Abstract
Wright’s F-statistics, and especially FST, provide important insights into the evolutionary processes that influence the structure of genetic variation within and among populations, and they are among the most widely used descriptive statistics in population and evolutionary genetics. Estimates of FST can identify regions of the genome that have been the target of selection, and comparisons of FST from different parts of the genome can provide insights into the demographic history of populations. For these reasons and others, FST has a central role in population and evolutionary genetics and has wide applications in fields that range from disease association mapping to forensic science. This Review clarifies how FST is defined, how it should be estimated, how it is related to similar statistics and how estimates of FST should be interpreted.
Nearly every plant or animal species includes many partially isolated populations. As a result of genetic drift or divergent natural selection, such populations become genetically differentiated over time. For example, recent analyses based on more than 370 short tandem repeat loci1 (microsatellites) and 600,000 SNPs2 suggest that only 5–10% of human genetic diversity is accounted for by genetic differences among populations from major geographical regions. These results indicate that there are far more similarities among geographically distinct human populations than differences. But what does it mean to say that 5–10% of diversity is accounted for by differences among populations, and how is this figure derived? The short answer is that the estimate of FST among human populations sampled from these regions is 0.05 for the microsatellite data and 0.10 for the SNP data. However, this answer helps only if one understands what FST is, how it is estimated from data and what it means to get two different estimates for the same set of populations when different genetic markers are used.
Working independently in the 1940s and 1950s, Sewall Wright3 and Gustave Malécot4 introduced F-statistics as a tool for describing the partitioning of genetic diversity within and among populations. In a paper published in 1931 (REF. 5), Wright had already provided a comprehensive account of the processes that cause genetic differentiation among populations. He showed that the amount of genetic differentiation among populations has a predictable relationship to the rates of important evolutionary processes (migration, mutation and drift). For example, large populations among which there is much migration tend to show little differentiation, whereas small populations among which there is little migration tend to be highly differentiated. FST is a convenient measure of this differentiation, and as a result FST and related statistics are among the most widely used descriptive statistics in population and evolutionary genetics.
But FST is more than a descriptive statistic and a measure of genetic differentiation. FST is directly related to the variance in allele frequency among populations and, conversely, to the degree of resemblance among individuals within populations. If FST is small, it means that the allele frequencies within each population are similar; if it is large, it means that the allele frequencies are different. If natural selection favours one allele over others at a particular locus in some populations, the FST at that locus will be larger than at loci in which among-population differences are purely a result of genetic drift. Genome scans that compare single-locus estimates of FST with the genome-wide background might therefore identify regions of the genome that have been subjected to diversifying selection6–8. Alternatively, if the demographic history of populations affects the genetic variation on sex chromosomes in a different way from the genetic variation on autosomes, the estimates of FST derived from sex chromosome markers might be different from those derived from autosomal markers9.
Estimates of FST are also important in association mapping of human disease genes and in forensic science. The same evolutionary processes that increase differentiation among populations also increase the similarity among individuals within populations. Therefore, FST must be considered when allele frequencies are compared between cases and controls to ensure that the differences between them are greater than expected by chance. Similarly, the probability of a match between a suspect and a crime scene sample is specific to the set of people who might reasonably be expected to be sources of the sample. However, defining this set is difficult, so a ‘θ correction’ is applied to population frequencies to accommodate variation among subpopulations. The θ correction depends on the value of FST.
In this review, we discuss how FST is defined, describe approaches for estimating it from data and illustrate several ways in which analysis of FST can provide insights into the genetic structure and evolutionary dynamics of populations. In addition, we discuss four statistics that are related to FST (GST, RST, ΦST and QST), clarify the differences among them and recommend when each should be used.
These additional statistics partition genetic diversity into within- and among-population components. of the four, GST is most closely related to FST, and it has been widely used as a measure of genetic differentiation among populations. However, as we describe below, GST is an appropriate measure of genetic differentiation only when the contribution of genetic drift to among-population differences is not of interest. As a result, the contexts in which it is useful are limited. By contrast, RST (for microsatellite data) and ΦST (for molecular sequence data) are useful in a wide range of contexts in which it is important to account for the mutational ‘distances’ among alleles, and QST is useful in the analysis of continuously varying traits.
Definitions
Wright introduced FST as one of three interrelated parameters to describe the genetic structure of diploid populations3. These parameters are: FIT, the correlation between gametes within an individual relative to the entire population; FIS, the correlation between gametes within an individual relative to the subpopulation to which that individual belongs; and FST, the correlation between gametes chosen randomly from within the same subpopulation relative to the entire population. We describe here how these parameters are defined in terms of the departure of genotype frequencies from Hardy–Weinberg proportions.
Deriving measures of genetic diversity
As an example of how to calculate genetic diversity, consider two populations that are segregating for two alleles at a single locus. The frequency of allele A1 in the first population is labelled as p1 and its frequency in the second population is labelled as p2. The frequency of genotype A1A1 in the first population is labelled as x11,1, the frequency of genotype A1A2 in the first population is labelled as x12,1, and so on. The genotype frequencies in the two populations are given by the following set of equations:
| (1) |
In this context, f1 and f2 are often referred to as the within-population inbreeding coefficients, but this term can be misleading. In practice, f is a measure of the frequency of heterozygotes compared with that expected when genotypes are in Hardy–Weinberg proportions. Inbreeding leads to a deficiency of heterozygotes relative to Hardy–Weinberg expectations, so when there is inbreeding in both populations, f1 and f2 will have positive values. But if individuals avoid inbreeding or if there is heterozygote advantage, then heterozygotes will be more common than expected under Hardy–Weinberg expectations, and f1 and f2 will be negative. In short, f1 and f2 are measures of how different the genotype proportions within populations are from Hardy–Weinberg expectations, and positive values of f indicate a deficiency of heterozygotes, whereas negative values indicate an excess.
Now consider the genotype frequencies in a combined sample that consists of a proportion c of individuals from the first population and a proportion 1 – c of individuals from the second population. Similar to the way in which the genotype frequencies in each population differ from Hardy–Weinberg expectations based on the allele frequency in each population, genotype frequencies in the combined sample differ from Hardy–Weinberg expectations based on the average allele frequency. The allele frequencies are given by:
| (2) |
in which π = cp1 + (1 – c)p2 is the average allele frequency for A1 in the combined sample and F is the total inbreeding coefficient10. F can be expressed as:
| (3) |
in which f = cf1 + (1 – c)f2 is the average within-population departure from Hardy–Weinberg expectations and θ is a measure of allele frequency differentiation among populations (see BOX 1 for a summary of the mathematical notation used in this review). We can define θ as:
| (4) |
in which is the variance in allele frequency among populations. π(1 – π) is the variance in the allelic state for an allele chosen randomly from the entire population, so it can be regarded as a measure of genetic diversity in the entire population. θ can therefore be interpreted as the proportion of genetic diversity that is due to the differences in allele frequency among populations.
Box 1. Mathematical notation.
In this box, we provide definitions for the mathematical symbols used throughout the Review.
| Parameter | Definition | |
|---|---|---|
| Among-population allele frequency distribution | ||
| π | Mean allele frequency | |
| Variance in allele frequency | ||
| Wright’s F-statistics and Cockerham’s θ-statistics | ||
| FIS | Correlation of alleles within an individual relative to the subpopulation in which it occurs; equivalently, the average departure of genotype frequencies from Hardy–Weinberg expectations within populations | |
| FST | Correlation of randomly chosen alleles within the same subpopulation relative to the entire population; equivalently, the proportion of genetic diversity due to allele frequency differences among populations | |
| FIT | Correlation of alleles within an individual relative to the entire population; equivalently, the departure of genotype frequencies from Hardy–Weinberg expectations relative to the entire population | |
| f | Co-ancestry for alleles within an individual relative to the subpopulation in which it occurs; equivalent to FIS | |
| θ | Co-ancestry for randomly chosen alleles within the same subpopulation relative to the entire population; equivalent to FST | |
| F | Co-ancestry for alleles within an individual relative to the entire population; equivalent to FIT | |
| Φ-statistics and RST* | ||
| ΦIS | Excess similarity of alleles within an individual relative to the subpopulation in which it occurs; analogous to FIS | |
| ΦST | Excess similarity among randomly chosen alleles within the same subpopulation relative to the entire population; equivalently, the proportion of genetic diversity (measured as the expected squared evolutionary distance between alleles) due to differences among populations; analogous to FST | |
| ΦIT | Excess similarity of alleles within an individual relative to the entire population; analogous to FIT | |
| RST | Excess similarity among randomly chosen alleles within the same subpopulation relative to the entire population; equivalently, the proportion of genetic diversity (measured as the expected squared difference in repeat numbers between alleles) due to differences among populations; analogous to FST | |
| Measuring genetic differentiation among populations in quantitative traits | ||
| Additive genetic variance within populations | ||
| Additive genetic variance among populations | ||
| QST | Proportion of additive genetic variation in the entire population due to differences among populations; analogous to FST | |
ΦST from analysis of molecular variance (AMOVA) is used for haplotype data (for example, nucleotide sequence data or mapped restriction site data) and requires a measure of evolutionary distance among all pairs of haploytpes. RST is used for microsatellite data and requires that alleles are labelled according to the number of repeat units that they contain.
Wright first developed these ideas in the context of a model of discrete populations, in which each population is the same size and receives immigrants from all other populations at the same rate5. However, the same statistical argument can be applied to any partitioning of genetic diversity in which the populations differ in allele frequency, whether or not those populations are discrete11. Therefore, when we use θ as a purely descriptive statistic that describes the partitioning of genetic diversity among ‘populations’, we do not need to make assumptions about whether the ‘populations’ we sample are discrete or about the evolutionary processes that might have led to differences among them. Nonetheless, other methods of analysis could be more informative in continuously distributed populations12–14.
Linking f, θ and F to Wright’s F-statistics
Using a different approach, Cockerham10,15 showed that f, θ and F can also represent intraclass correlation coefficients. He showed that f is the correlation between alleles within individuals relative to the population to which they belong, θ is the correlation between alleles within populations relative to the combined population and F is the correlation between alleles within individuals relative to the combined population. These are the definitions that Wright gave for FIS, FST and FIT, respectively. In short, f and FIS can be thought of either as the average within-population departure from Hardy–Weinberg expectations or as the correlation between alleles within individuals relative to the population to which they belong. θ and FST can be thought of either as the proportion of genetic diversity due to allele frequency differences among populations or as the correlations between alleles within populations relative to the entire population. F and FIT can be thought of either as the departure of genotype frequencies in the combined sample from Hardy–Weinberg expectations or as the correlation between alleles within individuals relative to the combined sample.
In Wright’s notation, subscripts refer to a comparison between levels in a hierarchy: IS refers to ‘individuals within subpopulations’, ST to ‘subpopulations within the total population’ and IT to ‘individuals within the total population’16. The hierarchy in equation 1 can be extended indefinitely to accommodate such structures. For example, Wright16 describes variation in the frequency of the Standard chromosome in Drosophila pseudoobscura in the western United States at the level of demes (D; local populations), regions (R; groups of several demes), subdivisions (S; groups of several regions) and the total range (T). The corresponding F-statistics are related in the same multiplicative way as f, θ and F:
| (5) |
In this scheme, FDR measures the differentiation among demes within a region, FRS measures the differentiation among regions within subdivisions and FST measures the differentiation among subdivisions within the total range.
If we return to the examples of genetic differentiation among human populations that were mentioned at the beginning of this review, we can now see that an estimate for FST or θ of 0.05 (from microsatellites) and 0.10 (from SNPs) suggests that only 5–10% of human genetic diversity is a result of genetic differentiation among human populations. What might be surprising is that the two estimates are derived from the same set of populations — this indicates that the amount of genetic differentiation among human populations is greater at SNP loci than at microsatellites.
Estimation
Statistical sampling
When Wright and Malécot introduced F-statistics, they did not distinguish between the parameters defined in the preceding section and the estimates of those parameters that we make from data. Not making this distinction is similar to confusing the mean height of the human population with an estimate of the mean height calculated from a sample of the population. Estimates of height must account for the variation associated with taking a finite sample from a population. New samples from the same population will have different characteristics. We refer to this variation as statistical sampling17 (BOX 2). In the context of F-statistics, statistical sampling refers to the variation associated with collecting genetic samples from a fixed set of populations that have fixed but unknown genotype frequencies. The magnitude of variation associated with statistical sampling can be reduced by increasing the size of within-population samples.
Box 2. Genetic sampling versus statistical sampling.
Genetic drift leads to differences among populations that are described by the distribution of allele frequencies among those populations. The variance of this distribution is directly related to FST (see equation 2), but in a typical study only a subset of populations is sampled. Therefore, in addition to accounting for the variation associated with sampling from populations, estimates of F-statistics must account for the variation associated with sampling sets of populations from the allele frequency distribution.
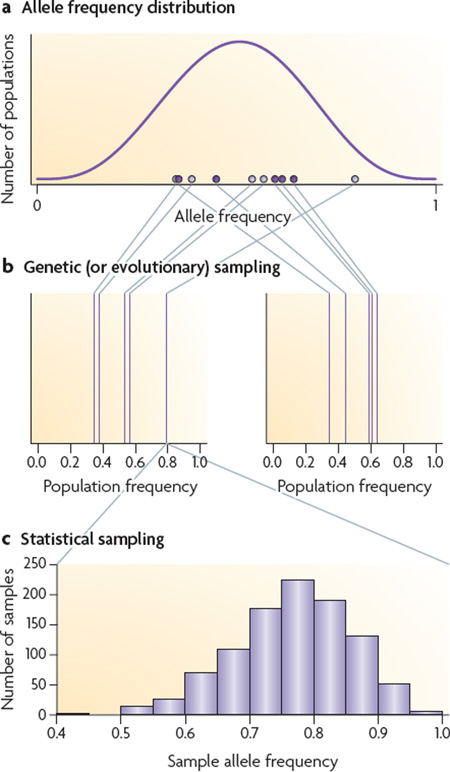
Genetic (or evolutionary) sampling
Part a of the figure shows the distribution of allele frequencies among populations corresponding to a mean allele frequency of π = 0.5 and FST = θ = 0.1. If two sets of populations (represented by dark and light circles) are sampled from this distribution, the allele frequencies in the first set of populations (light circles) will differ from those in the second set (dark circles). Part b provides an example in which two different sets of five population frequencies are drawn randomly from the distribution of allele frequencies shown in part a.
The variation in allele frequencies illustrated in part a reflects the effect of genetic or evolutionary sampling. The differences between the sets of samples in part b reflect the effect of sampling particular populations from the distribution of allele frequencies in part a and are analogous to the results that would be expected in an empirical study if it were repeated on a different set of populations.
Statistical sampling
Part c illustrates the more familiar idea of statistical sampling. It shows the distribution of sample allele frequencies obtained in 1,000 samples of 20 individuals from the population with the largest allele frequency in the population sample on the left in part b. Statistical sampling refers to the variation in sample composition that is expected when alleles are repeatedly sampled from a population with a particular allele frequency.
Investigators can control the amount of variation associated with statistical sampling by increasing the number of individuals sampled within populations: the larger the number of individuals sampled, the less that the sample allele frequencies will differ from the underlying population frequencies. By contrast, investigators cannot control the amount of variation associated with genetic sampling: the variation associated with genetic sampling is an intrinsic property of the underlying stochastic evolutionary process that contributes to the differentiation among populations.
The relationship between FST and GST
Nei introduced the statistic GST as a measure of genetic differentiation among populations33. It is defined in terms of the population frequencies in part b, not the allele frequency distribution in part a. By contrast, estimates of FST account for genetic sampling and they are intended to reflect the properties of the allele frequency distribution in part a. As a result, FST and GST measure different properties. Therefore, GST will be an appropriate measure only when interest focuses on characteristics of the particular samples illustrated in part b. In a typical population study, θ will be a more appropriate measure of differentiation.
It might seem that similar arguments should apply to exact tests of population differentiation102 because they also use permutations of sample configurations to determine whether populations are differentiated from one another. However, the permutation test is equivalent to determining whether the allele frequency distribution in part a has a variance greater than zero, so exact tests implicitly consider both statistical and genetic sampling effects.
Genetic sampling
There is an important difference between estimates made by F-statistics and estimates of height. In addition to accounting for statistical sampling, F-statistics must account for differences among the sets of populations that have been sampled. These differences might arise either because the populations that are sampled are only a subset of all of the populations that could be sampled (statistical sampling of populations rather than statistical sampling of genotypes within populations) or because the populations that are sampled represent only one possible outcome of an underlying stochastic evolutionary process. even if we could take the set of sampled populations back to a previous point in time and re-run the evolutionary process under all of the same conditions (the same population sizes, mutation rates, migration rates and selection coefficients), the genotype frequencies in the new set of populations would differ from those in the populations that were actually sampled18. This genetic sampling17 is an unavoidable consequence of genetic drift. The magnitude of variation associated with genetic sampling cannot be reduced by increasing either the number of individuals sampled within populations or the number of populations sampled. Indeed, the characteristics of genetic sampling are shown by estimates of F-statistics.
In simple cases, it might make sense to estimate statistical parameters using simple functions of the data, such as the sample mean. In more complicated cases, such as those presented by F-statistics, it is useful to have well-defined approaches for constructing estimates. Statisticians have developed several different approaches for estimating parameters from data19. Three widely used approaches are the method of moments, the method of maximum likelihood and Bayesian methods.
Approaches to estimating FST: method-of-moments estimates
The method of moments produces an estimate by finding an algebraic expression that makes the expected value of certain sample statistics equal to simple functions of the parameters that are being estimated (as explained in more detail below)19. Method-of-moments estimates are designed to have low bias in the sense that if samples are taken repeatedly from the same population, the average of the corresponding sample estimates will be close to the unknown population parameter. These estimates have the additional advantages that they are easy to calculate and do not require any assumptions about the shape of the distribution from which the sample is drawn, other than that it has a mean and variance.
For F-statistics, method-of-moments estimates17,20,21 are based on an analysis of variance (ANOVA) of allele frequencies. ANOVA is a statistical method that tests whether the means of two or more groups are equal and can therefore be used to assess the degree of differentiation between populations. Briefly, if the variance among populations is the same as the variance within populations, there is no population substructure. ANOVA calculations are framed in terms of mean squares. Therefore, in practice, one calculates the expected mean square among populations (that is, the variance of sample allele frequencies around the mean allele frequency over all populations) and the expected mean square within populations (that is, the heterozygosity within populations when genotypes are in Hardy–Weinberg proportions) averaged over all possible samples (statistical sampling) from all possible populations with the same evolutionary history (genetic sampling). These expected values are then equated to the observed mean squares that are calculated from a sample, and the resulting set of equations is solved for the corresponding variance components. Following the work of Cockerham10,22, F-statistics are defined in terms of these variance components (BOX 3).
Box 3. Comparing methods for estimating FST.
To illustrate the differences among calculating method-of-moments, maximum-likelihood and Bayesian estimates of F-statistics, we use data from a classic study on human populations that investigated the allele frequency differences at blood group loci (see the table). We use a subset of the data that were originally reported by Workman and Niswander103. Their data consist of genotype counts at several loci in Native American Papago and were collected from ten political districts in south-western Arizona. Estimates of FIS, FST and FIT derived from the MN blood group locus suggest that there is little departure of the genotype frequencies from Hardy–Weinberg expectations within each district and little genetic differentiation among the districts.
Method-of-moments analysis
Analysis of variance on the indicator variable yij,k, in which yij,k = 1 if allele i in individual j of population k is M, gives moment estimates for the variance components of , and , in which G stands for genotypes (alleles within individuals), I stands for individuals (individuals within populations) and P stands for populations (among populations). Following Cockerham10:
Therefore, the moment estimates are F = 0.0348, θ = 0.00402 and f = 0.0309. As expected for human populations, there is little evidence that the genotype proportions within each political district differ from Hardy–Weinberg expectations (f ≈ 0). Similarly, there is little evidence of genetic differentiation among political districts (θ ≈ 0).
Bayesian and likelihood analysis
By contrast, current implementations of a Bayesian approach to analysing these data typically assume independent uniform (0,1) prior distributions for both f and θ. The posterior mean of f and θ for these data are 0.0503 and 0.0189, respectively. The posterior distribution of f has a mode near 0 but is broad (with a 95% credible interval of 0.0033–0.123), which causes the posterior mean of f to be larger than the method-of-moments estimate. Similarly, the estimates of allele frequencies within each population are uncertain and the estimate of θ takes this uncertainty into account, suggesting that there is slightly more among-population differentiation than detected with moment estimates. For comparison, the maximum-likelihood estimates are F = 0.0408, θ = 0.00640 and f = 0.0346 (obtained by estimating the variance components in a Gaussian mixed model applied to the indicator variables and by using Cockerham’s definitions of F, f and θ in terms of the variance components).
| Parameter | Method of moments | Maximum likelihood | Bayesian |
|---|---|---|---|
| f | 0.0309 | 0.0346 | 0.0503 |
| θ | 0.00402 | 0.00640 | 0.0189 |
| F | 0.0348 | 0.0408 | 0.0683 |
To extend the method-of-moments approach to multiple alleles and multiple loci, calculations are done separately for every allele at every locus and the sums of squares are combined17,27. To extend the likelihood or Bayesian approaches, we make the assumption that f and θ have the same value at every locus and that genotype counts are sampled independently across loci and populations104,105.
Approaches to estimating FST: maximum-likelihood and Bayesian estimates
In contrast to method-of-moments estimates, likelihood and Bayesian estimates are difficult to calculate and require the specification of the probability distribution from which the sample was drawn. Once this probability distribution is specified, we can calculate a quantity called the likelihood, which is proportional to the probability of our observed data given those parameters. A maximum-likelihood estimate for the parameters is obtained by finding the values of the unknown parameters that maximize that likelihood19. In most cases, maximum-likelihood estimates are biased. Nonetheless, they typically have a smaller variance and deviate less from the unknown population parameter than the corresponding method-of-moments estimates19. For these and other reasons, the method of maximum likelihood is the most widely used technique for deriving statistical estimators23,24.
Bayesian estimates share many of the advantages associated with maximum-likelihood estimates because they use the same likelihood to relate the data to unknown parameters. However, they differ from maximum-likelihood estimates because the likelihood is modified by placing prior distributions on unknown parameters, and estimates are based on the posterior distribution, which is proportional to the product of the likelihood and the prior distributions. Both maximum-likelihood and Bayesian methods suffer the disadvantage that simple algebraic expressions for the estimates are rarely available. Instead, the estimates are obtained through computational methods. Because the Markov chain Monte Carlo methods (MCMC methods) used for analysis of Bayesian models do not require a unique point of maximum likelihood to be identified, Bayesian estimates can be obtained even in complex models with thousands or tens of thousands of parameters, for which numerical maximization of the likelihood would be difficult or impossible26.
For F-statistics, the likelihood approach27,28 specifies a probability distribution that describes the variation in allele frequencies among populations and a multinomial distribution that describes genotype samples within populations. θ is related to the variance of the probability distribution that describes the among-population distribution of allele frequencies, and the genotype frequencies are determined by the allele frequencies in each population and f. Estimates are obtained by maximizing the likelihood function with respect to θ, f and the allele frequencies. The Bayesian approach uses the same likelihood function, and after placing appropriate prior distributions on f, θ and allele frequencies, MCMC methods are used to sample from the posterior distributions of f and θ.
Comparing the methods
With more than 5,000 citations, the moments method described by Weir and Cockerham20 has been widely used, partly because of its robustness and partly because it is simple to implement. The maximum-likelihood methods also give simple equations when the distribution of allele frequencies among populations is assumed to be normal27, but only if the sample sizes are equal29. Bayesian methods allow probability statements to be made about F-statistics, and extensions of these methods allow the relationship between F-statistics and demographic or environmental covariates to be explored in the context of a single model30. However, implementations of Bayesian methods may be computationally demanding.
A simple data set is used in BOX 3 to illustrate the slightly different estimates obtained from each approach. Estimates of FST using moments and Bayesian methods have not been extensively compared, but our experience suggests that the differences in estimates are small when the average number of individuals per population is moderate to large (>20), when the number of populations is moderate to large (>10–15) and when most populations are polymorphic. When differences arise, they reflect differences in the treatment of allele frequency estimates when alleles are rare or sample sizes are small. The Bayesian approach ‘smooths’ population allele frequencies towards the mean24 and does so more aggressively when alleles are rare or sample sizes are small. The moments approach treats the sample frequencies as fixed quantities without such smoothing. The simulation results in REF. 31 are consistent with this interpretation, although they compare Bayesian estimates with estimates of GST32, which does not account for genetic sampling.
Related statistics
Population geneticists have proposed several statistical measures that are related to FST. Here, we describe four of them: GST, RST, ΦST and QST. Nei33 introduced GST as a measure of population differentiation. We discuss its relationship to FST in BOX 2. Haplotype and microsatellite data contain information not only about the frequency with which particular alleles occur but also on the evolutionary distance between them. Statistics such as ΦST (for haplotype data) and RST (for microsatellite data) are intended to take advantage of this additional information and to provide greater insight into the patterns of relationships among populations. Whereas FST, ΦST and RST all apply to discrete genetic data, QST is an analogous statistic for continuously varying traits. If the markers used to estimate FST can be presumed to be selectively neutral, comparing an estimate of QST with an estimate of FST can provide investigators with evidence that natural selection has shaped the pattern of variation in the quantitative trait.
RST, ΦST and AMOVA
The methods for estimating f, θ and F described above are appropriate for multi-allelic data when the alleles are regarded as equivalent to one another. However, when the data consist of variation at microsatellite loci or of nucleotide sequence (haplotype) information, related methods that allow mutation rates to differ between different pairs of alleles might be more appropriate. Excoffier et al.34 introduced analysis of molecular variance (AMOVA) for analysis of haplotype variation. AMOVA is based on an analysis-of-variance framework that is analogous to the one developed by Weir and Cockerham20. The mean squares in an AMOVA analysis are based on a user-specified measure of the evolutionary distance between haplotypes, and AMOVA leads to quantities that are analogous to classical F-statistics (BOX 1). Similarly, the mean squares used to calculate RST35,36 are based on differences in the number of repeats between alleles at each microsatellite locus. Although the result of both analyses is a partitioning of genetic variance into within- and among-population components analogous to FST, neither has a direct interpretation as a parameter of a statistical distribution. Instead, they estimate an index that is derived from two different statistical distributions: the distribution of allele (haplotype or microsatellite) frequencies among populations and the distribution of evolutionary distances among alleles. Nonetheless, such measures may be thought of as estimating the additional time since the common ancestry of randomly chosen alleles that accrues as a result of populations being subdivided37,38, provided that the measure of evolutionary distance between any two alleles is proportional to the time since their most recent common ancestor. Extensive simulation studies have shown that estimates of RST may be unreliable unless many loci are used39–41, but unlike FST the expected value of RST does not depend on the rate of mutation. Estimates of ΦST or RST may be useful when mutations have contributed substantially to allelic differences among populations, but their usefulness may be limited by the extent to which the mutational model underlying the statistics matches the actual mutational processes occurring in the system39.
QST and polygenic variation
Spitze42 noted that another quantity analogous to θ can be estimated for continuously varying traits. Specifically, we can define:
| (6) |
in which is the additive genetic variance among populations and is the additive genetic variance within populations. can be estimated from between-population crosses, and can be estimated from within-population crosses. Because the total variance in between-population crosses is , QST is the proportion of additive genetic variance in a trait that is due to among-population differences. If the trait is selectively neutral, if all genetic variation is additive and if the mutation rates at loci contributing to the trait are the same as those at other loci, we expect QST and FST to be equal43,44. Comparing the magnitude of QST and FST may therefore indicate whether a particular trait has been subject to stabilizing selection (QST<FST) or diversifying selection (QST>FST). However, because of the uncertainties associated with estimates of QST and FST, such comparisons are likely to be useful only when they are available for a moderately large number of populations (>20)45. Furthermore, caution is necessary when suggesting that a comparison of QST and FST provides evidence for stabilizing selection because non-additive genetic variation tends to change QST, even for a neutral trait46.
Applications
F-statistics include both FST, which measures the amount of genetic differentiation among populations (and simultaneously the extent to which individuals within populations are similar to one another), and FIS, which measures the departure of genotype frequencies within populations from Hardy–Weinberg proportions. Here, we focus on the applications of FST for several reasons (BOX 4).
Box 4. Why focus on FST?
We focus here on FST for several reasons. First, FIS is easier to interpret. It is defined with respect to the populations that are included in the sample, either through population-specific estimates or through the average of those estimates. By contrast, FST is defined and interpreted with respect to the distribution of allele frequencies among all populations that could have been sampled, not merely those that have been included in the sample. As a result, estimates of FST must account for genetic sampling, which introduces a level of complexity and subtlety that requires extra attention.
Second, the application of F-statistics to problems in population and evolutionary genetics often centres on estimates of FST. For example, when interpreting aspects of demographic history, such as sex-biased dispersal out of Africa in human populations9, detecting regions of the genome that might have been subject to stabilizing or diversifying selection8,58,61 or correcting the probabilities of obtaining a match in a forensic application for genetic substructure within populations106, estimates of FST often play a crucial part in interpretations of genetic data. Estimates of FIS reveal important properties of the mating system within populations, but estimates of FST reveal properties of the evolutionary processes that lead to divergence among populations.
Finally, in many populations of animals, and in human populations in particular, within-population departures from Hardy–Weinberg proportions are small. Where they are present, such departures may reveal more about genetic substructuring within populations than about departures from random mating. Moreover, although estimates of FIS may provide insights into the patterns of mating in inbred populations of plants or animals, the direct analysis of mother–offspring genotype combinations is usually more informative and reliable107,108.
Estimating migration rates
Wright5 showed that if all populations in a species are equally likely to exchange migrants and if migration is rare, then:
| (7) |
in which m is the fraction of each population composed of migrants (the backward migration rate)47 and Ne is the effective population size of local populations48. Because of this simple relationship, it is tempting to use estimates of FST from population data to estimate Nem.
Unfortunately, it has been recognized for many years that this simple approach to estimating migration rates might fail49. The most obvious reason for this failure is that populations are rarely structured so that all populations exchange migrants at the same rate, which causes some populations to resemble one another more than others. If differentiation between populations is solely a result of isolation by distance50, for example, then the slope of the regression of FST/(1 – FST) on either the logarithm of between-population distance (for populations distributed in two dimensions) or the between-population distance alone (for populations in a linear habitat) is proportional to Deδ2, in which De is the effective density of the population (De = Ne/area) and δ2 is the mean squared dispersal distance51. However, if differentiation is the result not only of isolation by distance but also of natural selection or if the drift–migration process has not reached a stationary point, the slope of this relationship cannot be interpreted as an estimate of migration. Moreover, a pure migration–drift process, a pure drift–divergence process or a combination of the two could produce the same distribution of allele frequencies. Indeed, migration–drift, drift–divergence or a combination of the two can account for any pattern of allele frequency differences among populations52. Therefore, although pairwise estimates of FST (or ΦST or RST) provide some insight into the degree to which populations are historically connected37,38, they do not allow us to determine whether that connection is a result of ongoing migration or of recent common ancestry.
There are additional difficulties with interpreting estimates of FST. Different genetic markers may give different estimates of FST for many reasons, and to derive an estimate of migration rates from FST, one must assume that the particular set of markers that are chosen have the expected relationship with Nem. This may often be problematic. For example, differences between FST estimates from human microsatellites (0.05) and SNPs (0.10) cannot reflect differences in migration rate because both estimates are derived from the same set of individuals and the same set of populations — the Human Genome Diversity Project–Centre d’Étude du Polymorphisme Humain sample1,2,53. The use of coalescent-based approaches (see later section) that incorporate models of the mutational process is one method of overcoming this difficulty54–56.
Inferring demographic history
Population-specific or pairwise estimates of FST may provide insights into the demographic history of populations when estimates are available from many loci. For example, Keinan et al.9 reported pairwise estimates of FST for 13,600–62,830 autosomal SNP loci and 1,100–2,700 X chromosome SNP loci in human population samples from northern Europe, East Asia and West Africa. Because there are four copies of each autosome in the human population for every three copies of the X chromosome, one would expect there to be greater differentiation at X chromosome loci than at autosomal loci. Specifically, for two populations that diverged t generations ago, one might expect:
| (8) |
in which Ne is the effective size of the local populations. Therefore, if Q is defined as:
| (9) |
Q is approximately:
| (10) |
Q is approximately 0.75 for comparisons between East Asians and northern Europeans (Q = 0.72 ± 0.05), but it is substantially smaller for comparisons between West Africans and other populations in the sample (Q = 0.58 ± 0.03 for the comparison with northern Europeans and Q = 0.62 ± 0.03 for the comparison with East Asians). These results suggest either sex-biased dispersal (long-range immigration of males from Africa after non-African populations were initially established) or selection on X chromosome loci after the divergence of African and non-African populations.
Identifying genomic regions under selection
Similarly, locus-specific estimates of FST may identify genomic regions that have been subject to selection. The logic is straightforward; the pattern of genetic differentiation at a neutral locus is completely determined by the demographic history of the populations (that is, the history of population expansions and contractions), the mutation rates at the loci concerned and the rates and patterns of migration among the populations6,57–60. In a typical multilocus sample, it is reasonable to assume that all autosomal loci have experienced the same demographic history and the same rates and patterns of migration. If the loci also have similar mutation rates and if the variation at each locus is selectively neutral, the allelic variation at each locus represents a separate sample from the same underlying stochastic evolutionary process. loci showing unusually large amounts of differentiation may indicate regions of the genome that have been subject to diversifying selection, whereas loci showing unusually small amounts of differentiation may indicate regions of the genome that have been subject to stabilizing selection58. Several groups have used such genome scans to examine patterns of differentiation in the human genome.
By comparing locus-specific estimates of FST with the genome-wide distribution, Akey et al.6 identified 174 regions (out of the 26,530 examined) that showed what they called ‘signatures of selection’ in the human genome. of these loci, 156 showed unusually large amounts of differentiation (suggesting diversifying selection) and 18 showed unusually small amounts of differentiation (suggesting stabilizing selection). By contrast, when Weir et al.7 examined the high-resolution Perlegen (~1 million SNPs) and phase I HapMap (~0.6 million SNPs) data sets in humans to examine locus-specific estimates of FST, they also found large differences in FST among loci, but their analyses suggested that the very high variance associated with single-locus estimates of FST precluded using these estimates to detect selection. Both sets of investigators noted a particular problem with single-locus estimates when using high-resolution SNP maps: the high correlation between FST estimates when loci are in strong gametic disequilibrium makes it difficult to determine whether the FST at any particular SNP is markedly different from expectation.
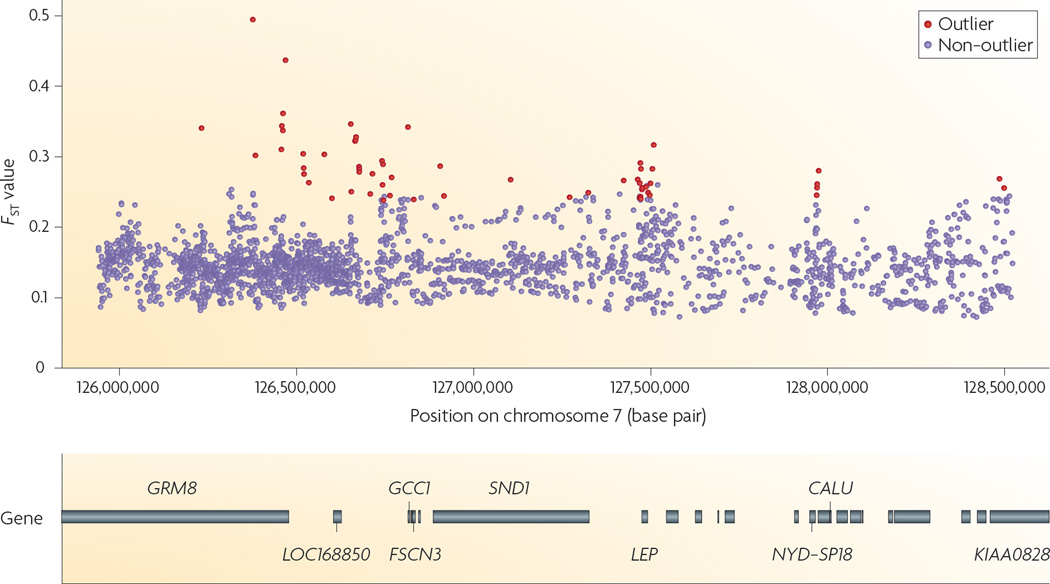
Although single-locus estimates of FST are highly uncertain, simulation studies suggest that when loci are inherited independently, background information about a few hundred loci is sufficient to allow the reliable identification of loci that are subject to selection when a suitable criterion for detecting ‘outliers’ is used8,58,61. Although few loci are falsely identified as being subject to selection when they are neutral, genome scans using FST may often fail to detect selection when it is present. For example, when a single allele is strongly favoured in all populations, not only is FST expected to be nearly zero but variation is also expected to be nearly non-existent, rendering estimates of FST either highly unreliable or unobtainable. Similarly, when selection is weak, data from many loci are needed to recognize that the estimate of FST at the locus involved is unusual. More importantly, as mentioned above, high-resolution genome scans must account for the statistical association between closely linked loci. Guo et al.8 used a conditional autoregressive scheme to identify 57 loci that showed unusually large amounts of among-population differentiation in a sample of 3,000 SNP loci on human chromosome 7 separated by only 860 nucleotides on average. Sixteen of these markers are associated with LEP, a gene encoding a leptin precursor that is associated with behaviours that influence the balance between food intake and energy expenditure62 (FIG. 1). Moreover, association studies in one French population had previously suggested a relationship between one of the SNPs identified as an outlier in this study and obesity63.
Figure 1. Locus-specific estimates of FST on human chromosome 7.
Estimates are as inferred from the phase II HapMap data set95. Horizontal bars indicate the locations of known genes. The red circles are posterior means for SNPs with estimates that are detectably different from the genomic background (purple circles). All ‘outliers’ show significantly more differentiation among the four populations in the sample than is consistent with the level of differentiation seen in the genomic background. The excess differentiation suggests that these SNPs are associated with genomic regions in which loci have been subject to diversifying selection among populations. CALU, calumenin; FSCN3, ascin homolog 3; GCC1, GRIP and coiled-coil domain containing 1; GRM8, glutamate receptor, metabotropic 8; LEP, leptin; SND1, staphylococcal nuclease and tudor domain containing 1. Figure is modified, with permission, from REF. 8 © (2009) American Statistical Association.
Forensic science and association mapping
In forensic science, matching a genetic profile taken from a suspect with a profile taken from a stain left at a crime scene serves as evidence linking the suspect to the crime. To quantify the strength of this evidence, it is useful to determine the probability of a random match — that is, the probability that the genetic profile at the crime scene matches that of the suspect if the suspect was not the source of the stain. In some cases, two people, the suspect and the person who left the crime sample, may belong to a subpopulation for which there is no specific allele frequency information. In such a case, we can use a θ correction64 to calculate the probability of a match based on allele frequency information from a larger population of which the subpopulation is a part. The probability of a random match takes into account the allele frequency variation among subpopulations within the wider population for which allele frequencies are available. For example, if the matching profile consisted of a homozygote AA at a single locus and if pA is the population frequency of allele A, the probability that the crime profile is AA given that the suspect is AA and the suspect is not the source of the stain is (REF. 65):
| (11) |
There is a similar equation for heterozygotes, and these θ-correction results are multiplied over loci. The 1996 National research Council report66 recommended using θ = 0.01 except for small isolated subpopulations, for which they suggested that a value of θ = 0.03 was more appropriate. The practical effect of the θ correction is that the numerical strength of the evidence against a suspect is reduced. If pA = 0.01, for example, the uncorrected probability of a match is 0.0001. However, with θ = 0.01, the probability of a match is an order of magnitude larger — 0.0012. With θ = 0.03, it is even larger — 0.0064. Therefore, it is much less surprising to see a match when we take account of the population substructure than when we ignore it.
In association mapping, case–control studies compare the allele frequencies at genetic markers (generally SNPs) between groups of people with a disease and groups who do not have the disease. When frequencies at a marker locus differ between the groups, it is interpreted as evidence for gametic disequilibrium between the marker and a disease-related gene. This in turn suggests that the marker and disease-related genes are in close proximity on the same chromosome. However, as many authors have pointed out, population substructure unrelated to disease status could cause the same kind of allele frequency difference67–70. The genomic control method is one way to account for population substructure. It uses background estimates of FST to control for subpopulation differences that are unrelated to disease status67,68. If cases and controls have different marker allele frequencies for reasons unconnected with the disease, as would be shown by frequency differences across the whole genome, an uncorrected case–control test would give spurious indications of marker–disease associations.
Relationship to coalescent-based methods
When Kingman introduced the coalescent process to population genetics just over 25 years ago71,72, it revolutionized the field. Many approaches to the analysis of molecular data, particularly molecular sequence and SNP data, now take advantage of the conceptual, computational and analytical framework that coalescent-based methods provide73–79. For example, whereas F-statistics provide only limited insight into the rates and patterns of migration, statistics based on the coalescent process can provide insights into the rates of mutation, migration and other evolutionary processes. Coalescent analysis is based on maximizing the likelihood of a given sample configuration or sampling from the corresponding Bayesian posterior distribution. The likelihood is constructed from the genealogical histories for the sample that are consistent with the unknown evolutionary parameters of interest; for example, the size of the population or populations from which the sample was taken, or the history of population size changes, mutation rates, recombination rates or migration rates55,80–86. Coalescent analyses are likely to provide precise estimates of effective population size, mutation rates and migration rates when certain conditions are met — that is, when the model used for analysis is consistent with the demographic history of populations from which samples are collected, with the migration patterns among populations in the sample and with the mutational processes that generated allelic differences in the sample, and also when it is reasonable to presume that the drift–mutation–migration process has reached an evolutionary equilibrium54,73. When these assumptions are not met it may not be reasonable to estimate the related evolutionary parameters, and the examples presented above show that analyses based on F-statistics may still provide substantial insights.
Conclusions
Sewall Wright5 provided a comprehensive account of the processes leading to genetic differentiation among populations nearly 80 years ago, but he did not provide the tools that empirical population geneticists needed to apply his insights to understanding variation in wild populations. During his work on isolation by distance in the plant Linanthus parryae in the 1940s50,87, the theory of F-statistics that he and Gustave Malécot later developed3,4,16,88 began to emerge. Because of the insights that F-statistics can provide about the processes of differentiation among populations, over the past 50 years they have become the most widely used descriptive statistics in population and evolutionary genetics. From the time population geneticists first began to collect data on allozyme variation89–94 to recent analyses of SNP variation in the human genome2,9,95–97, F-statistics, and FST in particular, have been used to investigate processes that influence the distribution of genetic variation within and among populations. Unfortunately, neither Wright nor Malécot distinguished carefully between the definition of F-statistics and the estimation of F-statistics. In particular, until Cockerham introduced his indicator formalism10,22, few if any population geneticists understood that estimators of F-statistics must take into account both statistical sampling and genetic sampling.
The statistical methodology for estimating F-statistics is now well established. With the availability of methods to estimate locus- and population-specific effects on FST7,8,27,58,61,98, geneticists now have a set of tools for identifying genomic regions or populations with unusual evolutionary histories. Through further extensions of this approach, it is even possible to determine the relationship between the recent evolutionary history of populations and environmental or demographic variables99. The basic principles of how population size, mutation rate and migration are related to the genetic structures of populations have been well understood for nearly 80 years. Analyses of F-statistics in populations of plants, animals and microorganisms have broadened and deepened this understanding, but these analyses have mostly been applied to data sets that contain a small number of loci. The age of population genomics is now upon us100,101. The 1,000 Genomes project and the International HapMap Project give a hint of what is to come. Despite the scale of these projects, much of the data can be understood fundamentally as allelic variation at individual loci. As a result, we expect F-statistics to be at least as useful in understanding these massive data sets as they have been in population and evolutionary genetics for most of the past century.
Acknowledgements
We thank R. Prunier and K. Theiss for their helpful comments on earlier versions of this Review. The work in the laboratories of the authors was supported in part by grants from the US National Institutes of Health (1 R01 GM 068449-01A1 to K.E.H; 1 R01 GM 075091 to B.S.W).
Glossary
- Genetic drift
The random fluctuations in allele frequencies over time that are due to chance alone.
- Short tandem repeat loci
Loci consisting of short sequences (2–6 nucleotides) that are repeated multiple times. Alleles at short tandem repeat loci differ from one another in their number of repeats.
- Variance
A measure of the amount of variation around a mean value.
- Diversifying selection
Selection in which different alleles are favoured in different populations. It is often a consequence of local adaptation (in which genotypes from different populations have higher fitness in their home environments owing to historical natural selection).
- Hardy–Weinberg proportions
When the frequency of each diploid genotype at a locus equals that expected from the random union of alleles. That is, the genotypes AA, Aa and aa will be at frequencies p2, 2pq and q2, respectively.
- Heterozygote advantage
A pattern of natural selection in which heterozygotes are more likely to survive than homozygotes.
- Likelihood
A mathematical function that describes the relationship between the unknown parameters of a statistical distribution — for example, the mean and variance of the allele frequency distribution among populations or the allele frequency in a particular population — and the data. It is directly proportional to the probability of the data given the unknown parameters.
- Prior distribution
A statistical distribution used in Bayesian analysis to describe the probability that parameters take on a particular value before examining any data. It expresses the level of uncertainty about those parameters before the data have been analysed.
- Posterior distribution
A statistical distribution used in Bayesian analysis to describe the probability that parameters take a particular value after the data have been analysed. It reflects both the likelihood of the data given particular parameters and the prior probability that parameters take particular values.
- Markov chain Monte Carlo methods
Methods that implement a computational technique that is widely used for approximating complex integrals and other functions. In this context, these methods are used to approximate the posterior distribution of a Bayesian model.
- Multinomial distribution
A statistical distribution that describes the probability of obtaining a sample with a specified number of objects in each of several categories. The probability is determined by the total sample size and the probability of drawing an object from each category. The binomial distribution is a special case of the multinomial distribution in which there are two categories.
- Additive genetic variance
The part of the total genetic variation that is due to the main (or additive) effects of alleles on a phenotype. The additive variance determines the degree of resemblance between relatives and therefore the response to selection.
- Stabilizing selection
Selection in which either the same allele or the same genotype is favoured in different populations.
- Effective population size
Formulated by Wright in 1931, the effective population size reflects the size of an idealized population that would experience drift in the same way as the actual (census) population. The effective population size can be lower than the census population size owing to various factors, including a history of population bottlenecks and reduced recombination.
- Coalescent-based approaches
Approaches that use statistical properties of the genealogical relationship among alleles under particular demographic and mutational models to make inferences about the effective size of populations and about rates of mutation and migration.
- Conditional autoregressive scheme
A statistical approach developed for analysis of data in which a random effect is associated with the spatial location of each observation. The magnitude of the random effect is determined by a weighted average of the random effects of nearby positions. In most applications, the weights of the averages are inversely related to the spatial distance between two sample points.
Footnotes
FURTHER INFORMATION
Kent E. Holsinger’s homepage: http://darwin.eeb.uconn.edu
1,000 Genomes project: http://www.1000genomes.org
ABC4F (approximate Bayesian computation for F-statistics): http://www-leca.ujf-grenoble.fr/logiciels.htm
Arlequin (an integrated software application for population genetics data analysis): http://cmpg.unibe.ch/software/arlequin3
BayeScan (BAYEsian genome SCAN for outliers): http://www-leca.ujf-grenoble.fr/logiciels.htm
Bayesian population genetic data analysis: http://darwin.eeb.uconn.edu/summer-institute/summer-institute.html
GDA (Genetic Data Analysis): http://www.eeb.uconn.edu/people/plewis/software.php
GenAlEx (integrated software for analysis of genetic data with an interface to Excel): http://www.anu.edu.au/BoZo/GenAlEx/genalex_download_6_1.php
Genepop: http://kimura.univ-montp2.fr/~rousset/Genepop.htm
GESTE (GEnetic STructure inference based on genetic and Environmental data): http://www-leca.ujf-grenoble.fr/logiciels.htm
Hickory (software for the analysis of geographic structure in genetic data): http://darwin.eeb.uconn.edu/hickory/hickory.html
Hierfstat (Weir & Cockerham F-statistics for any number of levels in a hierarchy): http://www2.unil.ch/popgen/softwares/hierfstat.htm
International HapMap Project: http://www.hapmap.org
Nature Reviews Genetics series on Fundamental Concepts in Genetics: http://www.nature.com/nrg/series/fundamental/index.html
The genetic structure of populations: http://darwin.eeb.uconn.edu/eeb348/lecture.php?rl_id=402
The genetic structure of populations: a Bayesian approach: http://darwin.eeb.uconn.edu/eeb348/lecture.php?rl_id=403
The Wahlund effect and Wright’s F-statistics: http://darwin.eeb.uconn.edu/eeb348/lecture.php?rl_id=445
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Kent E. Holsinger, Email: kent@darwin.eeb.uconn.edu.
Bruce S. Weir, Email: bsweir@u.washington.edu.
References
- 1.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 2.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 3. Wright S. The genetical structure of populations. Ann. Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. This paper develops the explicit framework for the analysis and interpretation of F-statistics in an evolutionary context.
- 4. Malecot G. Les Mathématiques de l’Hérédié. Masson, Paris: 1948. This book develops a framework — equivalent to Wright’s F-statistics — for the analysis of genetic diversity in hierarchically structured populations.
- 5. Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. A landmark paper in population genetics in which the effect of population size, mutation and migration on the abundance and distribution of genetic variation in populations is first quantitatively described.
- 6.Akey JM, Zhang G, Khang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Dey DK, Holsinger KE. A Bayesian hierarchical model for analysis of SNP diversity in multilocus, multipopulation models. J. Am. Stat. Assoc. 2009;164:142–154. doi: 10.1198/jasa.2009.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nature Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cockerham CC. Variance of gene frequencies. Evolution. 1969;23:72–84. doi: 10.1111/j.1558-5646.1969.tb03496.x. This paper develops the first approach for the analysis of F-statistics that recognizes the effect of genetic sampling on estimates of F-statistics from population data.
- 11.Wahlund S. Zusammensetzung von Population und Korrelationserscheinung vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas. 1928;11:65–106. [Google Scholar]
- 12.Sokal RR, Oden NL, Thomson BA. A simulation study of microevolutionary inferences by spatial autocorrelation analysis. Biol. J. Linn. Soc. 1997;60:73–93. [Google Scholar]
- 13.Sokal RR, Oden NL. Spatial autocorrelation analysis as an inferential tool in population genetics. Am. Nat. 1991;138:518–521. [Google Scholar]
- 14.Epperson BK. Geographical Genetics. Princeton Univ. Press; 2003. [Google Scholar]
- 15.Weir BS, Cockerham CC. Mixed self- and random-mating at two loci. Genet. Res. 1973;21:247–262. doi: 10.1017/s0016672300013446. [DOI] [PubMed] [Google Scholar]
- 16.Wright S. Evolution and the Genetics of Populations. Vol. 4. Univ. Chicago Press; 1978. [Google Scholar]
- 17.Weir BS. Genetic Data Analysis II. Methods for Discrete Population Genetic Data. Sunderland, USA: Sinauer Associates; 1996. [Google Scholar]
- 18.Rousset F. Inbreeding and relatedness coefficients: what do they measure? Heredity. 2002;88:371–380. doi: 10.1038/sj.hdy.6800065. [DOI] [PubMed] [Google Scholar]
- 19.Casella G, Berger RL. Statistical Inference. Duxbury, Pacific Grove: 2002. [Google Scholar]
- 20. Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. This paper develops the ANOVA framework to apply Cockerham’s approach to F-statistics and provides method-of-moments estimates for F-statistics.
- 21.Excoffier L. In: Handbook of Statistical Genetics. Balding DJ, Bishop M, Cannings V, editors. Chichester: John Wiley & Sons; 2001. pp. 271–307. [Google Scholar]
- 22.Cockerham CC. Analyses of gene frequencies. Genetics. 1973;74:679–700. doi: 10.1093/genetics/74.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger JO. Statistical Decision Theory and Bayesian Analysis. New York: Springer; 1985. [Google Scholar]
- 24.Robert CP. The Bayesian Choice: From Decision-Theoretic Foundations to Computational Implementation. New York: Springer; 2001. [Google Scholar]
- 25.Lee PM. Bayesian Statistics: An Introduction. London: Edward Arnold; 1989. [Google Scholar]
- 26.Gelfand AE, Smith AFM. Sampling-based approaches to calculating marginal densities. J. Am. Stat. Assoc. 1990;85:398–409. [Google Scholar]
- 27.Weir BS, Hill WG. Estimating F-statistics. Annu. Rev. Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 28.Wehrhahn C. Proceedings of the ecological genetics workshop. Genome. 1989;31:1098–1099. [Google Scholar]
- 29.Samanta S, Li YJ, Weir BS. Drawing inferences about the coancestry coefficient. Theor. Popul. Biol. 2009;75:312–319. doi: 10.1016/j.tpb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaggiotti OE, et al. Patterns of colonization in a metapopulation of grey seals. Nature. 2002;13:424–427. doi: 10.1038/416424a. [DOI] [PubMed] [Google Scholar]
- 31.Levsen ND, Crawford DJ, Archibald JK, Santos-Geurra A, Mort ME. Nei’s to Bayes’: comparing computational methods and genetic markers to estimate patterns of genetic variation in Tolpis (Asteraceae) Am. J. Bot. 2008;95:1466–1474. doi: 10.3732/ajb.0800091. [DOI] [PubMed] [Google Scholar]
- 32.Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 33. Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl Acad. Sci. USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. This article introduces GST as a measure of genetic differentiation among populations.
- 34. Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. This paper introduces ΦST and AMOVA for the analysis of haplotype data.
- 35. Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. This article introduces RST for the analysis of microsatellite data.
- 36.Rousset F. Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics. 1996;142:1357–1362. doi: 10.1093/genetics/142.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin M. Inbreeding coefficients and coalescence times. Genet. Res. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- 38.Holsinger KE, Mason-Gamer RJ. Hierarchical analysis of nucleotide diversity in geographically structured populations. Genetics. 1996;142:629–639. doi: 10.1093/genetics/142.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balloux F, Lugon-Molin N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- 40.Balloux F, Brunner F, Goudet J. Microsatellites can be misleading: an empirical and simulation study. Evolution. 2000;54:1414–1422. doi: 10.1111/j.0014-3820.2000.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 41.Gaggiotti OE, Lange O, Rassman K, Gliddon C. A comparison of two indirect methods for estimating average levels of gene flow using microsatellite data. Mol. Ecol. 1999;8:1513–1520. doi: 10.1046/j.1365-294x.1999.00730.x. [DOI] [PubMed] [Google Scholar]
- 42. Spitze K. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics. 1993;135:467–374. doi: 10.1093/genetics/135.2.367. This paper introduces QST for the analysis of continuously varying trait data.
- 43.Lande R. Neutral theory of quantitative genetic variance in an island model with local extinction and colonization. Evolution. 1992;46:381–389. doi: 10.1111/j.1558-5646.1992.tb02046.x. [DOI] [PubMed] [Google Scholar]
- 44.McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 2002;17:285–291. [Google Scholar]
- 45.O’Hara RB, Merila J. Bias and precision in QST estimates: problems and some solutions. Genetics. 2005;171:1331–1339. doi: 10.1534/genetics.105.044545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goudet J, Martin G. Under neutrality QST ≤ FST when there is dominance in an island model. Genetics. 2007;176:1371–1374. doi: 10.1534/genetics.106.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Notohara M. The coalescent and the genealogical process in geographically structured population. J. Math. Biol. 1990;29:59–75. doi: 10.1007/BF00173909. [DOI] [PubMed] [Google Scholar]
- 48.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nature Rev. Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 49.McCauley DE, Whitlock MC. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm + 1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- 50.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felsenstein J. How can we infer geography and history from gene frequencies? J. Theor. Biol. 1982;96:9–20. doi: 10.1016/0022-5193(82)90152-7. [DOI] [PubMed] [Google Scholar]
- 53.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 54.Beerli P. Comparison of Bayesian and maximum-likelihood estimation of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- 55.Kuhner MK. Coalescent genealogy samplers: windows into population history. Trends Ecol. Evol. 2009;24:86–93. doi: 10.1016/j.tree.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- 57.Fu R, Gelfand A, Holsinger KE. Exact moment calculations for genetic models with migration, mutation, and drift. Theor. Popul. Biol. 2003;63:231–243. doi: 10.1016/s0040-5809(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 58.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 59.Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B. 1996;263:1619–1626. [Google Scholar]
- 61.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 63.Mammès O, et al. Association of the G2548A polymorphism in the 5′ region of the LEP gene with overweight. Ann. Hum. Genet. 2000;64:391–394. doi: 10.1017/s0003480000008277. [DOI] [PubMed] [Google Scholar]
- 64.Balding DJ, Donnelly P. How convincing is DNA evidence? Nature. 1994;368:285–286. doi: 10.1038/368285a0. [DOI] [PubMed] [Google Scholar]
- 65.Balding DJ, Nichols RA. DNA match probability calculation: how to allow for population stratification, relatedness, database selection, and single bands. Forensic Sci. Int. 1994;64:125–140. doi: 10.1016/0379-0738(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 66.Council NR. The Evaluation of Forensic DNA Evidence. Washington DC: National Academy Press; 1996. [Google Scholar]
- 67.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 68.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 69.Pritchard JK, Donnelly P. Case–control studies of association in structured or admixed populations. Theor. Popul. Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 70.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kingman JFC. On the genealogy of large populations. J. Appl. Prob. 1982;19A:27–43. [Google Scholar]
- 72.Kingman JFC. The coalescent. Stoch. Proc. Appl. 1982;13:235–248. [Google Scholar]
- 73.Kuhner MK, Smith LP. Comparing likelihood and Bayesian coalescent estimation of population parameters. Genetics. 2007;175:155–165. doi: 10.1534/genetics.106.056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J. A coalescent-based estimator of admixture from DNA sequences. Genetics. 2006;173:1679–1692. doi: 10.1534/genetics.105.054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Innan H, Zhang K, Marjoram P, Tavare S, Rosenberg NA. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics. 2005;169:1763–1777. doi: 10.1534/genetics.104.032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wall JD, Hudson RR. Coalescent simulations and statistical tests of neutrality. Mol. Biol. Evol. 2001;18:1134–1135. doi: 10.1093/oxfordjournals.molbev.a003884. [DOI] [PubMed] [Google Scholar]
- 77.Nordborg M. Structured coalescent processes on different time scales. Genetics. 1997;146:1501–1514. doi: 10.1093/genetics/146.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donnelly P, Tavaré S. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 1995;29:401–421. doi: 10.1146/annurev.ge.29.120195.002153. [DOI] [PubMed] [Google Scholar]
- 79.Griffiths RC, Tavare S. Simulating probability distributions in the coalescent. Theor. Popul. Biol. 1994;46:131–159. [Google Scholar]
- 80.Fearnhead P, Donnelly P. Estimating recombination rates from population genetic data. Genetics. 2001;159:1299–1318. doi: 10.1093/genetics/159.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuhner MK, Beerli P, Yamato J, Felsenstein J. Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics. 2000;156:439–447. doi: 10.1093/genetics/156.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of recombination rates from population data. Genetics. 2000;156:1393–1401. doi: 10.1093/genetics/156.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhner MK, Felsenstein J. Sampling among haplotype resolutions in a coalescent-based genealogy sampler. Genet. Epidemiol. 2000;19(Suppl. 1):15–21. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 84.Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright S. An analysis of local variability of flower color in Linanthus parryae. Genetics. 1943;28:139–156. doi: 10.1093/genetics/28.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malécot G. The Mathematics of Heredity. San Francisco: W. H. Freeman; 1969. [Google Scholar]
- 89.Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B. 1996;351:1291–1298. [Google Scholar]
- 90.Hamrick JL. In: Isozymes in Plant Biology. Soltis DE, Soltis PS, editors. Portland: Dioscorides; 1989. pp. 87–105. [Google Scholar]
- 91.Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst. 1984;15:65–95. [Google Scholar]
- 92.Hamrick JL, Linhart YB, Mitton JB. Relationships between life history characteristics and electrophoretically detectable genetic variation in plants. Annu. Rev. Ecol. Syst. 1979;10:173–200. [Google Scholar]
- 93.Gottlieb LD. In: Progress in Phytochemistry. Reinhold L, Harborne JB, Swain T, editors. Vol. 7. Oxford: Pergamon; 1981. pp. 1–46. [Google Scholar]
- 94.Brown AHD. Enzyme polymorphism in plant populations. Theor. Popul. Biol. 1979;15:1–42. [Google Scholar]
- 95.International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He M, et al. Geographical affinities of the HapMap samples. PLoS ONE. 2009;4:e4684. doi: 10.1371/journal.pone.0004684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Balding DJ. Likelihood-based inference for genetic correlation coefficients. Theor. Popul. Biol. 2003;63:221–230. doi: 10.1016/s0040-5809(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 99.Foll M, Gaggiotti O. Identifying the environmental factors that determine the genetic structure of populations. Genetics. 2006;174:875–891. doi: 10.1534/genetics.106.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 102.Goudet J, Raymond M, de Meeus T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Workman PL, Niswander JD. Population studies on southwest Indian tribes. II. Local genetic differentiation in the Papago. Am. J. Hum. Genet. 1970;22:24–49. [PMC free article] [PubMed] [Google Scholar]
- 104.Holsinger KE. In: Hierarchical Modeling for the Environmental Sciences. Clark JS, Gelfand AE, editors. Oxford Univ. Press; 2006. pp. 25–37. [Google Scholar]
- 105.Holsinger KE. Analysis of genetic diversity in hierarchically structured populations: a Bayesian perspective. Hereditas. 1999;130:245–255. [Google Scholar]
- 106.Weir BS. The rarity of DNA profiles. Ann. Appl. Stat. 2007;1:358–370. doi: 10.1214/07-AOAS128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ritland KR. Joint maximum-likelihood estimation of genetic and mating system structure using open-pollinated progenies. Biometrics. 1986;42:25–43. [Google Scholar]
- 108.Thompson SL, Ritland K. A novel mating system analysis for modes of self-oriented mating applied to diploid and polyploid arctic Easter daisies (Townsendia hookeri) Heredity. 2006;97:119–126. doi: 10.1038/sj.hdy.6800844. [DOI] [PubMed] [Google Scholar]