Abstract
Objectives
We aimed to determine the frequency of pH1N1 transmission between humans and swine on backyard farms in Tumbes, Peru.
Design
Two‐year serial cross‐sectional study comprising four sampling periods: March 2009 (pre‐pandemic), October 2009 (peak of the pandemic in Peru), April 2010 (1st post‐pandemic period), and October 2011 (2nd post‐pandemic period).
Sample
Backyard swine serum, tracheal swabs, and lung sample were collected during each sampling period.
Main outcome measures
We assessed current and past pH1N1 infection in swine through serological testing, virus culture, and RT‐PCR and compared the results with human incidence data from a population‐based active surveillance cohort study in Peru.
Results
Among 1303 swine sampled, the antibody prevalence to pH1N1 was 0% pre‐pandemic, 8% at the peak of the human pandemic (October 2009), and 24% in April 2010 and 1% in October 2011 (post‐pandemic sampling periods). Trends in swine seropositivity paralleled those seen in humans in Tumbes. The pH1N1 virus was isolated from three pigs during the peak of the pandemic. Phylogenetic analysis revealed that these viruses likely represent two separate human‐to‐swine transmission events in backyard farm settings.
Conclusions
Our findings suggest that human‐to‐swine pH1N1 transmission occurred during the pandemic among backyard farms in Peru, emphasizing the importance of interspecies transmission in backyard pig populations. Continued surveillance for influenza viruses in backyard farms is warranted.
Keywords: Antibodies, backyard pig farms, human–animal transmission, influenza
Introduction
The 2009 influenza pandemic emphasized the continued threat that zoonotic influenza viruses pose to global health. Avian and swine influenza viruses present a particular threat to humans because of their potential for reassortment and emergence of genetically novel strains.1 In particular, influenza viruses of swine origin have demonstrated their pandemic potential.2 The influenza A(H1N1)pdm09 virus (pH1N1) was first identified among humans in March 2009 and generated the first pandemic of the 21st century.3 Since 2009, pH1N1 has become endemic in human populations globally and there have been numerous reports of human‐to‐swine transmission.1, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 Most of these reports, however, have focused on larger scale industrial farms,4, 7, 11, 18 as opposed to smaller scale backyard farms, despite the fact that large numbers of pigs are raised in backyard settings, particularly in developing countries, providing considerable opportunity for influenza virus transmission between humans and livestock.21, 22
In Peru, are no data exist regarding influenza viruses among swine populations. Similar to many low‐ and middle‐income countries, the vast majority of swine (3·8 million, 80%) are found on backyard farms.23 We conducted a 2‐year serial cross‐sectional study of the prevalence of pH1N1 infection and past exposure among swine from community backyard farms in northern Peru. In addition, we were able to compare the data on swine with incidence data on human influenza from a population‐based active surveillance cohort study conducted at the same time in the region.24
Methods
Study settings and design
Tumbes, a region of approximately 210 000 people on the northern coast of Peru, is largely comprised of small semirural communities where pigs and poultry are commonly raised for personal consumption (Figure 1). We conducted a serial cross‐sectional study in Tumbes comprising four sampling periods: March 2009 (pre‐pandemic), October 2009 (peak of the pandemic in Peru), April 2010 (1st post‐pandemic period) and October 2011 (2nd post‐pandemic period) (Figure 2).25
Figure 1.
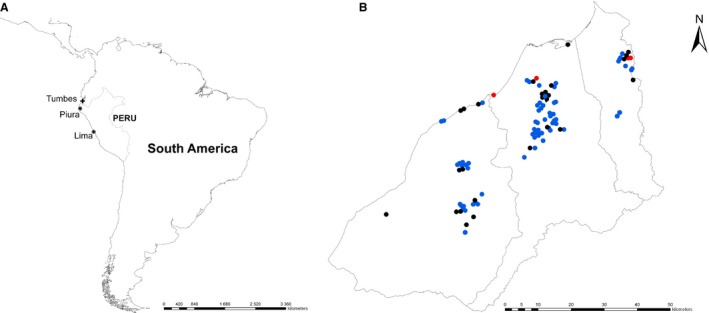
(A) Map of South America showing the study site region (indicated by a cross) and the geographic provenance of the three viruses (indicated by stars) which clustered most closely on phylogenetic analysis with the viruses obtained from swine in Tumbes. (B) Map of the Tumbes region, Peru, showing sites of backyard farms with pigs positive for antibody to influenza A(H1N1)pdm09 virus during the peak pandemic period (black circles), the 1st post‐pandemic period (blue circles), and during the 2nd post‐pandemic pandemic period (red circles).
Figure 2.
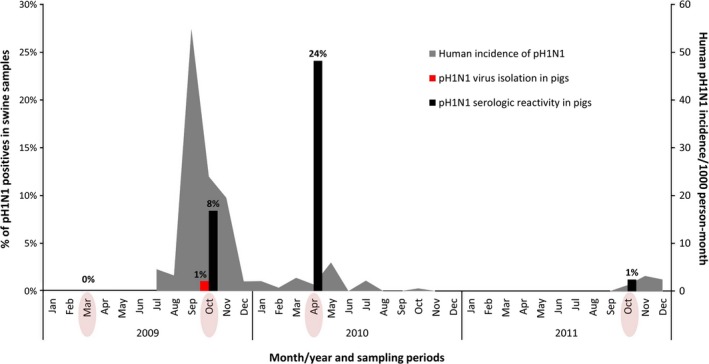
Antibody prevalence and isolation of influenza A(H1N1)pdm09 virus (pH1N1) from sera and tracheal swabs of backyard pigs collected in Tumbes, Peru, 2009–2011. The dates of the four sampling periods are circled on the x‐axis. The incidence of pH1N1 influenza in humans in Tumbes is shown in gray, obtained from surveillance through an influenza cohort study conducted by our research team which began on July 27, 2009.24 There was no evidence of pH1N1 influenza activity during November 2010 to September 2011. The first case of pH1N1 influenza in humans in Tumbes was reported by the Peruvian Ministry of Health on July 1, 2009.
Animals and sample collection
Serum samples for influenza virus testing were collected and made available by the Cysticercosis Elimination Program (CEP) in Peru.26 As part of the CEP's yearly surveillance activities in Tumbes, cysticercosis seronegative and seropositive backyard swine were identified by Western blot testing27 from sera collected previously and swine were purchased directly from their household owners at each time period as detailed previously.26 They were collected from 965 total backyard farms and transferred to the CEP facility where they were kept in pens separated by age and size for 1–5 days before being euthanized and slaughtered. A veterinarian evaluated the animals' general health condition and recorded daily observations. Swine slaughter was conducted under an animal protocol approved for the CEP study.26 Sampling and testing for influenza virus were approved by the U.S. Naval Medical Research Unit No. 6 (NAMRU‐6, Lima, Peru) Institutional Animal Care and Use Committee.
At the time of slaughter, in addition to the serum samples obtained, we collected samples of the respiratory tract (trachea and lungs) of pigs. Tracheal samples were obtained by vigorous swabbing of the tracheal wall followed by placement of swabs in viral transport medium.28 Lung samples were obtained by biopsying the cranial lobes of each lung, where lesions from pH1N1 infection are most frequent identified,29 and placing the tissues in cryovials. Respiratory tract samples were transported at approximately 4°C to the CEP in Tumbes between 30 and 180 minutes following collection where they were stored in liquid nitrogen until sent to the San Marcos University Veterinary School in Lima, Peru, for testing. All samples collected by the CEP were tested for influenza regardless of cysticercosis infection status.
Human influenza data
Data on incidence of human influenza were from a population‐based active surveillance cohort study begun in 2009 by NAMRU‐6 with support from the Peruvian Ministry of Health, the U.S. Centers for Disease Control and Prevention in Atlanta, GA, and the Armed Forces Health Surveillance Center in Silver Spring, MD.24, 30 In the study, over 2500 randomly selected households comprising more than 10 000 people in five sites in Peru, including approximately 450 households and 1800 people in Tumbes, were visited up to three times a week to screen household members for influenza‐like illness. Nasopharyngeal swabs were collected from identified cases and tested for influenza viruses by PCR. The study was approved by the NAMRU‐6 Institutional Review Board (NMRCD.2009.0005).
Laboratory methods
Hemagglutination inhibition
Sera were tested for antibodies to pH1N1 by hemagglutination inhibition (HI) assay as previously described31 using an influenza virus (A/swine/Peru/2010729235/2009) isolated from a pig in this study via culture in fertilized specific‐pathogen‐free (SPF) chicken eggs. Sera were pre‐treated with receptor‐destroying enzyme (Denka Seiken Co. Ltd., Tokyo, Japan) and heme‐absorbed with turkey erythrocytes. Titer results are reported as the reciprocal of the highest dilution of serum that inhibited virus‐induced hemagglutination of a 0·5% (v/v) solution of erythrocytes. The HI titer was expressed as the highest reciprocal serum dilution that completely inhibited hemagglutination of one hemagglutinin (HA) unit. We chose ≥1:10 as threshold of antibody titer to define seropositivity to pH1N1 infection, based on previous findings7 and the finding of absence of HI antibody against pH1N1 in the pre‐pandemic samples.
Virus isolation
Virus isolation was conducted by standard methods.32 Pools of five individual swabs were made according to date, sample type, and community. Pools were homogenized and filtered before inoculation into the allantoic cavity of five SPF 9‐day‐old embryonated chicken eggs. Eggs were incubated for 6 days with daily survival checks. Allantoic fluid of each egg was tested for hemagglutinating agents by direct HI. Negative pools were passaged a second time to confirm negativity. HI‐positive allantoic fluids were confirmed by antigen presence using the QuickVue Influenza Test (Quidel Corp., San Diego, CA, USA), after which each individual sample was cultured again following similar procedures.
Influenza virus PCR, gene sequencing, and analyses
For swine specimens, RNA extracts were prepared from 100 μl of allantoic fluid with the MagNA Pure Compact automated RNA extraction system (Roche Applied Science, Indianapolis, IN, USA). One‐step RT‐PCR (Invitrogen, Carlsbad, CA, USA) was used to amplify HA and neuraminidase (NA) genes with universal HA and NA oligonucleotide primers.33 Specimens from humans were tested for influenza virus by RT‐PCR using standard methods.34
Amplicons were purified by agarose gel electrophoresis followed by purification using the MinElute Gel Extraction Kit (QIAGEN, Valencia, CA, USA), and then sequenced on an automated Applied Biosystems 3730 system (Foster City, CA, USA) using cycle sequencing dye terminator chemistry. Ultimately, our study yielded five complete HA sequences (three from pigs in northern Peru, two from humans in northern Peru), five complete NA sequences (three from pigs, two from humans), five complete MP sequences (three from pigs, two from humans), and four complete whole‐genome sequences (two from pigs, two from humans). Whole‐genome sequences from pH1N1 viruses available on GenBank from the Western Hemisphere were included as background. Additional HA and NA sequences from swine in Latin America (in this case, Brazil and Colombia) were added, for which whole‐genome sequences were not available. Separate alignments for each of the eight genome segments were made using ClustalW (v1.83).35 As there was no evidence of reassortment between the HA and NA for our data, the HA and NA segments were concatenated36 for greater phylogenetic resolution and visual clarity in Figure 3. A maximum‐likelihood (ML) tree was inferred for each segment separately using RAxML v.7.2.6 (GTR+gamma substitution model), with statistical support for individual nodes assessed by bootstrap analysis (500 replicates). GenBank accession numbers of the sequences used in this analysis are listed in Appendix 2.
Figure 3.
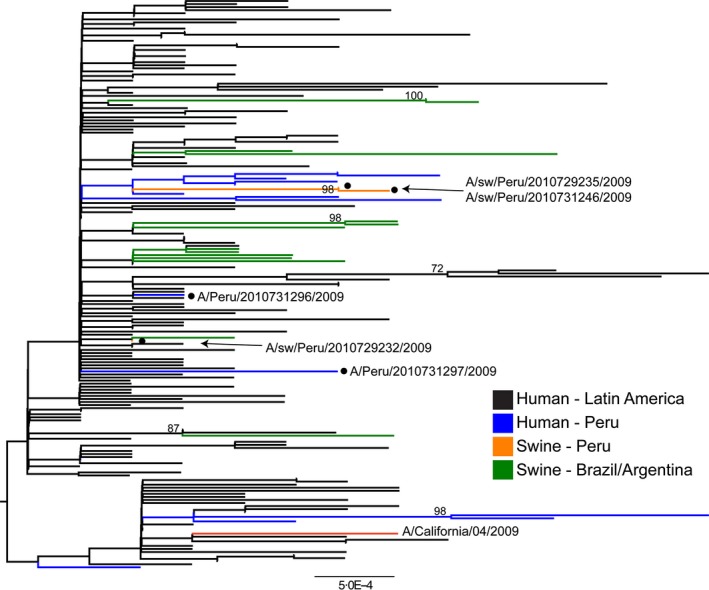
Maximum‐likelihood phylogeny of concatenated hemagglutinin and neuraminidase gene sequences from influenza A(H1N1)pdm09 viruses from Latin America, 2009–2011. Virus sequences shown were collected from three swine (orange with circles) and two humans in Tumbes (blue with circles) (see explanation in text), 13 swine in Argentina and Brazil (green), 14 humans in other sites in Peru (blue), 132 humans in other Latin American countries (black), and the red line: Influenza A/California/07/2009. Bootstrap values >70 are included for key nodes, and tree is midpoint rooted for clarity only. GenBank accession numbers of the sequences used in this analysis are listed in Appendix 2.
Statistical methods
The primary variable and outcome of interest was antibody to pH1N1 (≥1:10) assessed by the HI assay.7 We compared antibody prevalence between the 2009 and each other sampling periods using binary logistic regression adjusting for age and gender.
Results
Over the course of the study, we collected serum samples from 1303 backyard swine (310 pre‐pandemic, 322 peak of the pandemic, 328 1st post‐pandemic period, and 343 2nd post‐pandemic period). Tracheal and lung samples were available for all periods except for the 2nd post‐pandemic period. We collected 923 tracheal swabs and lung samples (310 pre‐pandemic, 288 peak of the pandemic, and 325 1st post‐pandemic period). None of the pigs showed symptoms of respiratory disease. The age of swine ranged from 2 to 60 months, and 75% were aged between 6 and 8 months (Table 1).
Table 1.
Demographic characteristics of swine sampled in Tumbes, Peru, 2009–2011
| Age, months | Sampling period | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre‐pandemic (March 2009) | Peak pandemic (October 2009) | 1st Post‐pandemic (April 2010) | 2nd Post‐pandemic (October 2011) | |||||||
| n = 310 | (%) | n = 322 | (%) | n = 328 | (%) | n = 343 | (%) | n = 1303 | (%) | |
| <6 | 154 | (50) | 0 | (0) | 0 | (0) | 64 | (19) | 218 | (17) |
| 6–8 | 54 | (17) | 322 | (100) | 328 | (100) | 279 | (81) | 983 | (75) |
| >8 | 102 | (33) | 0 | (0) | 0 | (0) | 0 | (0) | 102 | (8) |
| Sex | ||||||||||
| Male | 190 | (61) | 165 | (51) | 165 | (50) | 166 | (48) | 686 | (53) |
None of the 310 pre‐pandemic serum or respiratory samples was positive for antibodies against pH1N1 or pH1N1 virus (Table 2). The antibody prevalence of swine samples collected during the pandemic was 8% (27/322), 95% CI: 6–12, with pH1N1‐positive virus isolation and RT‐PCR testing from three tracheal swabs [1% (3/288), 95% CI: 0·2–3] and one lung sample [0·3% (1/288), 95% CI: 0–1·9], the latter from one of the animals with a positive tracheal swab (Figure 2). All three virus isolates were from pigs with HI antibody titers <1:10. Additionally, we also evaluated cutoff points of antibody titers <1:40 and observed similar trends, just a change in magnitude of positivity. Twenty‐four percent of samples (79/328, 95% CI: 20–29) were seropositive 6 months following the pandemic peak and 1·2% (4/343), 95% CI: 0·3–3, 2 years following the peak (Figure 2). Even though highest seroprevalence was reported during the 1st post‐pandemic period, antibody titers were highest during the pandemic period (i.e., 48% of positive swine (13/27) had HI titers of 80–160) and decreased progressively during the two post‐pandemic samplings [17% (13/79) and 0% (0/4), respectively] (Table 2).
Table 2.
Hemagglutination titers against influenza A(H1N1)pdm09 virus in pig sera in Tumbes, Peru, 2009–2011
| HI titera | No. of sera positive (%) by period | |||
|---|---|---|---|---|
| Pre‐pandemic (March 2009) | Peak pandemic (October 2009) | Post‐pandemic 1 (April 2010) | Post‐pandemic 2 (October 2011) | |
| 10 | 0 (0) | 3 (11) | 16 (20) | 1 (25) |
| 20–40 | 0 (0) | 10 (37) | 50 (63) | 3 (75) |
| 80–160 | 0 (0) | 13 (48) | 13 (17) | 0 (0) |
| 640 | 0 (0) | 1 (4) | 0 (0) | 0 (0) |
| Total | 0 (0) | 27 (100) | 79 (100) | 4 (100) |
The limit of detection for the hemagglutination assay was set to ≥1:10.
The HA and NA gene nucleotide sequences from the three pH1N1‐positive animals were ≥99% homologous compared with those of viruses identified during the same time period among humans in Tumbes (Appendix 1). Phylogenetic analysis of the HA and NA segments, however, indicated that the three swine viruses collected in our study and two viruses collected from humans in Tumbes in October 2009 were not monophyletic, indicative of more than one human‐to‐swine introduction (Figure 3). Two of the pH1N1 viruses found in swine (A/swine/Peru/2010729235/2009 and A/swine/Peru/2010731246/2009) clustered together with high bootstrap support on the NA tree (98%), suggesting that they could represent a single human‐to‐swine transmission event with onward transmission in swine (the viruses were collected 2 days apart: October 17, 2009, and October 19, 2009). These viruses were most closely related to human pH1N1 viruses collected from humans from our study in Tumbes.
Despite being closely related phylogenetically, A/swine/Peru/2010729235/2009 and A/swine/Peru/2010731246/2009 were collected on farms in different small communities of Tumbes (approximately 18·5 km apart) (Figure 1). This pair of swine viruses therefore represents one of three scenarios: long‐distance swine–swine transmission of a single viral introduction of human origin, two separate introductions of very similar pH1N1 viruses from humans, or viral transmission within the swine holding facility prior to sampling. The possibility of two separate human‐to‐swine transmissions cannot be ruled out due to the low number of background viruses available from humans in the Tumbes region. The third swine virus (A/swine/Peru/2010729232/2009), although collected at nearly the same time (October 15, 2009), is positioned in a separate part of the tree and likely represents another separate human‐to‐swine introduction within northern Peru (Figure 3).
Discussion
Our 2‐year cross‐sectional study revealed a high rate of infection of backyard swine in Tumbes, Peru, with pH1N1 viruses of human origin, with a peak of 24% seropositive during April 2010. We found phylogenetic evidence for two likely viral introductions into swine in Tumbes, as well as limited swine‐to‐swine transmission, although it is not clear whether this occurred in the backyard farm setting or subsequently in the swine holding pen. It would be of great interest to determine how extensively pH1N1 spread swine‐to‐swine within these backyard farm settings, or whether the virus mainly was reseeded in pigs via human introductions with little onward transmission in swine. However, such fine‐scale transmission dynamics will require further surveillance and sequencing.
During 2009–2011, pH1N1 infection in pigs on backyard farms closely paralleled pH1N1 activity in humans living in the same communities. The pH1N1 virus was introduced into the swine population within 3 months of the first reported human case in the region by the Peruvian Ministry of Health on July 1, 2009. Similar time lags between human and swine infection have been noted in studies of humans and swine on backyard farms in Vietnam, Sri Lanka, and Cameroon.5, 8, 14 The increase in antibody prevalence (24%) in swine during the first post‐pandemic period, in which most pigs sampled were 6–8 months old, likely represents human‐to‐swine infections that occurred during the peak of the human influenza pandemic and possibly limited onward transmission of pH1N1 in swine. Antibodies against classical H1 swine influenza virus in pigs can last for more than 1 year after primary infection,37 and we report the majority of HI titers in pigs were <1:80, 8 months after the peak pandemic period, indicating a decay in antibodies consistent with prior exposure. Additionally, although transfer of maternal antibody typically lasts 1–2 months in pigs, it could also account for some seropositive animals.38, 39
By the second post‐pandemic period (October 2011), the antibody prevalence in swine had fallen to 1%, indicating that pH1N1 had not become endemic in the swine population in Tumbes. The small number of pigs that remained positive likely represents some latent immunity among a small proportion of swine population or occasional transmission from swine or humans infected with pH1N1 during a period with very low incidence of pH1N1 in humans. Interestingly, the 1% antibody prevalence in swine corresponds to a slight increase in pH1N1 influenza in humans during the second post‐pandemic period at this time (Figure 2).
Our study reports the introduction of human pH1N1 virus into a low‐density backyard swine population in a rural community where influenza viruses have not been previously detected in swine. Our low rate of viral isolation does not allow us to determine whether the majority of swine infections were the result of contact with pH1N1‐infected humans or transmission between swine, although our findings do suggest at least limited transmission in backyard swine. The fact that most of these animals were raised in separate small backyard farms until days before slaughter indicates that influenza viruses can still infect swine in settings with low animal contact rates owing to high contact with humans.
The high incidence of human pH1N1 influenza (up to 55/1000 person‐months) during the peak pandemic period likely afforded ample opportunity for human‐to‐swine transmission. Although experimental infections in the laboratory confirm the potential for pH1N1 virus shedding and contact transmission between swine,20, 40 most field investigations suggest that sustained transmission among swine is limited in small‐scale farm settings5, 14 although, as infection is often subclinical or associated with mild disease,4, 9 it may go undetected.40 Consistent with this, none of the swine in our study were noted to be sick prior to slaughter and no abnormalities were noted on gross anatomic examination of the lungs of the three virus‐positive animals. The primary impediment to virus transmission between swine in backyard farms is likely to be more situational than biological, that is, a relatively small number of pigs are raised together, with limited opportunity for contact with animals from other household farms before slaughter. The typically brief period of time that animals are brought together for slaughter is probably inconsequential to transmission between swine, although the potential for swine‐to‐human infection remains.
The fact that the three pH1N1 viruses found in swine in Tumbes were not genetically identical to those found in humans in the region at the same time reflects how rapidly influenza virus disseminates through communities, such that it is difficult to infer the spatial origins of the direct progenitors of the swine viruses detected in our study. Furthermore, the comingling of swine prior to slaughter undermines the ability to assess the geographic origins of all three swine isolates. The most parsimonious explanation is that the swine in our study acquired pH1N1 viruses directly from humans or during close contact with other pigs within the CEP facility pens. However, we cannot rule out the possibility of longer distance transmission. Further isolation and sequencing of viruses from both swine and humans in our study would be required to further delineate routes of transmission.
Limitations
We used a relatively low HI titer (1:10) to define pH1N1 seropositivity. Nevertheless, as none of the pre‐pandemic period samples were positive at this cutoff, we believe it to be valid.7 Furthermore, although the magnitude of the prevalence noted at each time period would change if a higher cutoff point were used, the noted trends and general conclusions from our study would still be the same (data not shown). Our study was focused specifically on pH1N1 in swine and humans. Our diagnostic approach was therefore focused on this virus and would not have detected evidence of other influenza virus infection in swine. Virus culture was performed only in eggs, limiting our ability to isolate viruses that grow better in cell culture. We did not attempt direct detection by PCR before virus isolation. This diagnostic approach and the lack of tissue samples in the 2nd post‐pandemic period may explain the low number of viruses isolated. We were able to perform whole‐genome sequencing on only two of the viruses from swine and two from humans collected in our study, and there were limited pH1N1 sequence data available from swine in Latin America in GenBank. Furthermore, given the low genetic diversity of the newly emerged pH1N1 virus,1 phylogenies of individual segments had a low resolution and relatively few bootstrap values >70, limiting the inference of detailed transmission dynamics. Finally, this study consisted of four serial cross‐sectional samplings, and therefore, it provided prevalence per each period that may have differed if other periods have been evaluated.41
Backyard farms provide ample opportunity for contact between humans, swine, wild and domestic birds, and a range of other animals.22, 23 This setting would thus seem to be especially suitable for the circulation of diverse strains of human as well as zoonotic influenza viruses, with the potential for virus coinfection and production of reassortants. Indeed, numerous reassortants between pH1N1 and other influenza viruses have been reported.10, 16, 17, 18, 19, 20, 42, 43, 44 Although the small number of animals on backyard farms and limited transference of animals between farms may limit the potential for virus transmission between swine, continued intensive surveillance for influenza virus reassortants in backyard farms is warranted.
Addendum
Marc‐Alain Widdowson and John Barnes, U.S. Centers for Disease Control and Prevention, Atlanta, GA, U.S.A., involved in interpretation of the data and revising the intellectual content. Hugo Razuri and Maria C. Guezala, U.S. Naval Medical Research Unit No. 6, Lima, Peru, involved in interpretation of the data and critical writing of the manuscript.
Disclosures
The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of the Army, Department of Defense, Centers for Disease Control and Prevention, nor the U.S. Government. Several authors of this manuscript are military service members or employees of the U.S. government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.' Title 17 U.S.C. §101 defines a U.S. government work as a work prepared by a military service member or employee of the U.S. government as part of that person's official duties. The authors declare they have no competing interests.
Supporting information
Figure S1. Maximum‐likelihood phylogeny of PB2 gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S2. Maximum‐likelihood phylogeny of PB1 gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S3. Maximum‐likelihood phylogeny of PA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S4. Maximum‐likelihood phylogeny of HA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S5. Maximum‐likelihood phylogeny of NP gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S6. Maximum‐likelihood phylogeny of NA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S7. Maximum‐likelihood phylogeny of MP gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S8. Maximum‐likelihood phylogeny of NS gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011. Virus sequences shown were collected from swine (blue with asterisk) and humans in Tumbes (black with asterisk), swine strains from the Western Hemisphere (red), and human strains (black) (see explanation in text). Bootstrap values >70 are included for key nodes, and tree is midpoint rooted for clarity only. GenBank accession numbers of the sequences used in this analysis can be found at http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi.
Acknowledgements
We thank Hugo Garcia and Ayvar Viterbo from CEP in Peru and Maria Silva, Karen Segovia, and Bruno Ghersi from NAMRU‐6, for their valuable assistance. Work performed in this study was supported by funding from the Bill and Melinda Gates Foundation (23981), the Centers for Disease Control and Prevention, the Armed Forces Health Surveillance Center Global Emerging Infections Surveillance and Response System, the NIH/NSF ‘Ecology and Evolution of Infectious Diseases' award from the Fogarty International Center at the U.S. National Institutes of Health (3R01‐TW005869), and a training grant from the Fogarty International Center (D43‐TW001140)002E.
Appendix 1. Genetic relationship (based on percent nucleotide homogeneity of hemagglutinin and neuraminidase genes) between influenza A(H1N1)pdm09 viruses isolated from swine and humans in Tumbes, Peru, 2009–2011
| Strain | A/sw/Peru/2010729235/2009 (swine) | A/sw/Peru/2010731246/2009 (swine) | A/sw/Peru/2010729232/2009 (swine) | A/Peru/2010731296/2009 (human) | A/Peru/2010731297/2009 (human) |
|---|---|---|---|---|---|
| Hemagglutinin | |||||
| A/sw/Peru/2010729235/2009 (swine) | 100·0 | 99·9 | 99·8 | 99·8 | 99·6 |
| A/sw/Peru/2010731246/2009 (swine) | 99·9 | 100·0 | 99·7 | 99·7 | 99·5 |
| A/sw/Peru/2010729232/2009 (swine) | 99·8 | 99·7 | 100·0 | 99·9 | 99·7 |
| A/Peru/2010731296/2009 (human) | 99·8 | 99·7 | 99·9 | 100·0 | 99·7 |
| A/Peru/2010731297/2009 (human) | 99·6 | 99·5 | 99·7 | 99·7 | 100·0 |
| Neuraminidase | |||||
| A/sw/Peru/2010729235/2009 (swine) | 100·0 | 100·0 | 99·8 | 99·7 | 99·7 |
| A/sw/Peru/2010731246/2009(swine) | 100·0 | 100·0 | 99·8 | 99·7 | 99·7 |
| A/sw/Peru/2010729232/2009 (swine) | 99·8 | 99·8 | 100·0 | 99·9 | 99·9 |
| A/Peru/2010731296/2009 (human) | 99·7 | 99·7 | 99·9 | 100·0 | 99·9 |
| A/Peru/2010731297/2009 (human) | 99·7 | 99·7 | 99·9 | 99·9 | 100·0 |
Appendix 2. Influenza virus strains and GenBank accession numbers used to create the phylogenetic tree in Figure 3
| N | Accession | Strain | Host | Country |
|---|---|---|---|---|
| 1 | KM289084 | A/swine/Peru/2010729235/2009 (H1N1) | Swine | Peru |
| 2 | KM289087 | A/swine/Peru/2010729235/2009 (H1N1) | Swine | Peru |
| 3 | KM289085 | A/swine/Peru/2010731246/2009 (H1N1) | Swine | Peru |
| 4 | KM289088 | A/swine/Peru/2010731246/2009 (H1N1) | Swine | Peru |
| 5 | KM289086 | A/swine/Peru/2010729232/2009 (H1N1) | Swine | Peru |
| 6 | KM289089 | A/swine/Peru/2010729232/2009 (H1N1) | Swine | Peru |
| 7 | KM289090 | A/Peru/2010731296/2009(H1N1) | Human | Peru |
| 8 | KM289092 | A/Peru/2010731296/2009(H1N1) | Human | Peru |
| 9 | KM289091 | A/Peru/2010731297/2009(H1N1) | Human | Peru |
| 10 | KM289093 | A/Peru/2010731297/2009(H1N1) | Human | Peru |
| 11 | CY044256 | A/swine/Argentina/SAGiles‐31215/2009(H1N1) | Swine | Argentina |
| 12 | JQ666845 | A/swine/Brazil/1/2009(H1N1) | Swine | Brazil |
| 13 | JQ666855 | A/swine/Brazil/11/2009(H1N1) | Swine | Brazil |
| 14 | JQ666856 | A/swine/Brazil/12/2009(H1N1) | Swine | Brazil |
| 15 | JQ666857 | A/swine/Brazil/13/2009(H1N1) | Swine | Brazil |
| 16 | JQ666860 | A/swine/Brazil/16/2009(H1N1) | Swine | Brazil |
| 17 | JQ666861 | A/swine/Brazil/17/2009(H1N1) | Swine | Brazil |
| 18 | JQ666846 | A/swine/Brazil/2/2009(H1N1) | Swine | Brazil |
| 19 | JQ666847 | A/swine/Brazil/3/2009(H1N1) | Swine | Brazil |
| 20 | JQ666848 | A/swine/Brazil/4/2009(H1N1) | Swine | Brazil |
| 21 | JQ666849 | A/swine/Brazil/5/2009(H1N1) | Swine | Brazil |
| 22 | JQ666850 | A/swine/Brazil/6/2009(H1N1) | Swine | Brazil |
| 23 | JQ666851 | A/swine/Brazil/7/2009(H1N1) | Swine | Brazil |
| 24 | HM569666 | A/Argentina/07‐09GP/2009(H1N1) | Human | Argentina |
| 25 | HM569674 | A/Argentina/08AR/2009(H1N1) | Human | Argentina |
| 26 | HM569682 | A/Argentina/19527/2009(H1N1) | Human | Argentina |
| 27 | HM569690 | A/Argentina/19618/2009(H1N1) | Human | Argentina |
| 28 | HM569698 | A/Argentina/19656/2009(H1N1) | Human | Argentina |
| 29 | CY047776 | A/Argentina/7649/2009(H1N1) | Human | Argentina |
| 30 | CY047784 | A/Argentina/7785/2009(H1N1)/ | Human | Argentina |
| 31 | CY047808 | A/Argentina/7967/2009(H1N1) | Human | Argentina |
| 32 | CY047800 | A/Argentina/7953/2009(H1N1) | Human | Argentina |
| 33 | CY047816 | A/Argentina/7980/2009(H1N1) | Human | Argentina |
| 34 | CY047824 | A/Argentina/8019/2009(H1N1) | Human | Argentina |
| 35 | CY047840 | A/Argentina/8574/2009(H1N1) | Human | Argentina |
| 36 | CY047848 | A/Argentina/8673/2009(H1N1) | Human | Argentina |
| 37 | CY047856 | A/Argentina/8989/2009(H1N1) | Human | Argentina |
| 38 | CY047864 | A/Argentina/8994/2009(H1N1) | Human | Argentina |
| 39 | CY047872 | A/Argentina/9004/2009(H1N1) | Human | Argentina |
| 40 | CY047880 | A/Argentina/9180/2009(H1N1) | Human | Argentina |
| 41 | CY047888 | A/Argentina/9333/2009(H1N1) | Human | Argentina |
| 42 | CY047896 | A/Argentina/9384/2009(H1N1) | Human | Argentina |
| 43 | CY047920 | A/Argentina/9579/2009(H1N1) | Human | Argentina |
| 44 | CY047952 | A/Argentina/9705/2009(H1N1) | Human | Argentina |
| 45 | CY047968 | A/Argentina/9721/2009(H1N1) | Human | Argentina |
| 46 | CY083085 | A/Bogota/WRAIR0442T/2009(H1N1) | Human | Colombia |
| 47 | CY120722 | A/Brazil/AVS03/2009(H1N1) | Human | Brazil |
| 48 | CY120730 | A/Brazil/AVS04/2009(H1N1) | Human | Brazil |
| 49 | CY120770 | A/Brazil/AVS05/2009(H1N1) | Human | Brazil |
| 50 | CY120738 | A/Brazil/AVS06/2009(H1N1) | Human | Brazil |
| 51 | CY120746 | A/Brazil/AVS07/2009(H1N1) | Human | Brazil |
| 52 | CY075162 | A/Chile/158/2009(H1N1) | Human | Chile |
| 53 | CY075178 | A/Chile/1586/2009(H1N1) | Human | Chile |
| 54 | CY075186 | A/Chile/1598/2009(H1N1) | Human | Chile |
| 55 | CY075194 | A/Chile/1599/2009(H1N1) | Human | Chile |
Tinoco et al (2016) Transmission dynamics of pandemic influenza A(H1N1)pdm09 virus in humans and swine in backyard farms in Tumbes, Peru. Influenza and Other Respiratory Viruses 10(1), 47–56.
References
- 1. Nelson MI, Gramer MR, Vincent AL, Holmes EC. Global transmission of influenza viruses from humans to swine. J Gen Virol 2012; 93(Pt 10):2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vijaykrishna D, Smith GJ, Pybus OG et al Long‐term evolution and transmission dynamics of swine influenza A virus. Nature 2011; 473:519–522. [DOI] [PubMed] [Google Scholar]
- 3. Dawood FS, Jain S, Finelli L et al Emergence of a novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 4. Pasma T, Joseph T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect Dis 2010; 16:706–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trevennec K, Leger L, Lyazrhi F et al Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009–2010. Influenza Other Respir Viruses 2012; 6:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim SH, Moon OK, Lee KK et al Outbreak of pandemic influenza (H1N1) 2009 in pigs in Korea. Vet Rec 2011; 169:155. [DOI] [PubMed] [Google Scholar]
- 7. Moreno A, Di Trani L, Alborali L et al First pandemic H1N1 outbreak from a pig farm in Italy. Open Virol J 2010; 4:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Njabo KY, Fuller TL, Chasar A et al Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet Microbiol 2012; 156:189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forgie SE, Keenliside J, Wilkinson C et al Swine outbreak of pandemic influenza A virus on a Canadian research farm supports human‐to‐swine transmission. Clin Infect Dis 2011; 52:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitikoon P, Sreta D, Tuanudom R et al Serological evidence of pig‐to‐human influenza virus transmission on Thai swine farms. Vet Microbiol 2011; 148:413–418. [DOI] [PubMed] [Google Scholar]
- 11. Sreta D, Tantawet S, Na Ayudhya SN et al Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg Infect Dis 2010; 16:1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song MS, Lee JH, Pascua PN et al Evidence of human‐to‐swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J Clin Microbiol 2010; 48:3204–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Howden KJ, Brockhoff EJ, Caya FD et al An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J 2009; 50:1153–1161. [PMC free article] [PubMed] [Google Scholar]
- 14. Perera HK, Wickramasinghe G, Cheung CL et al Swine influenza in Sri Lanka. Emerg Infect Dis 2013; 19:481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pereda A, Cappuccio J, Quiroga MA et al Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg Infect Dis 2010; 16:304–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ducatez MF, Hause B, Stigger‐Rosser E et al Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 2011; 17:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howard WA, Essen SC, Strugnell BW et al Reassortant pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg Infect Dis 2011; 17:1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Starick E, Lange E, Fereidouni S et al Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J Gen Virol 2011; 92(Pt 5):1184–1188. [DOI] [PubMed] [Google Scholar]
- 19. Vijaykrishna D, Poon LL, Zhu HC et al Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 2010; 328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu H, Zhou B, Fan X et al Novel reassortment of Eurasian avian‐like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J Virol 2011; 85:10432–10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karlsson E, Ciuoderis KA, Freiden P et al Prevalence and characterization of influenza viruses in diverse species in Los Llanos, Colombia. Emerg Microbes Infect 2013; 2:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCune S, Arriola CS, Gilman RH et al Interspecies interactions and potential influenza A virus risk in small swine farms in Peru. BMC Infect Dis 2012; 12:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ministerio de Agricultura P. Sector Pecuario – Situación Porcina. Lima‐Peru 2014. Available at http://www.minag.gob.pe/portal/sector-agrario/pecuaria/situacion-de-las-actividades-de-crianza-y-produccion/porcinos (Accessed 9 October 2014).
- 24. Razuri H, Romero C, Tinoco Y et al Population‐based active surveillance cohort studies for influenza: lessons from Peru. Bull World Health Organ 2012; 90:318–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chowell G, Viboud C, Munayco CV et al Spatial and temporal characteristics of the 2009 A/H1N1 influenza pandemic in Peru. PLoS One 2011; 6:e21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia HH, Gonzalez AE, Rodriguez S et al [Epidemiology and control of cysticercosis in Peru]. Rev Peru Med Exp Salud Publica 2010; 27:592–597. [DOI] [PubMed] [Google Scholar]
- 27. Gilman RH, Gonzalez AE, Llanos‐Zavalaga F, Tsang VC, Garcia HH. Prevention and control of Taenia solium taeniasis/cysticercosis in Peru. Pathog Glob Health 2012; 106:312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. WHO . WHO Manual on Animal Influenza Diagnosis and Surveillance. Geneva: World Health Organization, Response DoCDSa, 2002. [Google Scholar]
- 29. Valheim M, Gamlem H, Gjerset B, Germundsson A, Lium B. Pathological findings and distribution of pandemic influenza A (H1N1) 2009 virus in lungs from naturally infected fattening pigs in Norway. Influenza Res Treat 2011; 2011:565787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tinoco Y, Razuri H, Ortiz EJ et al Preliminary population‐based epidemiological and clinical data on 2009 pandemic H1N1 influenza A (pH1N1) from Lima, Peru. Influenza Other Respir Viruses 2009; 3:253–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. CDC . Serologic detection of human influenza virus infections by hemagglutination‐inhibition assay using turkey RBCs [SOP]. SOP No LP‐003, 2009; 20.
- 32. Health WOfA . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds, and Bees). Paris: Health WOfA, 2008. [Google Scholar]
- 33. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full‐length amplification of all influenza A viruses. Arch Virol 2001; 146:2275–2289. [DOI] [PubMed] [Google Scholar]
- 34. WHO . CDC protocol of realtime RTPCR for influenza A(H1N1) 2009 updated (6 October 2009); 28 April 2009: Available at http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf (Accessed 15 November 2014).
- 35. Chenna R, Sugawara H, Koike T et al Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 2003; 31:3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson M, Spiro D, Wentworth D et al The early diversification of influenza A/H1N1pdm. PLoS Curr 2009; 1:RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Renshaw HW. Influence of antibody‐mediated immune suppression on clinical, viral, and immune responses to swine influenza infection. Am J Vet Res 1975; 36:5–13. [PubMed] [Google Scholar]
- 38. Fleck RBA. Evaluation of a maternal antibody decay curve for H1N1 swine influenza virus using the hemagglutination inhibition and the IDEXX ELISA tests. American Association of Swine Veterinarians.
- 39. Thacker B, editor. Vaccination strategies for swine influenza virus. Keeping pace with SIV: Proceedings from the Allen D Leman Swine Conference; 2000. Minneapolis, MN.
- 40. Lange E, Kalthoff D, Blohm U et al Pathogenesis and transmission of the novel swine‐origin influenza virus A/H1N1 after experimental infection of pigs. J Gen Virol 2009; 90(Pt 9):2119–2123. [DOI] [PubMed] [Google Scholar]
- 41. Levin KA. Study design III: cross‐sectional studies. Evid Based Dent 2006; 7:24–25. [DOI] [PubMed] [Google Scholar]
- 42. Pereda A, Rimondi A, Cappuccio J et al Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir Viruses 2011; 5:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiapponi C, Baioni L, Luppi A, Moreno A, Castellan A, Foni E. Full‐genome sequence of a reassortant H1N1 swine influenza virus isolated from pigs in Italy. Genome Announc 2013; 1:e00778–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fan X, Zhu H, Zhou B et al Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J Virol 2012; 86:2375–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum‐likelihood phylogeny of PB2 gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S2. Maximum‐likelihood phylogeny of PB1 gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S3. Maximum‐likelihood phylogeny of PA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S4. Maximum‐likelihood phylogeny of HA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S5. Maximum‐likelihood phylogeny of NP gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S6. Maximum‐likelihood phylogeny of NA gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S7. Maximum‐likelihood phylogeny of MP gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011.
Figure S8. Maximum‐likelihood phylogeny of NS gene sequence from A(H1N1)pdm09 viruses from Western Hemisphere, 2009–2011. Virus sequences shown were collected from swine (blue with asterisk) and humans in Tumbes (black with asterisk), swine strains from the Western Hemisphere (red), and human strains (black) (see explanation in text). Bootstrap values >70 are included for key nodes, and tree is midpoint rooted for clarity only. GenBank accession numbers of the sequences used in this analysis can be found at http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi.


