Skeletal muscles provide fascinating examples how humans have evolved and exercise. While humans developed superior cognition, metabolome evolution studies indicate an accelerated parallel decline in muscle energetic capacity and strength (1). New forms of locomotion including exceptional endurance were adapted ∼2 million years ago (2). Nowadays, we generally assume healthy aging and disease prevention depend on regular exercise. Contrasting with evolutionary adaptation, high-intensity interval training (HIIT) represents an ultrashort form of exercise and a promising intervention for disease prevention. However, the molecular mechanisms are incompletely understood. Studying recreationally active humans, Place et al. (3) used highly demanding HIIT cycling protocols. Importantly, these studies identify a novel molecular defect and putative mechanism of muscle adaptation, ryanodine receptor (RyR) fragmentation (3).
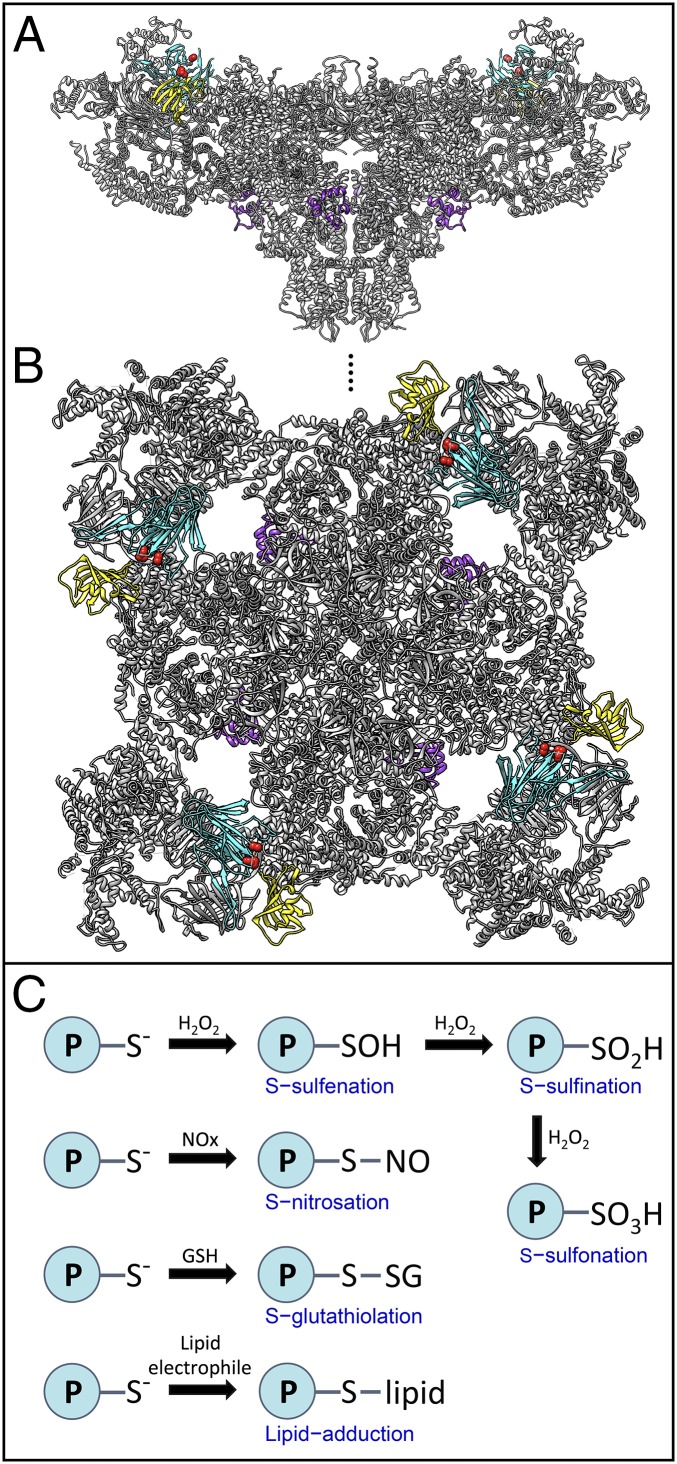
The human genome encodes three RyR isoforms. In skeletal and heart muscle RyR1 and RyR2 are highly expressed, respectively, each functioning as the major intracellular calcium (Ca2+) release channel. Approximately 50% of RyR1 channels in the sarcoplasmic reticulum (SR) are activated by protein–protein interactions with voltage-gated Ca2+ channels (VGCCs) in the plasma membrane (sarcolemma). Direct VGCC–RyR1 interactions enable rapid excitation–contraction (EC) coupling rates in myofibers necessary for sustained (tetanic) contractions. This is in contrast with diffusion-based RyR1 or RyR2 channel activation via Ca2+-induced Ca2+ release (CICR). Recent cryo-EM structures of RyR1 with near-atomic resolution (Fig. 1 A and B and refs. 4 and 5) provide potential clues about the putative RyR1 fragmentation versus allosteric activation mechanisms, which were correlated with depressed muscle function (3).
Fig. 1.
Structure of the RyR1 channel homotetramer. Views (A) from the plane of the membrane and (B) from the cytosol. Rabbit RyR1 structure indicated in gray, Calstabin1/2 (FKBP12/12.6) in yellow, EF-hands (amino acids 4072–4135) in purple, and SPRY3 (amino acids 1242–1614) in cyan (4). Red spheres are indicating the beginning and the end of an unresolved region (amino acids 1297–1436) within which the putative calpain cleavage site is located. (C) Oxidative modification reactions of protein-S− thiols include readily reversible (S-sulfenation, S-nitrosation, and S-thiolation), potentially reversible (S-sulfination and lipid adducts), and likely nonreversible (S-sulfonation) redox states.
Previous studies determined that calpain activation cleaves RyR1 channels into stably associated ∼375- and ∼150-kDa fragments sustaining subconductance channel open gating in vitro (6). Recombinant, N-terminally truncated RyR1 channels reconstituted in lipid bilayers exhibited CICR-like functions (7). Notably, 24 h after HIIT exercise recreationally active humans showed significant RyR1 fragmentation (re)producing a ∼375-kDa fragment in muscle biopsies similar to calpain-treated RyR1 (3, 6). Calpain is known to cleave RyR1 protomers at residues 1383–1400 (6), located within an unresolved region in the RyR1 model [Fig. 1 A and B; in both PDB ID code 3J8E and 3J8H (4, 5)], where VGCCs potentially bind RyR1 (8). Hence, it is plausible that CICR-like channel activity will remain intact whereas voltage-dependent RyR1 activation during EC coupling will be disrupted, and the truncated channels may sustain SR Ca2+ leak.
In skeletal muscle, calpain-3 mutations cause limb-girdle muscular dystrophy type 2A in patients. Place et al. (3) determined an approximately threefold increased calpain activity of the Ca2+-activated protease. In the case of calpain-3, specific enzyme domains enable rapid autolytic activation, however, the enzyme is thought to be inactive when bound to titin in sarcomeres. In analogy, in triad junctions the aldolase A scaffold forms a complex with RyR1 and inactive calpain-3 (9). Interestingly, loss of calpain-3 scaffold function was proposed to alter SR Ca2+ release, whereas calpain-3C129S knock-in, a structurally intact but enzymatically inactive mutant, would not alter SR Ca2+ release (10).
In addition, junctophilin isoforms are substrates of the ubiquitous calpain-1. In triad junctions, junctophilins stabilize nanodomain spacing between VGCC and RyR1 channels essential for regular EC coupling. Proteolytic junctophilin inactivation may result in impaired Ca2+ regulation and skeletal muscle weakness (11). Interestingly, 24 h after a single bout of eccentric exercise calpain-3 activation was observed in skeletal muscle by earlier studies; and recent studies also showed Ca2+-dependent calpain-1 activation (11). In agreement, Place et al. (3) observed RyR1 fragmentation in skeletal muscle biopsies 24 h after cycling bouts. Taken together, calpain-1 and calpain-3 are candidates for subcellular junctophilin and RyR1 proteolysis, respectively, and may contribute to SR Ca2+ leak. Place et al. (3) attribute the HIIT-induced calpain effects to selective degradation of RyR1, whereas junctophilin was not mentioned.
Intriguingly, RyR1 fragmentation occurred in a reactive oxygen species (ROS)-dependent manner following HIIT exercising (3). It is tempting to speculate that calpain-3 cleaves oxidized RyR1 selectively post-HIIT. Alternatively, calpain-3 may protect RyR1 from degradation by conventional calpains and RyR1 oxidation would diminish this protection. Consistent with protective calpain-3 roles, reduced expression of RyR1 and aldolase A was observed in calpain-3 knock-out but not in calpain-3C129S knock-in mice (9, 10).
Intense exercise induced oxidative posttranslational modification of RyR1 by the electrophile malondialdehyde (MDA) (3), which was perhaps causal for subsequent degradation. However, because basal MDA modification was already 50% of that achieved postexercise, this may argue against a causal relationship. A variety of additional nitro-oxidative cysteine modifications of RyR1 can occur (Fig. 1C), including S-nitrosation (12), S-glutathiolation, and sulfonation, as well as 3-nitrotyrosine formation (13), and these may mediate proteolysis.
Although the principal observation of RyR1 fragmentation during intense exercise is quite clear, many molecular and etiological details require follow-up. For instance, protective significance is attached to elevated levels of SOD2 and catalase in elite athletes. Catalase is normally located in peroxisomes, whereas other antioxidant enzymes such as glutathione- or thioredoxin-dependent peroxiredoxins and thioredoxin may also be important. If the molecular sources of the reactive oxygen species, such as NADPH oxidases (Noxs), mitochondria, uncoupled nitric oxide synthases or monoamine oxidase, mediating the fragmentation could be established, this would help define whether a 1 or 2 electron antioxidant may be rationally more effective in potentially modulating RyR1 proteolysis.
In myofibers, RyR1 fragmentation may further affect the subcellular state of RyR1 channel arrays in membrane nanodomains. Complex spatial RyR1 distributions were determined by superresolution and electron microscopy. VGCCs in transverse tubule invaginations interact locally with RyR1 clusters in triad junctions and synchronize cell-wide cluster activities (14). Superresolution microscopy showed nearly continuous RyR1 cluster distributions in fast-twitch, but not in slow-twitch, muscles (15). Importantly, RyR clusters represent extended protein networks with mutual physical and complex in situ behaviors of individual channels (16). Consequently, RyR channel organization in clusters emerged as an important control mechanism of local SR Ca2+ release. Whether RyR1 channel and cluster changes are locally linked to specific redox sources in cells is an unresolved question.
Supporting local RyR2 cluster changes, a decrease of the total dyadic coupling area was determined by EM in human cardiomyopathy samples (17). Recent superresolution microscopy in atrial fibrillation samples suggested RyR2 cluster fragmentation as additional cause of altered Ca2+ signaling (18). Hypothetical mechanisms of cluster fragmentation include loss of RyR protein interactions due to proteolytic domain clipping, oxidative modifications, and disruption through junctophilin loss. Although structure–function analysis at the nanometric scales of RyR clusters is challenging in native cells, superresolution Ca2+ spark modeling has become a vital option to characterize functional cluster changes, closely resembling local Ca2+ signaling behaviors (16).
The intriguing observation by Place et al. (3) that force depression in skeletal muscle is correlated with RyR1 fragmentation following intense cycling bouts stimulates important future questions. Proteolysis by calpain represents a putative mechanism of direct molecular uncoupling from voltage-dependent VGCC-RyR1 control (Fig. 1 A and B). In contrast, marathon running induced a distinct molecular instability of the channel complex evidenced by calstabin1 depletion (Fig. 1 A and B) (3), confirming earlier results (19). Further understanding may come from generation of mouse models expressing calpain-resistant RyR or specific proteolytic fragments, as achieved with myosin-binding protein C (20). Without doubt, the fascinating findings of Place et al. (3) will stimulate intense interest and hopefully lead to more effective means of disease prevention based on rationally founded future exercise interventions.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB1002-TP-B05 (to S.E.L.) and IRTG1816-RP2 (to P.E. and S.E.L.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 15492.
References
- 1.Bozek K, et al. Exceptional evolutionary divergence of human muscle and brain metabolomes parallels human cognitive and physical uniqueness. PLoS Biol. 2014;12(5):e1001871. doi: 10.1371/journal.pbio.1001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432(7015):345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- 3.Place N, et al. Ryanodine receptor fragmentation and sarcoplasmic reticulum Ca2+ leak after one session of high-intensity interval exercise. Proc Natl Acad Sci USA. 2015;112:15492–15497. doi: 10.1073/pnas.1507176112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zalk R, et al. Structure of a mammalian ryanodine receptor. Nature. 2015;517(7532):44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Z, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517(7532):50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoshan-Barmatz V, Weil S, Meyer H, Varsanyi M, Heilmeyer LM. Endogenous, Ca2+-dependent cysteine-protease cleaves specifically the ryanodine receptor/Ca2+ release channel in skeletal muscle. J Membr Biol. 1994;142(3):281–288. doi: 10.1007/BF00233435. [DOI] [PubMed] [Google Scholar]
- 7.Bhat MB, Zhao J, Takeshima H, Ma J. Functional calcium release channel formed by the carboxyl-terminal portion of ryanodine receptor. Biophys J. 1997;73(3):1329–1336. doi: 10.1016/S0006-3495(97)78166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J Mol Biol. 2004;342(1):145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 9.Kramerova I, et al. Novel role of calpain-3 in the triad-associated protein complex regulating calcium release in skeletal muscle. Hum Mol Genet. 2008;17(21):3271–3280. doi: 10.1093/hmg/ddn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojima K, et al. Non-proteolytic functions of calpain-3 in sarcoplasmic reticulum in skeletal muscles. J Mol Biol. 2011;407(3):439–449. doi: 10.1016/j.jmb.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy RM, et al. Ca2+-dependent proteolysis of junctophilin-1 and junctophilin-2 in skeletal and cardiac muscle. J Physiol. 2013;591(Pt 3):719–729. doi: 10.1113/jphysiol.2012.243279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun QA, Wang B, Miyagi M, Hess DT, Stamler JS. Oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor/Ca2+ release channel (RyR1): Sites and nature of oxidative modification. J Biol Chem. 2013;288(32):22961–22971. doi: 10.1074/jbc.M113.480228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanski J, Hong SJ, Schöneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem. 2005;280(25):24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 14.Appelt D, Buenviaje B, Champ C, Franzini-Armstrong C. Quantitation of ‘junctional feet’ content in two types of muscle fiber from hind limb muscles of the rat. Tissue Cell. 1989;21(5):783–794. doi: 10.1016/0040-8166(89)90087-6. [DOI] [PubMed] [Google Scholar]
- 15.Jayasinghe ID, Munro M, Baddeley D, Launikonis BS, Soeller C. Observation of the molecular organization of calcium release sites in fast- and slow-twitch skeletal muscle with nanoscale imaging. J R Soc Interface. 2014;11(99):20140570. doi: 10.1098/rsif.2014.0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker MA, et al. Superresolution modeling of calcium release in the heart. Biophys J. 2014;107(12):3018–3029. doi: 10.1016/j.bpj.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HB, et al. Ultrastructural uncoupling between T-tubules and sarcoplasmic reticulum in human heart failure. Cardiovasc Res. 2013;98(2):269–276. doi: 10.1093/cvr/cvt030. [DOI] [PubMed] [Google Scholar]
- 18.Macquaide N, et al. Ryanodine receptor cluster fragmentation and redistribution in persistent atrial fibrillation enhance calcium release. Cardiovasc Res. 2015:cvv231. doi: 10.1093/cvr/cvv231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellinger AM, et al. Remodeling of ryanodine receptor complex causes “leaky” channels: A molecular mechanism for decreased exercise capacity. Proc Natl Acad Sci USA. 2008;105(6):2198–2202. doi: 10.1073/pnas.0711074105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razzaque MA, et al. An endogenously produced fragment of cardiac myosin-binding protein C is pathogenic and can lead to heart failure. Circ Res. 2013;113(5):553–561. doi: 10.1161/CIRCRESAHA.113.301225. [DOI] [PMC free article] [PubMed] [Google Scholar]