Abstract
This protocol is to describe the methodology to characterize mitochondria DNA (mtDNA) heteroplasmy with parallel sequencing. Mitochondria play a very important role in important cellular functions. Each eukaryotic cell contains hundreds of mitochondria with hundreds of mitochondria genomes. The mutant mtDNA and the wild type may co-exist as heteroplasmy, and cause human disease. The purpose of this methodology is to simultaneously determine mtDNA sequence and to quantify the heteroplasmy level. The protocol includes two-fragment mitochondria genome DNA PCR amplification. The PCR product is then mixed at an equimolar ratio. The samples will be barcoded and sequenced with high-throughput next-generation sequencing technology. We found that this technology is highly sensitive, specific, and accurate in determining mtDNA mutations and the degree of heteroplasmic level.
Keywords: mitochondria, next-generation sequencing, heteroplasmy
INTRODUCTION
Mitochondria play a critical role in many fundamental cellular functions. First, mitochondria are the “powerhouses” of cells, which produce over 90% of the energy required by a cell through the process of oxidative phosphorylation (OXPHOS), an oxygen-requiring process that converts food to adenosine triphosphate (ATP) (Wallace 1992; Wallace 1992; Keeney, Xie et al. 2006; Wallace 2008). In this process, hydrogen derived from food is passed along the mitochondrial electron transport chain (ETC), generating ATP in the final step. In addition to producing ATP, mitochondria also generate reactive oxygen species (ROS) and participate in apoptosis and other important cellular functions.
Genetically, mitochondria are assembled with proteins encoded in the mitochondrial genome as well as the nuclear genome. An estimated 1,500 genes are required for mitochondrial assembly. Therefore, mitochondrial genetics is a combination of Mendelian autosomal dominant, autosomal recessive, X-linked, and maternal inheritance. In addition, mtDNA and mitochondria have the following major features: 1) maternal inheritance, 2) replicative segregation, 3) threshold expression, 4) high mutation rate, and 5) heteroplasmy. Because each cell has hundreds to thousands of mtDNAs, each cell can contain different proportions of mutant and normal (wild-type) mtDNAs. This condition is known as heteroplasmy. New mtDNA mutations also arise in cells, coexist with wild-type mtDNAs (heteroplasmy), and segregate randomly during cell division (Wallace 2007).
The human mitochondrial genome, which is a circular DNA molecule consisting of 16,569 base pairs (bp), encodes 13 polypeptides that are components of the ETC, as well as 22 tRNAs and two rRNAs that contribute to mitochondrial protein synthesis. A variety of human diseases are directly associated with mitochondrial DNA (mtDNA) mutations and hundreds of putative pathogenic mtDNA variants have been identified (Wallace 2007; Wallace 2008). Mutations in genes in the mitochondrial genome cause a large spectrum of clinical phenotypes.
In contrast to the nuclear genome, which is composed of two copies of each gene, eukaryotic cells have hundreds of mitochondria with hundreds of copies of mtDNA. Heteroplasmy is a condition in which wild type and mutant mitochondria genome co-exist. The degree of heteroplasmy is used to describe the ratio of the mutant mtDNA as compared to the wild type. It has been shown that many human diseases are caused by heteroplasmic mtDNA mutations. The degree of heteroplasmy also influences phenotype. The degree of heteroplasmic mtDNA mutations may vary significantly among different tissues, even in the same subject. Moreover, different percentages of mutant mtDNA can be associated with completely distinct clinical manifestations (Wallace 2008). It has been a challenge to identify all of the mutations in the mitochondrial genome and simultaneously quantify the mtDNA heteroplasmy levels. In addition to the molecular diagnosis of mitochondrial diseases, there is a rapidly growing need for methods of analyzing mtDNA variants for other applications, including evolutionary and forensic studies (Wallace 2005; Wallace 2005). Therefore, it is critical that mitochondrial genome sequences can be acquired and detected in a reliable, high-throughput, and cost-effective manner, especially in samples with clinically relevant levels of mtDNA heteroplasmy.
Currently, there are many mitochondrial genome-sequencing methods, including direct sequencing and the MitoChip. However, these methods are neither sensitive nor specific enough to detect mtDNA heteroplasmy (Hartmann, Thieme et al. 2009). Methods used for mitochondrial genome-wide heteroplasmic position screening include denaturing high-performance liquid chromatography (HPLC) (Meierhofer, Mayr et al. 2005), Surveyor Nuclease digestion (Bannwarth, Procaccio et al. 2005), and high-resolution melt (HRM) profiling (Dobrowolski, Gray et al. 2009). Although these methods can be used to detect mtDNA heteroplasmy, they cannot localize or quantify the heteroplasmic position(s). Several other techniques have been developed for the specific quantification of mtDNA heteroplasmy levels. These methods include polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis (Holt, Harding et al. 1990), allele-specific oligonucleotide dot-blot analysis (Liang, Johnson et al. 1998), real-time amplification refractory mutation system quantitative PCR (Bai and Wong 2004), and pyrosequencing (White, Durston et al. 2005). However, these methods can only be used to analyze a known mutation and are labor-intensive (Please see Unit 19.6 and/or 19.7).
Using parallel sequencing, we are able to identify mitochondrial genome-wide heteroplasmies, and quantify mtDNA heteroplasmy levels. The deep sampling inherent in the “next-generation” sequencing systems should enable the efficient detection of low-level DNA heteroplasmies and address this unmet need. Herein, we use the Illumina Genome Analyzer to re-sequence human mtDNA samples from two subjects that were combined at five different ratios of mtDNA (1:99, 5:95, 10:90, 20:80, and 50:50). The different high-throughput parallel sequencing platforms may have advantages and disadvantages. However, we expect other platforms will also work well. We assessed the sensitivity, specificity, and accuracy of this system. Our results showed that mtDNA heteroplasmies of ≥ 5% were detected 100%, with virtually no false positives, and that the estimates of mtDNA heteroplasmy levels were remarkably close to the theoretical values (correlation coefficient = 0.96). Therefore, parallel sequencing provides a simple, high-throughput, and cost-effective platform for mitochondrial genome sequencing with extraordinary sensitivity and specificity for mtDNA heteroplasmy detection.
BASIC PROTOCOL 1: PCR AMPLIFICATION OF mtDNA FOR ILLUMINA PAIRED-END SEQUENCING
This protocol was described previously and has been in use in the authors’ laboratory and others for routine analysis (Tang, Batra et al.; Tang and Huang). In principle, mtDNA is amplified by PCR amplification in two fragments with Takara PCR kits. The human mitochondrial genome is amplified in two overlapping fragments of 9,289 bp (Fragment I) and 7,626 bp (Fragment II) in length. The primer pair used for amplification of fragment I includes hmtF1 569 (5′-AACCAAACCCCAAAGACACC-3′) and hmtR1 9819 (5′-GCCAATAATGACGTGAAGTCC-3′), and the primer pair used for amplification of fragment II includes htmF2 9611 (5′-TCCCACTCCTAAACACATCC-3′) and hmtR2 626 (5′-TTTATGGGGTGATGTGAGCC-3′). PCR reactions are performed using TAKARA LA Taq polymerase. Sequencing is performed with Illumina GA II and data analyzed with NextGen software.
Equipment
Brinkmann Transferpette-8, 2.5–25 pL
Gilson Pipetman Rainin (Cat. P-20)
Fisherbrand Finnpipette 8 channel pipette 50–300 μl
Superfine Sephadex G-50
96-Well 45 μl MultiScreen Column Loader
MultiScreen 96-Well Filter Plates
96-Well Hydration Solution Collection Plate
Digital Balance PG802-S Mettler Toledo
Semi-Skirt 0.2 ml 96-Well PCR Plate E&K Scientific
Jouan GR412 Centrifuge
Savant SpeedVac Concentrator
Savant Refrigerated Condensation Trap
Savant OFP-400 Vacuum Pump
Thermal Cycler-Perkin Elmer (Cat. No. N801-0150)
Pipette tips and tubes
Materials
Genomic DNA isolated from blood was stored in Tris-EDTA buffer (TE, pH 7.4). Total genomic DNA was extracted from peripheral blood using the QIAamp DNA extraction kit (QIAGEN, Valencia, CA, http://www.qiagen.com/products/genomicdnastabilizationpurification/qiaampsystem/qiaampdnaminikit.aspx).
Water for PCR (Gibco DNAse, RNAse free, Ultra Pure)
Primers at 10 μM concentration
Deionized, distilled, RNAse free, DNAse free water (Gibco #10977-015, Milli-Q or equivalent).
-
Primers:
hmtF1 569 (5′-AACCAAACCCCAAAGACACC-3′)
hmtR1 9819 (5′-GCCAATAATGACGTGAAGTCC-3′)
htmF2 9611 (5′-TCCCACTCCTAAACACATCC-3′)
hmtR2 626 (5′-TTTATGGGGTGATGTGAGCC-3′)
100 mM 4dNTP mix (Roche; 25 mM each dNTP; also see APPENDIX 2D)
5 U/μl Takara Taq DNA polymerase (Takara)
10× PCR amplification buffer containing 15 mM MgCl2 (Roche)
Low-melting agarose (e.g., Fisher)
10× Tris/Borate/EDTA (TBE) buffer
10 mg/ml ethidium bromide
2× gel loading buffer
Additional reagents and equipment for PCR and agarose gel electrophoresis
mtDNA Amplification Reaction
-
1
Prepare PCR worksheet and check off ingredients as they are added to the mix. Prepare Master Mix in PCR hood. Make sure tubes are labeled appropriately (Table 1).
-
2
Remove two 46 μl aliquots of the above mixture for positive and negative controls. Add 2 μl of positive control DNA.
-
3
Add 1 μl of forward primer & 1 μl of reverse primer (@10 μM concentration) to each reaction. To facilitate this step, make a 1:1 mixture of forward and reverse primers in advance, so that 2 μl of the mixture can be added to each reaction (Table 2).
-
4
When PCR is completed, store labeled tubes at −20 °C.
Quality control: Always include a blank to confirm that there is no contamination during extraction & PCR amplification steps. A blank has all components of the reaction, but DNA is replaced by an equal volume of H2O
Table 1.
Components for PCR amplification (50 μl per reaction)
| Reagent | (1X) =Volume for 1 reaction | X 3 Reactions cocktail Per Patient (5 Reactions + 2 controls+ 1 extra) |
|---|---|---|
| PCR Water, Gibco | 39.5 μl | 120.5 μl |
| 10X PCR Buffer | 5.0 μl | 15.0 μl |
| dNTP, 10 mM, Takara | 1.0 μl | 3.0 μl |
| Forward Primer* @ 10 μM | 1.0 μl | n/a-add individually to reaction |
| Reverse Primer* @ 10 μM | 1.0 μl | n/a-add individually to reaction |
| Takara Taq, 5 U/μl | 0.5 μl | 1.5 |
| Total: | 48 μl |
Table 2.
Thermocycler program
| State | Temperature | Time |
|---|---|---|
| Denaturation | 95 °C | 2 min |
| Amplification (35 cycles) | 95 °C | 20 sec |
| 61 °C | 30 sec | |
| 68 °C | 10 min | |
| End of Amplification/Incubation | 68 °C | 20 min |
| 4 °C | ∞ |
Gel Analysis of PCR Product
-
5
Prepare a 1% agarose gel according to Table 3. Microwave the mixture of agarose and 1X TBE buffer until the agarose is dissolved (~1 min, in 30 sec increments).
-
6
Add ethidium bromide solution and swirl to mix. Pour gel into gel tray, with appropriate gel combs. Allow gel to solidify before removing combs.
-
7
Add 16 μl of amplicons from each PCR reaction to new tubes. Add 4 μl of 5X loading dye to each tube.
-
8
Load sample/dye mixture (10 μl) onto the gel. Also include a lane with 1kb ladder (5 μl of 1kb ladder + 5 μl of 2X loading dye) as a reference mark.
-
9
Perform electrophoresis at 150 V until the Orange G dye is at the bottom of the gel. This takes approximately 2 h. (To make 5X Orange G, dissolve 20g of Sucrose 100mg of Orange G in 40ml water and then add to total 50ml with water.
-
10
Use the lab gel documentation system, print out a picture of the gel and label all lanes appropriately.
Confirm that the PCR product results are clean single bands of the correct expected size.Quality Control: The PCR must be repeated when DNA appears in the reagent blank well. -
11
Measure the PCR product concentrations using a NanoDrop 2000 spectrophotometer (Thermo Scientific; see Appendix 3D).
-
12
Pool equimolar amounts of fragment I and II and use 500 ng of mtDNA as starting material for Illumina GA libraries.
-
13
Perform parallel DNA sequencing using the Illumina GA according to the manufacturer’s instructions (Illumina protocol: “Preparing Samples for Paired-End Sequencing,” Part# 1005063, http://qb3.berkeley.edu/gsl/Protocols_files/Paired-End_SamplePrep_Guide_1005063_B.pdf).
Briefly, samples are sheared to an average length of 150–200 bp using sonication. DNA fragment ends are repaired and phosphorylated using Klenow, T4 DNA Polymerase and T4 Polynucleotide Kinase. The resulting fragments are ligated to modified adapters that include 3 bp indexing tags. Following this “barcoding” step, samples are multiplexed at 16 samples per lane in the Illumina GA flowcell. See also Unit 18.4, Targeting Exon Sequencing by In-solution Hybrid Selection).
Table 3.
Example for Small Gel Electrophoresis Apparatus
| 1% Gel | Amount(Final Volume = 60 ml) |
|---|---|
| Agarose | 1 g |
| 1X TBE | 100 ml |
| 1% Ethidium Bromide | 5 μl |
BASIC PROTOCOL 2: ANALYSIS OF SEQUENCING DATA TO DETERMINE DEGREE OF HETEROPLASMY
Equipment
Computer minimum requirements: Windows 7, 64 bit, RAM 8GB, 2 CPUs.
NextGene Software is V2.1 to align to the genome. 6 GB of RAM or above to align to multiple mitochondrial genomes with 1 GB of data file. The more is required for assembly assemble larger data sets.
Process data
-
1
Process initial data and base calling, including extraction of cluster intensities, using real-time analysis (RTA) software 1.6.47.1 (SCS version 2.6.26).
-
2
Execute sequence quality filtering script using Illumina CASAVA software (ver 1.6.0, Illumina, Hayward, CA).
-
3
Produce quality metric data and analyze through html files, including the summary.html file provided with the data.
-
4
Examine the data quality using the perfect.html file, which shows the approximate proportion of sequences with 1, 2, 3 or 4 errors.
-
5
Run other quality metrics including intensity versus cycle (IVC) plots or visualization of cluster intensity over the duration of the sequencing run.
-
6
Use NextGENe (Softgenetics, State College, PA) to analyze the reads and align them against the revised Cambridge reference sequence (rCRS) of human mtDNA (Andrews, Kubacka et al. 1999).
-
7
The summary.html field “Lane Yield” is calculated as the number of sequence clusters that pass quality filters (PF, this value is also displayed as % PF Clusters) multiplied by the number of tiles per lane (120) multiplied by the number of sequencing cycles. (Note that the lane yield is displayed as kilobases of sequence. First cycle intensity and % intensity after 20 cycles are measurements of fluorophore intensity at cycles 1 and 20, respectively. Typical first cycle intensities are approximately 150 and % intensity after 20 cycles should be approximately 75%. Small variations in intensity are well tolerated, while larger variations can be used for troubleshooting purposes when required).
Variant and indel analysis
-
8
NextGENe software is used for next generation sequence alignment (URL: http://softgenetics.com/NextGENe.html Version 1.94).
-
9
NextGENe software is also used to generate two types of single nucleotide polymorphism (SNP) reports before (raw report) and after condensation (final report). Positions with SNP percentages less than 2% are removed from the final SNP report.
-
10
Perform the following for Format Conversion and Quality Filtering
Convert FASTQ files to FASTA format with the Format Conversion Tool
Quality Filtering to trim 3′ end at the position of <Q16 for 3 continuing bases.
Read Median Score ≥ 20
Paired End Data (Filtered Reads are Listed as “N” in Order to Preserve Pair Associations)
Alignment Settings (to human genome hg18, version 36)
Matching Base Number >= 12
Matching Base Percentage >= 85.0
Mutation Percentage Filter <= 2
Forward and reverse balance filter <=0.15.
Coverage Filter <= 7
Load Paired Reads
Min Pair End Gap:0
Max Pair End Gap:250
-
11
Mutation Report Note:
-
12
Reference Position: gives the numeric representation of the reference genome.
-
13
Reference Nucleotide: refers to the wildtype nucleotide at a certain position.
-
14
Coverage: refers to the total number of reads at that position.
-
15
Score is based on the concept of Phred scores, with a maximum value of 30, meaning the probability that the mutation call is wrong is 1/1000; 20, 1/100; and 10, 1/10. A low overall mutation score, however, does not mean that the mutation is more than likely a false mutation. The low score implies only that the mutation cannot be called a true mutation with absolute certainty.
-
16
Mutation call denotes the nucleotide change.
-
17
Example: T > TG indicates a heterozygous change from the reference nucleotide T to T and G.
-
18
Example: T > G indicates a homozygous change from the reference nucleotide T to G.
CAUTION: Ethidium bromide is toxic; wear gloves at all times.
COMMENTARY
Background Information
Disturbances in mitochondrial function have been implicated in a wide range of human diseases, including cancer (Park, Sharma et al. 2009), heart disease (Fan, Waymire et al. 2008), diabetes (Wallace), Alzheimer’s disease (Coskun, Beal et al. 2004), Parkinson’s disease (Wallace 2005) and hypertension (Wang, Li et al.). Diseases caused by mutations of the mitochondrial genome, also known as mtDNA disease, can result from either mtDNA base substitution mutations or rearrangement mutations. mtDNA base substitution mutations can either affect OXPHOS protein (mRNA) or protein synthesis (rRNA or tRNA) genes. Either way, the ultimate affect is to cause mitochondrial dysfunction. mtDNA are present in hundreds to thousands of copies per cell. Hence, mutant and normal mtDNAs segregate stochastically during cell division. These alternative genetic rules provide a more rational explanation of the diversity of mitochondrial diseases.
mtDNA mutations can affect the amino acid sequence of any of the mtDNA polypeptides. These mutations can range from very mild to severe, and present with diverse symptoms. The milder missense mutations are frequently close to homoplasmic in the proband and tend to be associated with more stereotyped phenotypes. By contrast, the more severe mutations tend to cause more variable phenotypes, in part due to variation in the percentage of mutant mtDNAs between individuals. The following are some common conditions associated with heteroplasmic mutations of mtDNA: A) Myoclonic epilepsy and ragged red fiber disease (MERRF) syndrome caused by the tRNALys np 8344 Mutation: This moderately severe protein synthesis mutation is virtually always heteroplasmic. Symptom severity is in proportion with the percentage of heteroplasmy. Most patients with more than 75% mutant mtDNA exhibit mitochondrial myopathy, and patients with 90–95% mutation frequently manifest myoclonic epilepsy. Other symptoms can include lactic acidosis, deafness, cardiomyopathy, and occasionally diabetes. B) Mitochondrial encephalomyopathy, lactic acid, and stroke-like episodes (MELAS) syndrome caused by the tRNALeu(UUR) np 3243 Mutation: This relatively severe mutation is always heteroplasmic. The severity of the symptoms is proportional to the percentage mutation, with most patients over 75% mutant exhibiting mitochondrial myopathy. Many of the most severe cases exhibit MELAS. Other symptoms of more severely affected patients can include cardiomyopathy, cardiac conduction defects, or short stature. C) Diabetes and Deafness caused by the tRNALeu(UUR) np 3243 Mutation: While high mutation percentages of the tRNALeu np 3243 mutation result in severe degenerative disease, 10 to 30 % of this same mutation results only in diabetes mellitus, frequently adult-onset (Type II), with or without deafness. D) Mitochondrial Myopathy caused by tRNALeu np 3303 Mutation: This relatively severe, rare mutation is always heteroplasmic and can be associated with mitochondrial myopathy, preferentially affecting the neck muscles. Characterization of human mitochondrial genome sequences is important for the molecular diagnosis of mitochondrial diseases, especially in samples with a low level of mitochondrial DNA (mtDNA) heteroplasmy which often missed by Sanger Sequencing.
The recently developed parallel sequencing method (Bentley 2006) has the capacity for massive-scale sequencing and offers a highly robust and less labor-intensive system for genome-wide scale sequencing. Currently, there are many next-generation sequencing platforms, including the Illumina Genome Analyzer (GA), the Roche 454 Genome Sequencer FLX system, the Applied Biosystems SOLiD system, and the Helicos True Single Molecule Sequencing system. The small size of the human mitochondrial genome and the resulting high coverage for each nucleotide position by parallel sequencing should enable the detection of low levels of mtDNA heteroplasmy. Previously, 454 sequencing was used to generate 34.9-fold coverage of mtDNA from ~0.3 g of bone from a 38,000 year-old Neanderthal individual (Green, Malaspinas et al. 2008). The Illumina GA, coupled with target microarray-based capture, was successfully employed to re-sequence the entire mitochondrial genome (coverage > 2,900 bp) and the exons of 362 nuclear genes encoding mitochondrial proteins (Vasta, Ng et al. 2009). However, neither of these studies investigated the capability of technologies used for heteroplasmy identification and quantification. In this protocol, we utilized Illumina GA to sequence the entire human mitochondrial genome and determined the sensitivity and specificity of this platform for the analysis of heteroplasmic mtDNA samples. In this protocol, we mainly focus on the detection of heteroplasmic mutations of the mitochondrial genome by next generation sequencing.
Critical Parameters
PCR Product
The human mitochondria are the product of symbiosis of nucleated cells and bacteria. Since the initial event three billion years ago, the bacteria have lost most of their genome. Some of the bacterial genes have been integrated into the nucleus and many of them remain as pseudogenes. Therefore, it is very important to test primers for the mitochondria genome to make sure that the PCR product is not derived from nuclear pseudogenes, but rather from true mtDNA. In this method, we have tested our primers with rho-0 cells that have no mitochondria genome as a negative control. Our results show that the two pairs of primers that use this protocol produce two distinct bands without background. In addition, no pseudogenes are amplified with this primer. Another consideration is the quality of the PCR product. It is very important to have no background without overloading the template, which may increase background.
In terms of coverage of the mitochondria genome, as parallel sequencing advances, the sequencing capacity has increased dramatically. With increased sequencing capacity, coverage becomes deeper and deeper. The sensitivity of detection of heteroplasmy is coverage-dependent. With Illumina GAII, we barcode 16 mitochondria genomes and get on average 1,700-fold coverage. Theoretically this gives high sensitivity to detect heteroplasmic mutations of mtDNA. With developments of new barcodes, we may be able to barcode more samples in a single run. However, minimal coverage will be 100-fold for any single nucleotide.
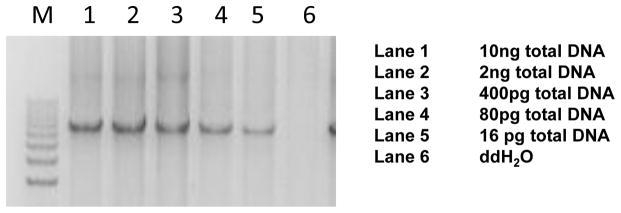
To test the sensitivity of this method, we performed a serial dilution of genomic DNA. We were able to obtain a clear PCR product at 16 pg, as shown in Figure 1, which allowed us to perform high quality parallel sequencing. Theoretically, if mitochondrial genomes number around 1,000 per cell, high quality DNA from one cell should be sufficient for this type of study.
Fig. 1. Serial deletion of genomic DNA to test PCR sensitivity.
Lane 1, 10 ng; lane 2, 2 ng; lane 3, 400 pg; lane 4, 80 pg; lane 5, 16 pg; lane 6, H20. Note that clear PCR results were obtained at the 16 pg dilution.
Level of mtDNA Heteroplasmy
Low-level mtDNA heteroplasmy detection and quantification based on the short reads from parallel sequencing requires powerful analysis tools capable of distinguishing between instrumental errors and true low frequency mutations. Different in silico algorithms can yield divergent results. We first used the DNASTAR algorithm to quantify the heteroplasmic variants and found that the estimated ratios deviated dramatically from the theoretical values and many false positives were annotated (data not shown). In contrast, NextGENe was highly specific and accurate for mtDNA heteroplasmy analysis. NextGENe employs a condensation tool to solve the three critical problems associated with parallel sequencing: short reads, high system error rates, and large volumes of data. Specifically, the condensation tool clusters similar short reads from Illumina GA, containing a unique anchor sequence. Therefore, data of adequate coverage are condensed, short reads are lengthened, and instrument errors are filtered from the analysis. The reads used for each condensed read are recorded to maintain allelic frequency information (Tang and Huang).
Anticipated Results
To test the sensitivity and specificity of parallel sequencing, we recently established an artificial system. In our recent publication, we selected two DNA samples (NS01 and NS09) from a group of subjects with known mitochondrial genome sequences. Pair-wise comparisons of the number of divergent nucleotides revealed that this pair had 56 divergent nucleotide positions throughout the mitochondrial genome, enabling both intra- and inter-genome comparisons of mtDNA heteroplasmy detection (Tang and Huang).
Depth of coverage by parallel sequencing of mtDNA
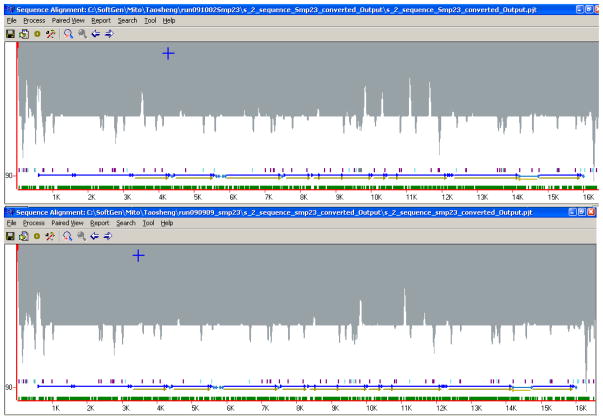
Using the barcoding protocol to pool 16 samples in the same lane, each variant nucleotide position was covered by 655 to 6,368 reads (average = 1,785; Figure 2). The fold coverage was fairly even across the mitochondrial genome with a few peaks and troughs. The pattern of the coverage map was very reproducible in replicate runs of the same samples (Fig. 2) and in different samples (data not shown).
Fig. 2. Coverage maps for 5% mixture sample in Test 1 (top) and Test 2 (bottom).
The X-axis represents the position of nucleotide on the mitochondrial genome (rCRS as reference genome, 16.568 kb), and the Y-axis stands for the fold of coverage for each nucleotide position.
Sensitivity of parallel sequencing for mtDNA heteroplasmy detection
To determine the detection threshold of parallel sequencing for mtDNA heteroplasmy, we generated mtDNA mixtures of NS01 and NS09 at five different ratios (ranging from 1% to 50%). In the 56 known variant positions between NS01 and NS09, we found that all heteroplasmies ≥ 5% were detected when using this method. The Condensation tool developed in the NextGENe software combines similar sequence reads to yield new consensus sequences. The consensus sequence gives more weight to the 5′ nucleotides and less weight to the 3′ nucleotides, so that sequence errors at the 3′ end can be reduced. Consensus sequences are longer than the original reads and the numbers of reads in both the forward and reverse directions that make up the consensus are recorded. Lower frequency SNPs can be determined from the original read number for each of the two heteroplasmy alleles determined by the two consensus sequences. The PCR amplification errors or PCR primer reads in the system often show up as imbalances, and these reads are eliminated from the consensus sequence. The white peaks pointing up in the shadow regions show the locations of the primers, while the grey peaks pointing down show locations of SNPs (Fig. 2).
Among the 56 variant positions, 54 were substitutions and the remaining two were insertions/deletions. Compared to rCRS, the NS01 sample has 523delA and 523delC. The Illumina GA system was able to detect this dinucleotide deletion down to the 5% level, and the estimated variant load was very close to the theoretical ratio.
For the 54 substitutions, all were called in the 5% test mixture, indicating that parallel sequencing is extremely sensitive to heteroplasmies of 5% and above (Tang and Huang). Theoretically, 1% mtDNA heteroplasmy is equivalent to 10 variant calls for a position with 1,000-fold coverage. With the 1,785-fold average coverage in the current study, the parallel sequencing method is expected to detect 1% mtDNA heteroplasmy. Indeed, in the raw report from the 1% test mixture, 49 out of 54 expected substitutions were identified. However, more than 100 possible false positive variants were also listed in the raw report from the 1% test mixture. Taken together, these observations suggest that the parallel sequencing system used was sensitive at 1%, but that the specificity was low at this level. As the parallel sequencing technology advances, the coverage will become deeper, and we expect that the sensitivity will further improve without sacrificing specificity. The clinical significance at <5% level of heteroplasmy is yet to determined.
Accuracy of parallel sequencing for mtDNA heteroplasmy detection
To determine the accuracy of the parallel sequencing system, we analyzed the detection levels of all 54 expected substitutions and plotted the observed estimates against theoretical values. We found that the observed mtDNA heteroplasmy ratios were very consistent with the theoretical levels, except for rCRS positions 9,596 and 16,086 (Tang and Huang). In addition, we found an excellent correlation between detected and theoretical levels at the different ratios (5:95, 10:90, 20:80, and 50:50) for individual positions, as shown by the almost identical pattern of scattered points for each test mixture (Fig. 2). The average detected mtDNA heteroplasmy level in each test mixture was nearly equal to the theoretical ratio, and the mean values from the different test mixtures exhibited a near-perfect linear regression (R2 = 0.9997) with a correlation coefficient of 0.96 for the predicted ratio (Tang and Huang).
Two outliers, rCRS positions 9,596 and 16,086, were associated with significantly lower detected mtDNA heteroplasmy levels than the theoretical values. However, there was no ambiguity in the alignment of the reads covering these two positions. It is possible that fragments with specific nucleotides were preferentially amplified in the GA library and resulted in the deviated ratio of the two mtSNPs.
Specificity and reproducibility of parallel sequencing for mtDNA heteroplasmy detection
While detection of all predicted variations is essential, a low number of false positives is also important. We found no false positives in test mixtures ≥ 5% in the final report. Such low noise levels in the machine-generated results did not require manual input to eliminate noise, which is especially crucial and advantageous for the application of this high-throughput platform in large-scale mtDNA sequencing projects. To determine the reproducibility of the system for mtDNA heteroplasmy analysis, replicate runs were carried out and analyzed for each test mixture. The results from the two replicate assays were highly reproducible, with the exception of rCRS position 9,596 in the 50% test mixture, demonstrating the superior reproducibility of the system(Tang and Huang).
Comparison with dideoxy-terminator sequencing and conventional PCR-RFLP methods
To compare the sensitivity of parallel sequencing to those of the widely-used PCR-RFLP and dye terminator sequencing methods for low-level mtDNA heteroplasmy detection, a mtDNA fragment (rCRS positions 15,991 – 626) was PCR amplified from the two different samples. The purified PCR products were mixed at different ratios and the mtDNA mixtures were digested with EcoRV, as one mtDNA has a C16278T transition, which creates an EcoRV recognition site. We found the sensitivity to be around 20%. The same PCR product mixtures were also directly sequenced with BigDye Terminator and the detection level for heteroplasmy was about 20%.
Time Considerations
The first step is to amplify mtDNA with 2 fragments. Depending on the starting material, the time can vary. Typically a blood sample is used. We will need a half day to isolate genomic DNA and another half day to complete the PCR amplification. Therefore, the first step of the experiment, PCR product amplification, is expected to be completed in the first day.
The second step of this protocol is to run parallel sequencing. This step includes fragmentation of the mitochondria genome, addition of adaptors, and also library preparation. Depending on the number of samples that will be run simultaneously, two to three days time is required for less than 12 samples. An additional week is required to run Illumina GAII. Generally, SNP calling is expected to be done in one or two days. The total protocol is expected to be completed in two to three weeks.
Acknowledgments
We would like to thank Dr Changsheng (Jonathan) Liu at SoftGenetics LLC for his input on data analysis and Mariella Simon for her critical reading this manuscript. This project is supported by the Darren J. Carroll Family. TH is also supported by the National Eye Institute (NEI 1R01EY018876).
Literature Cited
- Andrews RM, Kubacka I, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Bai RK, Wong LJ. Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: a single-step approach. Clin Chem. 2004;50(6):996–1001. doi: 10.1373/clinchem.2004.031153. [DOI] [PubMed] [Google Scholar]
- Bannwarth S, Procaccio V, et al. Surveyor Nuclease: a new strategy for a rapid identification of heteroplasmic mitochondrial DNA mutations in patients with respiratory chain defects. Hum Mutat. 2005;25(6):575–82. doi: 10.1002/humu.20177. [DOI] [PubMed] [Google Scholar]
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16(6):545–52. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Beal MF, et al. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101(29):10726–31. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski SF, Gray J, et al. Identifying sequence variants in the human mitochondrial genome using high-resolution melt (HRM) profiling. Hum Mutat. 2009;30(6):891–8. doi: 10.1002/humu.21003. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319(5865):958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Malaspinas AS, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134(3):416–26. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Thieme M, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30(1):115–22. doi: 10.1002/humu.20816. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, et al. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46(3):428–33. [PMC free article] [PubMed] [Google Scholar]
- Keeney PM, Xie J, et al. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci. 2006;26(19):5256–64. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang MH, Johnson DR, et al. Preparation and validation of PCR-generated positive controls for diagnostic dot blotting. Clin Chem. 1998;44(7):1578–9. [PubMed] [Google Scholar]
- Meierhofer D, Mayr JA, et al. Rapid screening of the entire mitochondrial DNA for low-level heteroplasmic mutations. Mitochondrion. 2005;5(4):282–96. doi: 10.1016/j.mito.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Park JS, Sharma LK, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18(9):1578–89. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Batra A, et al. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion. 10(4):350–7. doi: 10.1016/j.mito.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 48(4):287–96. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- Vasta V, Ng SB, et al. Next generation sequence analysis for mitochondrial disorders. Genome Med. 2009;1(10):100. doi: 10.1186/gm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 51(5):440–50. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256(5057):628–32. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–80. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria as chi. Genetics. 2008;179(2):727–35. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li R, et al. Maternally Inherited Essential Hypertension Is Associated With the Novel 4263A>G Mutation in the Mitochondrial tRNAIle Gene in a Large Han Chinese Family. Circ Res. 108(7):862–70. doi: 10.1161/CIRCRESAHA.110.231811. [DOI] [PubMed] [Google Scholar]
- White HE, V, Durston J, et al. Accurate detection and quantitation of heteroplasmic mitochondrial point mutations by pyrosequencing. Genet Test. 2005;9(3):190–9. doi: 10.1089/gte.2005.9.190. [DOI] [PubMed] [Google Scholar]