cause a change in macrocycle conformation, and permits the retention of channel-forming activity yet changes the ion channel conductance by 3-fold. These findings are consistent with structural4 and theoretical4d models for the AmB ion channel that place the C3 alcohol in the path of ion conductance. They also suggest that other modifications at the C3-position, many of which should be accessible from intermediate 2, might enable further understanding and/or targeted optimization of this archetypical small molecule-based ion channel.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge the NIH (R01GM080436, R01GM112845) and HHMI for financial support. We gratefully acknowledge computational assistance from M. Hallock (School of Chemical Sciences) and C. Williams (CCG). T.V.P. was an NCSA Faculty Fellow when this work was completed.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Detailed synthesis, spectral data, and electrophysiological protocols. This material is available free of charge via the Internet at http://pubs.acs.org.
UIUC has filed a patent application on C3deOAmB.
REFERENCES
- 1.(a) Bolard J. Biochim. Biophys. Acta. 1986;864:257. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]; (b) Hartsel SC, Hatch C, Ayenew W. J. Liposome Res. 1993;3:377. [Google Scholar]; (c) Cereghetti DM, Carreira EM. Synthesis. 2006;6:914. [Google Scholar]; (d) Andreoli TE, Monahan M. J. Gen. Physiol. 1968;52:300. doi: 10.1085/jgp.52.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Cass A, Finkelstein A, Krespi V. J. Gen. Physiol. 1970;56:100. doi: 10.1085/jgp.56.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Ermishkin LN, Kasumov KM, Potzeluyev VM. Nature. 1976;262:698. doi: 10.1038/262698a0. [DOI] [PubMed] [Google Scholar]; (g) Ermishkin LN, Kasumov KM, Potseluyev VM. Biochim. Biophys. Acta, Biomembr. 1977;470:357. doi: 10.1016/0005-2736(77)90127-4. [DOI] [PubMed] [Google Scholar]; (h) Borisova MP, Ermishkin LN, Silberstein AY. Biochim. Biophys. Acta, Biomembr. 1979;553:450. doi: 10.1016/0005-2736(79)90300-6. [DOI] [PubMed] [Google Scholar]; (i) Kasumov KM, Borisova MP, Ermishkin LN, Potseluyev VM, Silberstein AY, Vainshtein VA. Biochim. Biophys. Acta. 1979;551:229. doi: 10.1016/0005-2736(89)90001-1. [DOI] [PubMed] [Google Scholar]; (j) Borisova MP, Brutyan RA, Ermishkin LN. J. Membr. Biol. 1986;90:13. doi: 10.1007/BF01869681. [DOI] [PubMed] [Google Scholar]; (k) Mickus DE, Levitt DG, Rychnovsky SD. J. Am. Chem. Soc. 1992;114:359. [Google Scholar]
- 2.Cioffi AG, Hou J, Grillo AS, Diaz KA, Burke MD. J. Am. Chem. Soc. 2015;137:10096. doi: 10.1021/jacs.5b05765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Finkelstein A, Holz R. Aqueous pores created in thin lipid membranes by the polyene antibiotics nystatin and AmB membranes. Lipid Bilayers and Antibiotics. New York: Dekker; 1973. [PubMed] [Google Scholar]; (b) DeKruijf B, Demel RA. Biochim Biophys Acta. 1974;339:57. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]; (c) Andreoli TE. Ann. N. Y. Acad. Sci. 1974;235:448. doi: 10.1111/j.1749-6632.1974.tb43283.x. [DOI] [PubMed] [Google Scholar]
- 4.(a) Khutorsky VE. Biochim. Biophys. Acta, Biomembr. 1992;1108:123. doi: 10.1016/0005-2736(92)90015-e. [DOI] [PubMed] [Google Scholar]; (b) Khutorsky VE. Biophys. J. 1996;71:2984. doi: 10.1016/S0006-3495(96)79491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Baginski M, Resat H, McCammon JA. Mol. Pharmacol. 1997;52:560. doi: 10.1124/mol.52.4.560. [DOI] [PubMed] [Google Scholar]; (d) Baginski M, Resat H, Borowski E. Biochim. Biophys. Acta. 2002;1567:63. doi: 10.1016/s0005-2736(02)00581-3. [DOI] [PubMed] [Google Scholar]
- 5.(a) Taylor AW, Costello BJ, Hunter PA, Maclachlan WS, Shanks CT. J. Antibiot. 1993;46:486. doi: 10.7164/antibiotics.46.486. [DOI] [PubMed] [Google Scholar]; (b) W. Taylor A, T. MacPherson D. Tetrahedron Lett. 1994;35:5289. [Google Scholar]
- 6.(a) Byrne B, Carmody M, Gibson E, Rawlings B, Caffrey P. Chem. Biol. 2003;10:1215. doi: 10.1016/j.chembiol.2003.12.001. [DOI] [PubMed] [Google Scholar]; (b) Power P, Dunne T, Murphy B, Lochlainn LN, Rai D, Borissow C, Rawlings B, Caffrey P. Chem. Biol. 2008;15:78. doi: 10.1016/j.chembiol.2007.11.008. [DOI] [PubMed] [Google Scholar]; (c) Murphy B, Anderson K, Borissow C, Caffrey P, Griffith G, Hearn J, Ibrahim O, Khan N, Lamburn N, Lee M, Pugh K, Rawlings B. Org. Biomol. Chem. 2010;8:3758. doi: 10.1039/b922074g. [DOI] [PubMed] [Google Scholar]
- 7.(a) Preobrazhenskaya MN, Olsufyeva EN, Solovieva SE, Tevyashova AN, Reznikova MI, Luzikov YN, Terekhova LP, Trenin AS, Galatenko OA, Treshalin ID, Mirchink EP, Bukhman VM, Sletta H, Zotchev SB. J. Med. Chem. 2008;52:189. doi: 10.1021/jm800695k. [DOI] [PubMed] [Google Scholar]; (b) Brautaset T, Sletta H, Nedal A, Borgos SEF, Degnes KF, Bakke I, Volokhan O, Sekurova ON, Treshalin ID, Mirchink EP, Dikiy A, Ellingsen TE, Zotchev SB. Chem. Biol. 2008;15:1198. doi: 10.1016/j.chembiol.2008.08.009. [DOI] [PubMed] [Google Scholar]; (c) Tevyashova AN, Olsufyeva EN, Solovieva SE, Printsevskaya SS, Reznikova MI, Trenin AS, Galatenko OA, Treshalin ID, Pereverzeva ER, Mirchink EP, Isakova EB, Zotchev SB, Preobrazhenskaya MN. Antimicrob. Agents Chemother. 2013;57:3815. doi: 10.1128/AAC.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Palacios DS, Anderson TM, Burke MD. J. Am. Chem. Soc. 2007;129:13804. doi: 10.1021/ja075739o. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Palacios DS, Dailey I, Siebert DM, Wilcock BC, Burke MD. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6733. doi: 10.1073/pnas.1015023108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2234. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wilcock BC, Endo MM, Uno BE, Burke MD. J. Am. Chem. Soc. 2013;135:8488. doi: 10.1021/ja403255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahoney WS, Brestensky DM, Stryker JM. J. Am. Chem. Soc. 1988;110:291. [Google Scholar]
- 10.Jarzembska KN, Kaminski D, Hoser AA, Malinska M, Senczyna B, Wozniak K, Gagos M. Cryst. Growth Des. 2012;12:2336. [Google Scholar]
- 11.Delaglio F, Wu Z, Bax A. J. Magn. Reson. 2001;149:276. doi: 10.1006/jmre.2001.2297. [DOI] [PubMed] [Google Scholar]
- 12.Labute P. J. Chem. Info. Model. 2010;50:792. doi: 10.1021/ci900508k. [DOI] [PubMed] [Google Scholar]
- 13.(a) Silverstein RM, Webster FX. Chemical Classes and Chemical Shifts. In: Rose N, Swain E, editors. Spectrometric Identification of Organic Compounds, 6. Vol. 222. New York: John Wiley& Sons, Inc.; 1998. [Google Scholar]; (b) Bible RH. Interpretation of NMR Spectra. New York: Plenum Press; 1966. [Google Scholar]; (c) Lee J, Kobayashi Y, Tezuka K, Kishi Y. Org. Lett. 1999;1:2181. doi: 10.1021/ol990379y. [DOI] [PubMed] [Google Scholar]
- 14.For both AmB and C3deOAmB, single channel formation was observed at concentrations between 0.25 and 0.5 nM. See Supporting Information for experimental details.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.