Significance
Yakutia is among the coldest regions in the Northern Hemisphere, showing ∼40% of its territory above the Arctic Circle. Native horses are particularly adapted to this environment, with body sizes and thick winter coats minimizing heat loss. We sequenced complete genomes of two ancient and nine present-day Yakutian horses to elucidate their evolutionary origins. We find that the contemporary population descends from domestic livestock, likely brought by early horse-riders who settled in the region a few centuries ago. The metabolic, anatomical, and physiological adaptations of these horses therefore emerged on very short evolutionary time scales. We show the relative importance of regulatory changes in the adaptive process and identify genes independently selected in cold-adapted human populations and woolly mammoths.
Keywords: ancient genomics, adaptation, population discontinuity, regulatory changes, horse
Abstract
Yakutia, Sakha Republic, in the Siberian Far East, represents one of the coldest places on Earth, with winter record temperatures dropping below −70 °C. Nevertheless, Yakutian horses survive all year round in the open air due to striking phenotypic adaptations, including compact body conformations, extremely hairy winter coats, and acute seasonal differences in metabolic activities. The evolutionary origins of Yakutian horses and the genetic basis of their adaptations remain, however, contentious. Here, we present the complete genomes of nine present-day Yakutian horses and two ancient specimens dating from the early 19th century and ∼5,200 y ago. By comparing these genomes with the genomes of two Late Pleistocene, 27 domesticated, and three wild Przewalski’s horses, we find that contemporary Yakutian horses do not descend from the native horses that populated the region until the mid-Holocene, but were most likely introduced following the migration of the Yakut people a few centuries ago. Thus, they represent one of the fastest cases of adaptation to the extreme temperatures of the Arctic. We find cis-regulatory mutations to have contributed more than nonsynonymous changes to their adaptation, likely due to the comparatively limited standing variation within gene bodies at the time the population was founded. Genes involved in hair development, body size, and metabolic and hormone signaling pathways represent an essential part of the Yakutian horse adaptive genetic toolkit. Finally, we find evidence for convergent evolution with native human populations and woolly mammoths, suggesting that only a few evolutionary strategies are compatible with survival in extremely cold environments.
Yakutia (Sakha Republic, Russian Federation) is the coldest country in the whole Northern Hemisphere, showing annual thermal amplitudes over 100 °C and its entire range covered by permafrost (1). Despite such extreme conditions, a group of Turkic-speaking horse-riders, likely originating from the Altai-Sayan and/or Baïkal area, migrated into this region between the 13th and 15th centuries, pressed by the expansion of Mongolic tribes (2–4). The Yakut people successfully developed a unique economy based on horse and cattle breeding, with Yakutian horses mostly exploited as sources of meat and milk.
The Yakutian horse is the most northerly distributed horse on the planet and certainly the most resistant to cold. In contrast to cattle, which are kept in stables during winter, horses live in the open air all year round, grazing on vegetation that is buried under deep snow cover for 7–8 mo (5). Yakutian horses exhibit unique morphoanatomical adaptations to the subarctic climate, being extraordinarily hairy and, as predicted by Allen’s rule, compactly built, with short limbs. Their metabolic needs are in phase with seasonal conditions, because they accumulate important fat reserves during the extremely brief period of vegetation growth and lower their activities during winter (6). Yakutian horses also show an increased carbohydrate metabolism in the spring, likely supporting higher energy expenditure and fetal growth (7).
The evolutionary origins of Yakutian horses, however, still remain contentious. The most commonly accepted hypothesis proposes that they descend from a founding population brought by the Yakut people (2, 4). Some authors have suggested that at the time of their arrival, founding horses admixed with local populations descending from wild Late Pleistocene horses (8–10). In contrast, others have proposed that modern Yakutian horses exclusively originate from native Late Pleistocene populations, and were secondarily domesticated by the Yakut people upon their arrival (11).
To date, the genomic diversity of Yakutian horses is mostly unknown, and genetic analyses are limited to few microsatellites and mtDNA sequences (12–14). In this study, we have sequenced and analyzed the complete genomes of 11 Yakutian horses, including nine present-day horses and two ancient specimens that lived in the region ∼5,200 y ago and in the early 19th century. The results revealed a genetic discontinuity between the Pleistocene and mid-Holocene genomic landscapes, showing that the contemporary population fully descends from a stock of domesticated horses. Taken together, these findings support the predominant hypothesis that the Yakut people introduced the breed in the 13th–15th centuries, and implies that the unique genetic adaptations of Yakutian horses, which we reveal here for the first time to our knowledge, were selected on an extremely short evolutionary time scale.
Results and Discussion
Genome Sequencing of Ancient Yakutian Horses.
We extracted DNA from nine horse fossil specimens excavated in Yakutia and spanning a Late Pleistocene to historical temporal range (SI Appendix, Table S1.2). We constructed and sequenced low-depth Illumina DNA libraries to identify extracts showing DNA preservation levels compatible with a cost-effective genome characterization. For most of the specimens, we found a predominance of microbial sequences closely resembling soil microbial communities (SI Appendix, section S3). However, two specimens showed relatively large fractions of high-quality hits against the horse reference genome EquCab2.0 (15) (38.2–43.4% after quality filtering) and were sequenced further to 18.3-fold and 20.2-fold mean coverage (SI Appendix, Table S2.6). The first specimen, hereafter referred to as Batagai, was excavated near Verkhoyansk (Fig. 1A), and radiocarbon dated to 4,450 ± 35 uncalibrated radiocarbon years [2,939–3,337 calibrated years B.C. (i.e., ∼5,200 y ago)]. The second specimen (CGG101397) was excavated in the same region as part of French archeological expeditions (MAFSO 2008), within an autochthonous burial from the early 19th century. The depth of coverage of the X chromosome, which was approximately half the depth of coverage of autosomes (SI Appendix, Table S2.6), indicated that the two specimens were stallions. Sequencing data were also compatible with the retrieval of the complete mitochondrial genome of two additional ancient specimens (ODJ6 from the 18th century and the ∼5,450-y-old Yukagir).
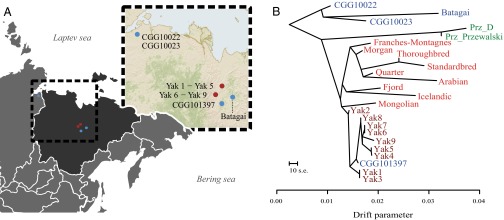
Fig. 1.
Geographic distribution and population affinities. (A) Geographic location of ancient (blue) and modern (dark red) Yakutian horse samples (SI Appendix, Table S1.1). (B) TreeMix relationships between modern Yakutian horses (brown), ancient horses (blue), Przewalski's horses (green), and our panel of nine domestic breeds (red).
The size distribution of the Batagai and CGG101397 DNA inserts showed a strong ∼10-bp periodicity (SI Appendix, Fig. S2.1), likely reflecting nucleosome protection (16), as observed for other permafrost-preserved (17, 18) and temperate samples (19). Additionally, typical signatures of postmortem DNA damage were found, with DNA library types based on blunt-end ligation (20) showing decreasing C→T misincorporation rates from read starts paralleled by increasing G→A toward read ends (SI Appendix, Figs. S2.3 and S2.5). This pattern was also observed for TruSeq DNA libraries (Illumina), together with a range of other misincorporation types toward read ends, likely resulting from palindromic artifacts (21). We thus trimmed read ends to limit their impact on downstream analyses and used mapDamage2 (22) to downscale the quality scores of bases inferred to be damage-driven nucleotide misincorporations, which resulted in final estimates of sequencing error rates of 0.0735% and 0.0404% errors per base for the Batagai and CGG101397 genomes, respectively (SI Appendix, Fig. S2.9).
Genome Sequencing of Present-Day Yakutian Horses.
We collected hair samples from 12 living animals originating from distant locations in Yakutia, stretching from the Yana River to the Kolyma River (SI Appendix, section S2.3). Hair DNA extracts were compatible with complete genome sequencing at ninefold to 21.6-fold mean coverage for nine individuals from the Eveno-Bytantaj district (Yak1–Yak9; Fig. 1A). With error rates ranging between 0.0386% and 0.0694%, the quality of these genomes is similar to the quality of our ancient genomes (SI Appendix, Fig. S2.9).
Population Genetic Discontinuity from Mid-Holocene to Present Times.
We first compared the 16 complete mitochondrial sequences characterized here (SI Appendix, section S5.1) with complete mitochondrial sequences from 27 ancient and 78 present-day horses (14, 18, 23–28). Maximum likelihood (ML) and Bayesian phylogenetic inference showed that the Yakutian mitochondrial haplotypes of specimens Yak1–Yak7, Yak9, and CGG101397 are closely related to the mitochondrial haplotypes of domesticated breeds (SI Appendix, Figs. S5.1 and S5.2). In contrast, specimen Yak8 clustered within a now-extinct group of Late Pleistocene horses. Finally, the mitochondrial haplotypes of the Batagai and Yukagir specimens (∼5,200 and 5,450 y old, respectively) grouped together within a cluster absent from all living horses hitherto characterized, and thus possibly extinct.
We took advantage of the presence of nine stallions in our genome dataset to analyze the diversity present in the Y chromosome. We found that modern Yakutian horses display four haplotypes closely related to the haplotypes of domesticated horses and the CGG101397 historical specimen (SI Appendix, Fig. S5.4). This close relationship indicates paternal continuity since the early 19th century and that diverse stallions were incorporated into the makeup of present-day Yakutian horses, in contrast to previous observations in other domesticated horse breeds (29, 30). Interestingly, the mid-Holocene specimen Batagai presented a haplotype not described in any present-day horse but closely related to the haplotype of an ∼16,000-y-old stallion from the Taymir Peninsula (Russia; specimen CGG10023) (18), which not only suggests paternal continuity between ancient Yakutian and Taymir populations but also the later extinction and/or replacement of ancient local males by domestic patrilineages.
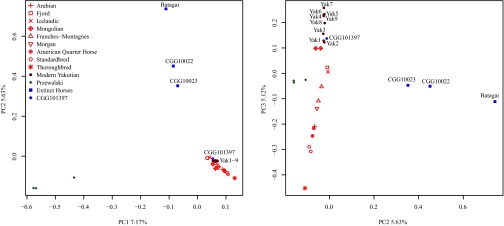
An exome-based ML tree confirmed the phylogenetic affinities of present-day Yakutian horses and modern domesticated breeds (represented here by Arabian, Franches-Montagnes, Icelandic, Mongolian, Morgan, Norwegian Fjord, American Quarter Horse, and Standardbred breeds) (15, 18, 27, 31, 32) (SI Appendix, section S5.3 and Fig. S5.5). Additionally, contemporary Przewalski’s horses represent a distinct group, separated from a third clade consisting of the two Late Pleistocene horses (CGG10022 and CGG10023) (18) and the ∼5,200-y-old Yakutian sample Batagai (100% bootstrap support). The same population affinities were recovered in TreeMix using 356,720 high-quality SNPs (Fig. 1B) and in principal component analysis (PCA) (Fig. 2 and SI Appendix, Fig. S6.7). The genetic affinities between the Taymir Peninsula and Yakutia possibly reflect an ancient population surviving in Yakutia at least until ∼5,200 y ago and stretching over thousands of kilometers across the Holarctic region.
Fig. 2.
PCA. The analysis was based on genotype likelihoods and 29 genomes representative of present-day Yakutian horses, nine domestic breeds, the Przewalski’s horse population, CGG101397, and extinct horses. The fraction of the total variance explained by each of the three principal components is indicated on the corresponding axes.
Projections (33) of ancient genomes onto the Yakutian horse genome panel allowed further characterization of the genetic continuity of Yakutian horses in the past ∼5,200 y (SI Appendix, section S6.2). Projection values (w) lower than 1 indicate that the derived alleles are less frequent in the test genome, and thus that they have further diverged from the Yakutian horse population. Projection of the CGG101397 genome onto the panel of modern Yakutian horses approaches the w = 1 line, suggesting the former as part of the same population background (SI Appendix, Fig. S6.6). The specimens Batagai, CGG10022, and CGG10023 tested against the same panel show remarkably lower genome projections across the whole frequency spectrum of derived mutations. Their minimum projection values (MPVs) were significantly lower (MPV < 0.6067) than the MPVs of CGG101397 (MPV = 0.8719) and Przewalski’s horse genomes (mean MPV = 0.6926, SD = 0.0009), confirming phylogenetic results where Batagai (and Late Pleistocene horses) appeared more distantly related to contemporary Yakutian horses than to Przewalski’s horses (Fig. 1B). Taken together, these results support that present-day Yakutian horses were introduced later than ∼5,200 y ago and earlier than the 19th century, likely following the migration of the Yakut people in the region around the 13th–15th centuries (2, 4).
Demographic History of Yakutian Horses.
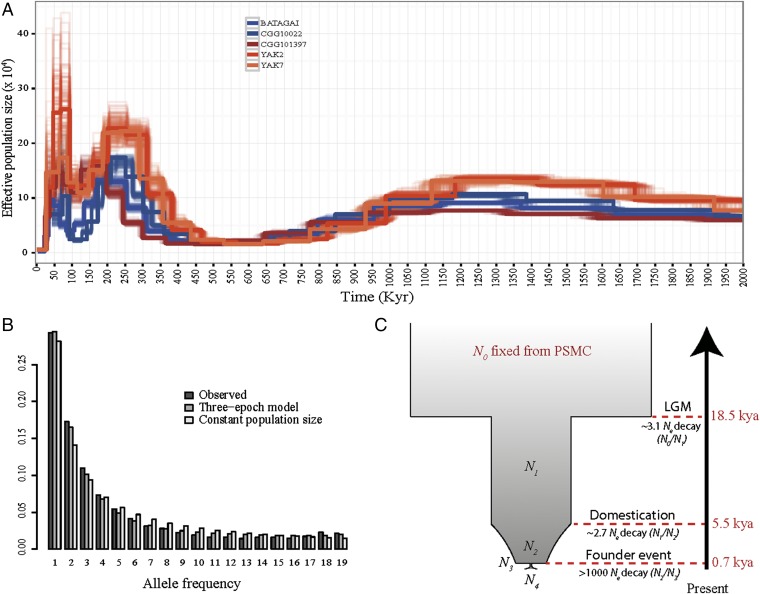
We next investigated the demographic trajectory of Yakutian horses. The pairwise sequential Markov coalescent (PSMC) profile of contemporary Yakutian horses was similar to the PSMC profile of CGG101397 and to the profiles previously reconstructed for domesticated horses (18, 27). PSMC trajectories of Late Pleistocene and domesticated horses diverge ∼160,000 y ago, in agreement with the split times previously inferred for both populations (Fig. 3A). However, they exhibit a common dramatic decline in effective population size (Ne) ∼20,000 y ago, likely reflecting the major climatic change from the end of the Late Pleistocene (34). Because the statistical power of PSMC profiling is reduced toward recent times (35), we applied the diffusion approximation implemented in ∂a∂i to gain insights on the recent demography of Yakutian horses (SI Appendix, section S6.1.2). Results confirmed a steady decline ∼20,000 y ago and an exponential decay following the onset of domestication (Fig. 3C). Furthermore, the inclusion of a severe population bottleneck within the past thousand years significantly increased the model likelihood (SI Appendix, Table S6.2). This population bottleneck, and the extremely short branches found in TreeMix (Fig. 1B), represents the genomic signature of the founder event associated with the migration of the Yakut people.
Fig. 3.
Long- and short-term Ne changes of the Yakutian horse population. (A) Scaled PSMC profile tracking effective population size changes over the past 2 Mya. (B) Site frequency spectra estimated from the present-day population of Yakutian horses (including CGG101397), expected under the neutral model, and fitted under the best demographic scenario (three-epoch model). (C) Three-epoch demographic model for the Yakutian horse population. This model represents the best fit in ∂a∂i analyses. Kyr, thousand years; LGM, Last Glacial Maximum.
Despite the compelling evidence pointing to a recent origin of modern Yakutian horses, it is unclear whether the native population of horses was already extinct by the time of the arrival of the Yakut people. Interestingly, we found Late Pleistocene horses less inbred and more heterozygous than Batagai [inbreeding coverage = 0.083 vs. 0.000–0.020 and mean log(θw) (Watterson theta estimator) = 3.159 vs. 3.677–3.950, respectively; SI Appendix, Figs. S4.2–S4.4]. Together with the PSMC profile, these estimates may indicate that the now-extinct population of ancient Yakutian horses was already declining ∼5,200 y ago. Even so, this population could still have survived by the time of the arrival of Yakut people, partially introgressing genetic material into the present-day variability, as possibly suggested by the mitochondrial haplotype of the Yak8 specimen, which was closely related to the specimens of Late Pleistocene horses.
We therefore tested this introgression hypothesis using a combination of analyses, including admixture tests based on f3-statistics (36) and D-statistics (37). None of the f3-statistics in the form of (modern Yakutian; Batagai, domesticated) was significant (f3-statistics ≥ 3.23e−5, Z-score ≥ 0.086), providing no evidence for the genome of present-day Yakutian horses representing a merger from Batagai (or Late Pleistocene horses) and domesticated horse population backgrounds (SI Appendix, section S6.4). Using D-statistics and the following tree topology (outgroup, Batagai; modern Yakutian, domesticated), the specimen Batagai did not appear closer to modern Yakutian horses than to any other modern domesticated horse (SI Appendix, section S6.4.1 and Fig. S6.9), ruling out possible introgression of Batagai alleles in contemporary Yakutian populations. Interestingly, similar outcomes were obtained when Batagai was replaced by any of the two Late Pleistocene horses representing the same population background. However, the tree topology could be rejected when replacing Batagai with CGG101397 (Z-score ≥ 4.40), suggesting that specimen CGG101397 is closer to contemporary Yakutian horses than to other domesticated horses. The latter finding further supports genetic continuity from the early 19th century to the present day and suggests that the introduction of exogenous alleles from a range of domesticated breeds that was attempted during the Soviet period (5) massively failed.
Genetic Adaptations of Yakutian Horses.
Yakutian horses developed striking physiological and morphological adaptations to subarctic environments following their recent arrival in the area. This demographic context provides a unique opportunity to investigate the genetic basis and evolutionary mechanisms underlying these traits, including the relative contribution of coding and noncoding mutations to fast-evolving adaptations.
We estimated the fixation index of genetic differentiation (FST) within 50-kb sliding windows (step size of 10 kb) by comparing the genomes of modern Yakutian horses (YAK; including the closely related historical CGG101397 specimen; SI Appendix, section S7) against two different panels of domesticated genomes (SI Appendix, Tables S7.1 and S7.2). The first panel included all 27 domesticated horses across nine breeds (DOM), which alleviates possible breed-specific selective processes. The second panel included horses from the same breed [12 Franches-Montagnes (FM) horses], which minimizes the impact of the genetic structure within domesticated breeds on FST. We applied different approaches to identify FST outlier regions, reporting from 251 to 1,489 genes potentially undergoing positive selection (SI Appendix, Tables S7.2 and S7.3), whose biological significance is discussed below (SI Appendix, Tables S7.4–S7.7). We noticed that the FST outlier windows were depleted of protein-coding genes (SI Appendix, Fig. S7.3), suggesting a long-term effect of negative selection in reducing genetic differentiation, as well as ongoing adaptive pressures in noncoding, potentially regulatory regions.
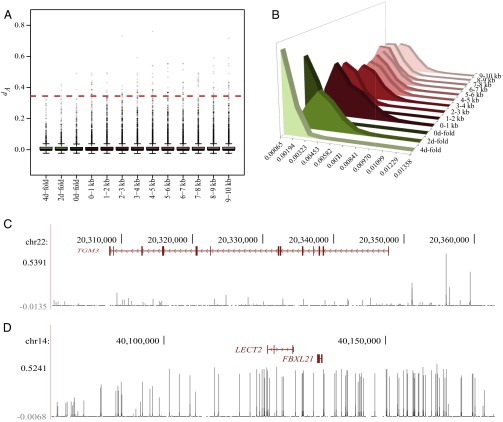
We further investigated the role played by noncoding regions at a finer scale, stratifying sites into different coding (i.e., 0d-fold, 2d-fold, 4d-fold degenerate coding positions) and noncoding [10 first 1-kb bins upstream of the translation start site (TSS)] categories. Because FST is not defined for monomorphic positions, we calculated the average net number of nucleotide differences (dA) between populations as a proxy for their differentiation (38). We used the dA distribution of 4d-fold sites to delineate a threshold for positions evolving under neutrality (SI Appendix, section S7.2). Nucleotide positions exceeding this threshold thus represent key adaptive candidates (Fig. 4A). We found that the proportion of adaptive candidates was not homogeneously distributed across all site categories but was enriched in noncoding positions, especially within the most proximal kilobase upstream of the TSS, where regulatory elements often concentrate (Fig. 4B). A similar pattern is observed for the YAK-DOM population pair (SI Appendix, Figs. S7.4 and S7.5), supporting a major role for transcriptional evolution in driving the adaptation of Yakutian horses to their extreme environment. Our results contribute to ongoing debates in the field of evolutionary biology regarding the adaptive potential of cis-regulatory elements and protein-coding regions (39–41). Cis-regulatory regions have indeed been proposed to offer more standing variation readily available for natural selection and to induce transcriptional changes in very specific spatiotemporal conditions. Therefore, and unlike mutations altering protein sequences, cis-regulatory changes may enable rapid attainment of the fitness optima by finely tuning gene expression. The opponents to such models argue that other evolutionary mechanisms can also minimize the evolutionary constraints at the protein-coding regions, mainly through gene duplication and subsequent subfunctionalization or neofunctionalization (41, 42). Our findings support the contention that cis-regulatory elements play a major role in fast adaptive processes, such as the adaptive process of Yakutian horses to the Arctic.
Fig. 4.
Relative contribution of coding and noncoding regions to adaptation. (A) Red dashed line delimits the neutrality threshold [i.e., the top-0.001% dA quantile of 4d-fold degenerate sites (dA = 0.3448)]. (B) Distribution of the proportion of adaptive mutations across categories, including coding (green, 0d-fold, 2d-fold, and 4d-fold degenerate positions) and noncoding (red, 10 first bins of 1 kb upstream from the TSS) sites. (C) dA pattern at the TGM3 gene (reverse strand). (D) dA pattern at the LECT2 and FBLX21 gene region. chr, chromosome.
Beyond an overrepresentation of nonspecific fast-evolving systems, such as sensory perception of smell (SI Appendix, Tables S7.10 and S7.11), a detailed inspection of the list of adaptive cis-regulatory candidates uncovered variants participating to minimize heat loss (SI Appendix, Tables S7.8–S7.14), such as an extreme hair density, s.c. fat accumulation, and a relatively small surface relative to body mass. Strikingly, this list is enriched in genes associated with cellulitis (adjusted P ≤ 0.0391) and bone and limb morphogenesis, including abnormal foot bone ossification and multiple sclerosis (adjusted P ≤ 0.0036). It also includes the TGM3 gene, whose loss of function in mice results in impaired and thinner hairs (43). The dA peak detected ∼8 kb from its TSS (Fig. 4C and SI Appendix, Tables S7.12 and S7.14) might thus be involved in the hairy phenotype of Yakutian horses.
Extreme temperatures and seasonal variations in daylight exposure (ca. 40% of Yakutia lies above the Arctic Circle) are also known to drive a range of endocrine responses, such as the release of steroid and glucocorticoid hormones by the hypothalamus–pituitary–adrenal axis (44–50). These hormones activate adrenoreceptors in the liver, and thus glycogenesis, which ultimately regulates thermogenic requirements and produces antifreezing compounds (51). We detected significant enrichment for hypothyroidism (adjusted P = 0.0090) and endocrine system diseases (adjusted P = 0.0057), including posterior pituitary disorders (adjusted P = 0.0420) and type I diabetes mellitus (adjusted P = 0.0077; SI Appendix, Tables S7.9–7.14). The latter might reflect adaptive signatures for regulating glycemia, in relation to the thermogenic antifreezing properties of glucose. More strikingly, our list of cis-regulatory candidates also includes the thyroid hormone receptor-associated gene (THRAP3), which physically interacts with the adrenoreceptor α1B (52) that regulates the vasoconstriction/vasodilatation reflex following cold exposure.
Another strategy to avoid or tolerate internal ice formation is indeed reducing the volume of circulating blood (53–55). This strategy, however, deprives internal organs of oxygen and triggers a burst of reactive oxygen species during seasonal temperature recovery (51). We found dA peaks at the upstream regions of genes involved in oxidative stress responses (ADAM9, CAT, JUN, SLC8A1, and SOD2), vasodilation (KCNJ8 and SOD2), and blood coagulation (F2R, F3, LPA, PAPSS2, PF4, and PLG). We consistently found enrichment in functionally associated terms, such as peripheral arterial disease (adjusted P = 0.0219); disorders of fluid, electrolyte, and acid-base balance (adjusted P = 0.0460); and abnormal cell blood morphology (adjusted P = 0.0460; SI Appendix, Tables S7.9–S7.14). Overall, our results suggest a major role of noncoding changes in the adaptive response of Yakutian horses.
The analyses of protein-coding sites confirmed the participation of functional categories similar to those categories overrepresented among cis-regulatory candidates. First, a selection of 130 genes harboring highly differentiated nonsynonymous alleles (SI Appendix, section S4.1.3 and Table S4.3) was significantly enriched for a number of pathways, including galactose, starch, and sucrose metabolisms (adjusted P ≤ 0.0169), as well as diseases like Nelson’s syndrome (adjusted P = 0.0214), which leads to an excess of the adrenocorticotropic hormone. Additionally, the lists of 251–1,489 genes located within FST outlier regions (SI Appendix, section S7.1.1) were overrepresented for a range of metabolic and endocrine disorders (SI Appendix, Tables S7.4–S7.7), including diabetes (adjusted P ≤ 0.0022), endochondral ossification (adjusted P = 0.0115), water reabsorption (adjusted P ≤ 0.0003), and oxidative stress (adjusted P ≤ 0.0172).
The functional consistency between protein-coding and upstream regions may be partly explained by the underlying demographic history, which imposes a short time scale whereby founder alleles remain physically linked in large haplotypes. We indeed observed some cases with large runs of high differentiation values, such as the ∼90-kb region covering LECT2 and FBXL21 (Fig. 4D), two genes involved in the growth of chondrocytes (56) and the regulation of the circadian clock (57), respectively. It is worth noting, however, that the population bottleneck erased a large fraction of the standing variation, so that differentiation is often restricted to the sole segregating positions remaining in the population (Fig. 4C).
Finally, because gene duplication represents another major source of functional innovation (41, 42), we screened for segmental duplication within Yakutian horse genomes. Overall, we identified a total number of 178 candidate regions for copy number variants (CNVs), showing patterns of depth-of-coverage variation in at least one Yakutian horse that are absent from the reference genome (Twilight; SI Appendix, Table S4.12). These CNV regions contained 91 annotated protein-coding genes enriched for a number of biological pathways (SI Appendix, Tables S4.12–S4.15), including steroid hormone biosynthesis, fatty acid metabolism, metabolic pathways, and olfactory transduction (adjusted P ≤ 0.0111). Interestingly, phenotype enrichment analyses revealed four genes (ACADSB, ATP1A2, CYP11B2, and HSPG2) associated with abnormality of temperature regulation (adjusted P = 0.02120) (SI Appendix, Table S4.14).
The functional intersection of noncoding, coding, and gene duplication lists suggests that positive selection is consistently targeting similar metabolic pathways and morphological traits. To investigate whether similar mechanisms underpinned cold adaptations in other mammals, we compared our dA list against the top adaptive candidates recently identified in woolly mammoths (58) and in human populations living in Siberia (59). Although no dA peak was found for TRPV3, a recently described temperature sensor in mammoths (58), we identified other genes with apparent signatures of convergent evolution, including BARX2 (involved in hair and epidermis development) and PHIP (a key regulator of the insulin metabolism). Convergent adaptation signatures were also apparent within human populations, including at the PRKG1 gene, which is key to minimize heat loss by regulating blood vessel constriction. Overall, this overlapping suggests that subarctic environments considerably restrict fitness landscapes, with only a minority of evolutionary strategies being compatible with survival.
Conclusions
Our diachronical genome dataset has enabled us to illuminate the origins and adaptations of Yakutian horses. We found that present-day Yakutian horses are a subgroup of domesticated horses, genetically distinct from the now-extinct group that populated Yakutia in the mid-Holocene and the Taymir Peninsula in the Late Pleistocene. Additionally, the comparison with other domesticated breeds unveiled the genetic makeup of present-day Yakutian horses. We found that cis-regulatory regions represented a larger proportion of the adaptive candidate loci than gene bodies, which might be a common evolutionary signature of fast adaptive processes. Finally, we found evidence of convergent adaptations among woolly mammoths and human and Yakutian horse populations currently living in the Arctic, suggesting that the adaptive landscape of mammals to such environments is likely restricted.
Methods
Detailed descriptions of samples and methods are provided in SI Appendix. The sequencing data are available from the European Nucleotide Archive, accession no. PRJEB10854.
Genome Sequencing.
DNA was extracted from nine ancient bone and tooth samples in dedicated ancient DNA facilities and prepared into indexed Illumina libraries, following the procedures described elsewhere (16, 18, 27, 60–62). Contamination was monitored through extraction, library, and amplification blanks, which were all negative. Indexed Illumina libraries were prepared from modern specimens using DNA extracts from the hair of 12 modern Yakutian horses following established methods (27, 60, 63). Illumina sequencing was performed at the Danish National High-Throughput DNA Sequencing Centre and the Kurchatov Institute (SI Appendix, sections S2.1–S2.3).
Read Mapping, Damage Signatures, Genotyping, and Microbial Profiling.
The PALEOMIX pipeline was used to process reads from trimming to genotyping (62) using default parameters, except that the minimal mapping quality and the maximum per-site depth were set to 25 and the top 5% quantile of the read-depth distribution. For ancient specimens, seeding was also disabled and uncollapsed paired-ended reads were excluded. Reads were independently aligned against the horse reference genome EquCab2.0 (15) and the mitochondrial genome (accession no. NC_001640.1) (64). Mitochondrial genomes were reconstructed using a majority rule and requiring a depth of coverage ≥3 and base qualities ≥30. We used mapDamage2.0 (22) to investigate DNA damage signatures based on 100,000 randomly selected nuclear read alignments per library and to rescale base qualities at read ends. Metagenomic analyses were carried out using the metagenomic module of PALEOMIX, as previously described (61, 62) (SI Appendix, sections S2.5, S2.6, and S3).
Heterozygosity and Inbreeding.
Genome-wide heterozygosity values were calculated as the average of the Watterson theta estimator (θw) within 50-kb windows following the method of Orlando et al. (27). Inbreeding coverage estimates were measured within present-day horses, exploiting the genome-wide distribution of the heterozygosity tracts calculated above (62) (SI Appendix, section 4.3).
Phylogenetic Analyses.
Mitochondrial haplotypes were aligned to a dataset of 105 available horse mitochondrial genomes, partitioned into rRNA; tRNA; control region; and the first, second, and third codon positions for coding DNA sequences. Y-chromosome haplotypes were reconstructed aligning reads using the procedure described by Schubert et al. (62). The best substitution models were determined by Modelgenerator version 0.85 (65) for each of the mitochondrial partitions, and for the full mitochondrial and Y-chromosome alignments. For mtDNA and Y-chromosome data, ML phylogenetic inference was performed in PhyML3.0 (66) using the best substitution models. For mtDNA, Bayesian analyses were also performed on the concatenated partitions in BEAST 1.8.0 (67). Exome-based phylogenetic inference was performed from a supermatrix based on the 50% longest protein-coding genes annotated in EquCab version 2.76 (68), using PALEOMIX (62) (SI Appendix, section S5).
Genetic Affinities and Population Structure.
PCA based on 356,720 high-quality variants called from whole-genome sequences of 31 horses was carried out with EIGENSTRAT version EIG5.0.1 (69), without outlier iterations (options “-numoutlieriter: 0”). PCA was also estimated in a genotype likelihood framework using Analysis of Next Generation Sequencing Data (ANGSD) and ngsTools (70, 71) (SI Appendix, section S6.3). To investigate genetic continuity from ancient to present-day Yakutian horses, we used genome projections (33), where the frequency spectrum of a derived allele in a test genome is compared with the frequency spectrum seen within a reference panel (SI Appendix, section 6.2).
Demographic Reconstructions.
PSMC (35) demographic profiles were scaled using the genome-wide substitution rate of 7.242e−9 per site per generation and a generation time of 8 y (27) (SI Appendix, section S6.1.1). The ∂a∂i program (version 1.6.3) (72) was used to reconstruct the recent demographic history of domesticated and Yakutian horses, exploiting the site frequency spectra calculated from 4d-fold protein-coding positions.
Admixture Analyses.
We ran a series of admixture tests, including NGSAdmix (73), f3-statistics (36) and D-statistics (74). In NGSAdmix, we considered a minimum minor allele frequency of 5% and a maximum number of expectation maximization iterations of 5,000. The matrix of 356,720 variants used in PCA analyses was used as input for “threepop” and TreeMix (75) software (-k 1,500 and considering up to four migration edges among population groups) (SI Appendix, section S6.4.2). The f3-statistics were calculated (domesticated horse, Batagai; present-day Yakutian horse). The D-statistics were calculated following the implementations of Schubert et al. (18), Orlando et al. (27), and Jónsson et al. (76), disregarding transitions for ancient genomes (SI Appendix, section S6.4.1).
CNVs and Nonsynonymous Mutations.
We identified Yakutian-specific CNVs by detecting regions showing an excess of reads mapping to the genome of the Thoroughbred horse (Twilight), in which duplications were masked beforehand using mrCaNaVaR version 0.31 and mrFAST version 2.0.0.5 (77). Potential Yakutian horse-specific markers were collected based on criteria described by Baye et al. (78). Nonsynonymous mutations with a difference in allele frequencies between Yakutian and other domesticated horses superior to 0.4 were selected (SI Appendix, sections S4.1.3–S4.2).
FST-Based Selection Scan.
The FST index (79) was estimated between the domesticated (represented either by 12 Franches-Montagnes or 27 mixed-breed horses) and Yakutian (n = 10) horses. The latter were determined within the genotype likelihood framework implemented in ANGSD (80) and ngsTools (71), in 50-kb sliding windows (step size = 10 kb), and disregarding windows where coverage was less than 90%. Genomic regions showing a local and significant increase of the FST value (candidates for positive selection) were identified following the method of Jónsson et al. (76) (SI Appendix, section S7.1).
dA-Based Selection Scan.
We used the mstatspop program to calculate the dA index (38) for different coding (i.e., 0d-fold, 2d-fold, and 4d-fold degenerate coding positions) and noncoding sites (10 first 1-kb bins located upstream of the TSS). We disregarded positions where the fraction of valid (nonmissing) alleles was less than 90%. We assumed that the 4d-fold dA distribution delineates the threshold for positions evolving under neutrality. The proportion of adaptive mutations for each site category was estimated by bootstrapping (SI Appendix, section S7.2).
Functional Enrichment.
Functional enrichment was tested for Yakutian-specific CNVs and nonsynonymous mutations, as well as for FST and dA outlier regions and positions. We used human and mouse orthologs and the online platform WebGestalt with the human and the mouse as model organisms (81) (SI Appendix, section S7.3).
Supplementary Material
Acknowledgments
We thank P. Selmer Olsen, T. Brand, and the staff of the Danish National High-Throughput DNA Sequencing Centre for technical assistance, and Haldja Viinalass and Erkki Sild from the Estonian University of Life Sciences (Tartu, Estonia) for laboratory work. This work was supported by the Danish Council for Independent Research, Natural Sciences (Grant 4002-00152B); the Danish National Research Foundation (Grant DNRF94); a Marie-Curie Career Integration grant (Grant FP7 CIG-293845); Initiative d'Excellence Chaires d'attractivité, Université de Toulouse (OURASI); and the International Research Group Program (Grant IRG14-08), Deanship of Scientific Research, King Saud University. P.L. was supported by a Villum Fonden Blokstipendier grant (primary investigator: L.O.); H.J. by a Marie-Curie Initial Training Network grant [EUROTAST (Exploring the History, Archeology, and New Genetics of the Transatlantic Slave Trade); Grant FP7 ITN-290344]; C.G. and L.E. by Marie-Curie Intra-European fellowships (FP7-IEF-328024 and FP7 IEF-302617); M. Schubert by a Lundbeck Foundation grant (Grant R52-A5062); M.A.Y. by a National Science Foundation Graduate Research fellowship; M.F. by a Human Frontier Science Program fellowship (LT000320/2014); A.A. by a Villum Fonden Blokstipendier grant; M. Schubert by an NIH grant (Grant R01-GM40282); and J.K. by the Academy of Finland (Grant 286040). Research work on the ancient Yakut population was supported by the French Archaeological Mission in Oriental Siberia (Ministry of Foreign and European Affairs, France), the North-Eastern Federal University (Yakutsk, Sakha Republic), and the Human Adaptation Program of the French Polar Institute Paul Emile Victor [HUMAD MAFSO (Missions Archéologiques Françaises en Sibérie Orientale) 1038].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB10854).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513696112/-/DCSupplemental.
References
- 1.Stepanova N. On the lowest temperature on Earth. Monthly Weather Review. 1958;86:6–10. [Google Scholar]
- 2.Pakendorf B, et al. Investigating the effects of prehistoric migrations in Siberia: Genetic variation and the origins of Yakuts. Hum Genet. 2006;120(3):334–353. doi: 10.1007/s00439-006-0213-2. [DOI] [PubMed] [Google Scholar]
- 3.Crubézy E, et al. Human evolution in Siberia: from frozen bodies to ancient DNA. BMC Evol Biol. 2010;10(1):25. doi: 10.1186/1471-2148-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keyser C, et al. The ancient Yakuts: A population genetic enigma. Philos Trans R Soc Lond B Biol Sci. 2015;370(1660):20130385. doi: 10.1098/rstb.2013.0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferret C. Une civilization du cheval. Editions Belin; Paris: 2009. [Google Scholar]
- 6.Solomonov NG, Anufriev AI, Yadrikhinskii VF, Isaev AP. Body temperature changes in purebred and hybrid Yakut horses under the conditions of Yakutia. Dokl Biol Sci. 2009;427(1):358–361. doi: 10.1134/s0012496609040164. [DOI] [PubMed] [Google Scholar]
- 7.Grigoreva NN, Koryakina LP. Dynamics of products of a metabolism glycoproteide and activity of enzymes AcAT, AAT at different type the Yakut breed of a horse. Doklady Rossiiskoi Akademii Sel'skokhozyaistvennykh Nauk. 2008;3:44–46. [Google Scholar]
- 8.Gabushev GM. Yakutian Horse. Yakutian Publishing House; Yakutsk, Russia: 1957. [Google Scholar]
- 9.Sieroszewski W, Stepanov SA, Zhornitskaia MIA, Mukhamediarov SF. 1993. Yakuts: Experience Ethnographic Research (Association “Russian Encyclopedia,” Moscow)
- 10.Alekseev AN. The Turkic World: Mongolia and Yakutia: Digest of Articles. Ulan-Bator, Mongolia: New Polygraph; 2007. About the time of settlement of the Middle Vilyu; pp. 197–201. [Google Scholar]
- 11.Lazarev PA. Anthropogene Horses of Yakutia. Nauka; Moscow, Russia: 1980. [Google Scholar]
- 12.Keyser-Tracqui C, et al. Mitochondrial DNA analysis of horses recovered from a frozen tomb (Berel site, Kazakhstan, 3rd Century BC) Anim Genet. 2005;36(3):203–209. doi: 10.1111/j.1365-2052.2005.01316.x. [DOI] [PubMed] [Google Scholar]
- 13.McGahern AM, et al. Mitochondrial DNA sequence diversity in extant Irish horse populations and in ancient horses. Anim Genet. 2006;37(5):498–502. doi: 10.1111/j.1365-2052.2006.01506.x. [DOI] [PubMed] [Google Scholar]
- 14.Lippold S, Matzke NJ, Reissmann M, Hofreiter M. Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol Biol. 2011;11:328. doi: 10.1186/1471-2148-11-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade CM, et al. Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326(5954):865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen JS, et al. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 2014;24(3):454–466. doi: 10.1101/gr.163592.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enk JM, et al. Ancient whole genome enrichment using baits built from modern DNA. Mol Biol Evol. 2014;31(5):1292–1294. doi: 10.1093/molbev/msu074. [DOI] [PubMed] [Google Scholar]
- 18.Schubert M, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci USA. 2014;111(52):E5661–E5669. doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507(7491):225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010(6):db.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 21.Star B, et al. Palindromic sequence artifacts generated during next generation sequencing library preparation from historic and ancient DNA. PLoS One. 2014;9(3):e89676. doi: 10.1371/journal.pone.0089676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto H, et al. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski’s horses. Genome Biol Evol. 2011;3:1096–1106. doi: 10.1093/gbe/evr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achilli A, et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci USA. 2012;109(7):2449–2454. doi: 10.1073/pnas.1111637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S, et al. High altitude adaptation and phylogenetic analysis of Tibetan horse based on the mitochondrial genome. J Genet Genomics. 2007;34(8):720–729. doi: 10.1016/S1673-8527(07)60081-2. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Q, et al. The complete mitochondrial genome and phylogenetic analysis of the Debao pony (Equus caballus) Mol Biol Rep. 2011;38(1):593–599. doi: 10.1007/s11033-010-0145-8. [DOI] [PubMed] [Google Scholar]
- 27.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499(7456):74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 28.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7(3):e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindgren G, et al. Limited number of patrilines in horse domestication. Nat Genet. 2004;36(4):335–336. doi: 10.1038/ng1326. [DOI] [PubMed] [Google Scholar]
- 30.Wallner B, et al. Identification of genetic variation on the horse y chromosome and the tracing of male founder lineages in modern breeds. PLoS One. 2013;8(4):e60015. doi: 10.1371/journal.pone.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Do K-T, et al. Genomic characterization of the Przewalski’s horse inhabiting Mongolian steppe by whole genome re-sequencing. Livest Sci. 2014;167(0):86–91. [Google Scholar]
- 32.Der Sarkissian C, et al. Evolutionary Genomics and Conservation of the Endangered Przewalski’s Horse. Curr Biol. 2015;25(19):2577–2583. doi: 10.1016/j.cub.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang MA, Harris K, Slatkin M. The projection of a test genome onto a reference population and applications to humans and archaic hominins. Genetics. 2014;198(4):1655–1670. doi: 10.1534/genetics.112.145359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark PU, et al. The Last Glacial Maximum. Science. 2009;325(5941):710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28(8):2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M. Molecular Evolutionary Genetics. Columbia Univ Press; New York: 1987. [Google Scholar]
- 39.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3(7):e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 41.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61(5):995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 42.Ohno S. Evolution by Gene Duplication. Springer; Berlin: 1970. [Google Scholar]
- 43.John S, et al. Epidermal transglutaminase (TGase 3) is required for proper hair development, but not the formation of the epidermal barrier. PLoS One. 2012;7(4):e34252. doi: 10.1371/journal.pone.0034252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handa RJ, Weiser MJ. Gonadal steroid hormones and the hypothalamo-pituitary-adrenal axis. Front Neuroendocrinol. 2014;35(2):197–220. doi: 10.1016/j.yfrne.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurberg P, Andersen S, Karmisholt J. Cold adaptation and thyroid hormone metabolism. Horm Metab Res. 2005;37(9):545–549. doi: 10.1055/s-2005-870420. [DOI] [PubMed] [Google Scholar]
- 46.Dimicco JA, Zaretsky DV. The dorsomedial hypothalamus: A new player in thermoregulation. Am J Physiol Regul Integr Comp Physiol. 2007;292(1):R47–R63. doi: 10.1152/ajpregu.00498.2006. [DOI] [PubMed] [Google Scholar]
- 47.Little AG, Kunisue T, Kannan K, Seebacher F. Thyroid hormone actions are temperature-specific and regulate thermal acclimation in zebrafish (Danio rerio) BMC Biol. 2013;11(1):26. doi: 10.1186/1741-7007-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva JE. Thyroid hormone control of thermogenesis and energy balance. Thyroid. 1995;5(6):481–492. doi: 10.1089/thy.1995.5.481. [DOI] [PubMed] [Google Scholar]
- 49.Houser DS, Yeates LC, Crocker DE. Cold stress induces an adrenocortical response in bottlenose dolphins (Tursiops truncatus) J Zoo Wildl Med. 2011;42(4):565–571. doi: 10.1638/2010-0121.1. [DOI] [PubMed] [Google Scholar]
- 50.Smith SM, Vale WW. The role of the hypothalamic-pituitary-adrenal axis in neuroendocrine responses to stress. Dialogues Clin Neurosci. 2006;8(4):383–395. doi: 10.31887/DCNS.2006.8.4/ssmith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storey JM, Storey KB. Cold hardiness and freeze tolerance. In: Storey KB, editor. Functional Metabolism. Wiley; New York: 2004. pp. 473–503. [Google Scholar]
- 52.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanhoutte PM. Comprehensive Physiology. Wiley; New York: 2011. Physical factors of regulation. [Google Scholar]
- 54.Flavahan NA. The role of vascular alpha-2-adrenoceptors as cutaneous thermosensors. Physiology (Bethesda) 1991;6(6):251–255. [Google Scholar]
- 55.Thompson GE. Physiological effects of cold exposure. Int Rev Physiol. 1977;15:29–69. [PubMed] [Google Scholar]
- 56.Yamagoe S, Kameoka Y, Hashimoto K, Mizuno S, Suzuki K. Molecular cloning, structural characterization, and chromosomal mapping of the human LECT2 gene. Genomics. 1998;48(3):324–329. doi: 10.1006/geno.1997.5198. [DOI] [PubMed] [Google Scholar]
- 57.Hirano A, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152(5):1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 58.Lynch VJ, et al. Elephantid Genomes Reveal the Molecular Bases of Woolly Mammoth Adaptations to the Arctic. Cell Reports. 2015;12(2):217–228. doi: 10.1016/j.celrep.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 59.Cardona A, et al. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One. 2014;9(5):e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seguin-Orlando A, et al. Ligation bias in illumina next-generation DNA libraries: Implications for sequencing ancient genomes. PLoS One. 2013;8(10):e78575. doi: 10.1371/journal.pone.0078575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Der Sarkissian C, et al. Shotgun microbial profiling of fossil remains. Mol Ecol. 2014;23(7):1780–1798. doi: 10.1111/mec.12690. [DOI] [PubMed] [Google Scholar]
- 62.Schubert M, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 2014;9(5):1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 63.Nedoluzhko AV, et al. Analysis of the Mitochondrial Genome of a Novosvobodnaya Culture Representative using Next-Generation Sequencing and Its Relation to the Funnel Beaker Culture. Acta Naturae. 2014;6(2):31–35. [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X, Arnason U. The complete mitochondrial DNA sequence of the horse, Equus caballus: Extensive heteroplasmy of the control region. Gene. 1994;148(2):357–362. doi: 10.1016/0378-1119(94)90713-7. [DOI] [PubMed] [Google Scholar]
- 65.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6(1):29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 67.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue) D1:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 70.Fumagalli M, et al. Quantifying population genetic differentiation from next-generation sequencing data. Genetics. 2013;195(3):979–992. doi: 10.1534/genetics.113.154740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fumagalli M, Vieira FG, Linderoth T, Nielsen R. ngsTools: Methods for population genetics analyses from next-generation sequencing data. Bioinformatics. 2014;30(10):1486–1487. doi: 10.1093/bioinformatics/btu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195(3):693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jónsson H, et al. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci USA. 2014;111(52):18655–18660. doi: 10.1073/pnas.1412627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41(10):1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baye TM, Tiwari HK, Allison DB, Go RC. Database mining for selection of SNP markers useful in admixture mapping. BioData Min. 2009;2(1):1. doi: 10.1186/1756-0381-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wright S. The genetical structure of populations. Ann Eugen. 1951;15(4):323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 80.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15(1):356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleics Acids Res. 2005;33(Web Server issue):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.