Significance
Water drives molecular interactions central to the chemistry of life. Critical to life’s metabolic machinery, enzymes catalyze reactions with remarkable selectivities and efficiencies resulting from noncovalent interactions between water, substrate, and the enzyme active site. Here, amide hydrogen–deuterium exchange kinetics provide quantification of the electronic and steric factors that influence exchange between water and a highly charged host molecule whose protective outer shell and hydrophobic cavity allow it to function like an enzyme. Investigations suggest that the host’s interior microenvironment reacts with encapsulated water in a manner that is correlated to the host’s ability to mediate acid catalysis, thereby providing a tool for developing enzyme-like microenvironments to mediate acid- or base catalysis.
Keywords: supramolecular, noncovalent, catalysis, proton, hydrogen–deuterium exchange
Abstract
The mechanism of proton exchange in a metal–ligand enzyme active site mimic (compound 1) is described through amide hydrogen–deuterium exchange kinetics. The type and ratio of cationic guest to host in solution affect the rate of isotope exchange, suggesting that the rate of exchange is driven by a host whose cavity is occupied by water. Rate constants for acid-, base-, and water-mediated proton exchange vary by orders of magnitude depending on the guest, and differ by up to 200 million-fold relative to an alanine polypeptide. These results suggest that the unusual microenvironment of the cavity of 1 can dramatically alter the reactivity of associated water by magnitudes comparable to that of enzymes.
Water-mediated proton transfer in molecular cavities plays an essential role in the function of nature’s remarkable metabolic machinery (1, 2). Of the broad diversity of enzyme-catalyzed reactions, those involving transfer of hydrogen are ubiquitous and of high fundamental importance (3). For instance, ATP synthase employs an internal chain of water molecules to mediate proton transfer across an electrochemical gradient (4, 5). This process drives ATP synthesis, which allows organisms to broadly meet their basic energy needs. Recent studies of tunneling in enzymatic hydron transfer reactions have other far-reaching and broad implications, namely the importance of the dynamic enzyme motions in driving catalysis (6, 7).
Of the methods used to study hydron transfer, kinetic studies of the mechanisms of amide hydrogen–deuterium exchange (HDX) have helped to elucidate the dynamic interactions between water and protein surfaces that are central to the unique capabilities of enzymes (8–13). Noncovalent interactions, such as electrostatic effects and hydrogen bonding, have also been shown to exert a significant influence on amide HDX rates. These properties are manifest in amide HDX rate constants that can vary by a factor of a billion for residues on the same protein (8). How these variations arise, and how such a property could be harnessed to drive enzyme-like reactivity, remain challenging questions.
In a growing effort to emulate the efficacy of biological receptors and enzymes, studies of the preparative, guest-binding, and catalytic properties of synthetic assemblies have been pursued (14–21). Whereas guest encapsulation has been reported in a variety of media, association processes of organic guests are generally more thermodynamically favorable in water (22, 23). Central to these studies is the notion that the displacement of a high-energy water cluster from a receptor drives guest association, and in some cases, subsequent catalysis. However, despite the abundance of structural data documenting diverse water clusters encapsulated in various host cavities, mechanistic descriptions of the dynamic processes between water and host are rare (24–32).
As part of the broader field of synthetic hosts previously mentioned, our group has developed a class of biologically inspired, anionic K12Ga4L6 assemblies that exhibit catalytic properties similar to those of enzymes (1; Figs. 1 and 2A) (33, 34). Assembly 1 exhibits 12 intramolecular amide hydrogen bonds which, in analogy to structurally important peptide bonds found in polypeptides, preferentially stabilize the desired tetrahedral supramolecular structure over other conformers (35). Compound 1 and related hosts have been shown to catalyze several important chemical reactions with sizable rate accelerations (up to 106) and unusual selectivity reminiscent of enzyme catalysis (34, 36–41). Unlike most enzymes, the reactions catalyzed by 1 are functionally and mechanistically very diverse. Despite these differences, those reactions proceeding with the largest rate enhancements in 1 all involve proton transfer steps. Spurred by this insight, we have investigated the fundamental mechanism of proton transfer in 1 through amide HDX kinetics. The purpose of these studies is twofold: first, to probe the fundamental mechanisms of acid-, base-, and water-mediated proton transfer in a host that is known to promote enzyme-like rate accelerations in acid-mediated processes; second, to explore the effect of guest binding on proton exchange, which provides insight into the interplay of solvent and guest exchange for a synthetic active site. The results of these studies are broadly relevant to the ligand–receptor binding events involving biomacromolecules and the development of synthetic enzyme mimics that emulate the utility of their biological counterparts.
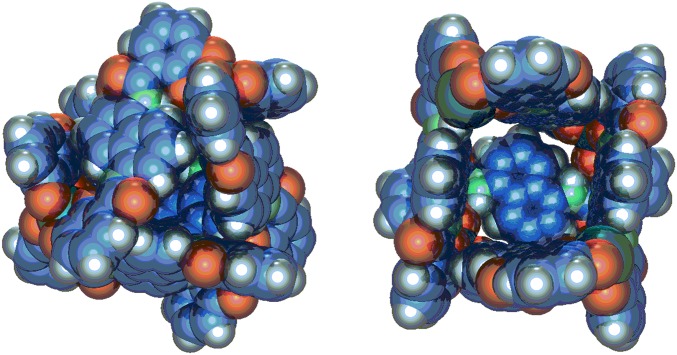
Fig. 1.
(Left) Space-filling model of K12Ga4L6 tetrahedron 1. (Right) Rotated image of 1, where a ligand has been removed to reveal the tetrahedron’s interior.
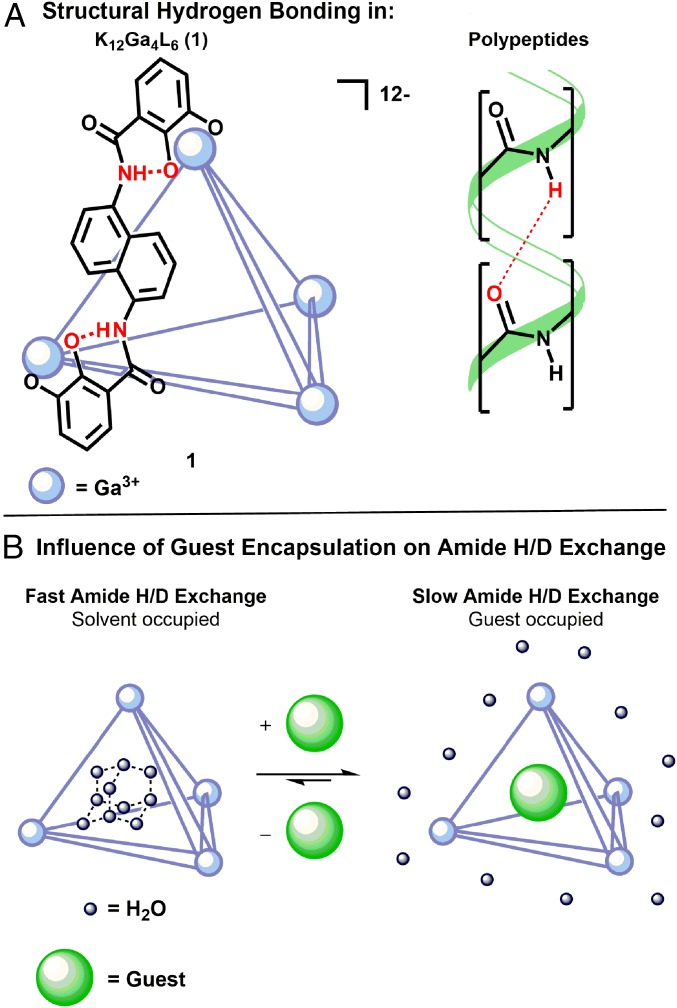
Fig. 2.
(A) Structurally important hydrogen bonding in tetrahedron 1 (lines represent ligands and spheres represent Ga3+ centers), and polypeptides. (B) General scheme of solvent displacement from 1 by a guest molecule.
Results and Discussion
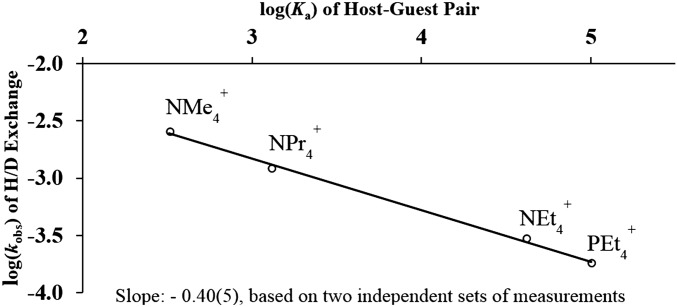
Initial studies revealed a puzzling relationship between the observed rate of amide HDX and the identity of the encapsulated guest. In the absence of guest, 1H NMR analysis revealed that 1 undergoes first-order amide HDX relatively rapidly, as evidenced by the loss of a single protio amide resonance at 13.1 parts per million (ppm) (100 mM phosphate buffer, pD 8.40, 25 °C). In the presence of a cationic guest, this rate decreases by a magnitude that is commensurate with the binding affinity of the host–guest pair. This relationship is represented in Fig. 3, where with excess guest, the log(kobs) of amide HDX correlates linearly with log(Ka) (42) of the encapsulated guest. Thus, although it was clear that the presence of a strongly bound cationic guest inhibits host amide HDX, further investigations were necessary to explain this relationship.
Fig. 3.
Linear free-energy relationship between log(Ka) and log(kobs) of amide HDX. All measurements were obtained using two equivalents of guest.
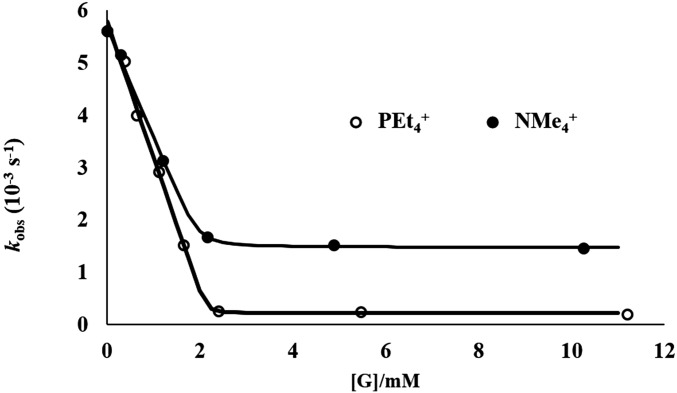
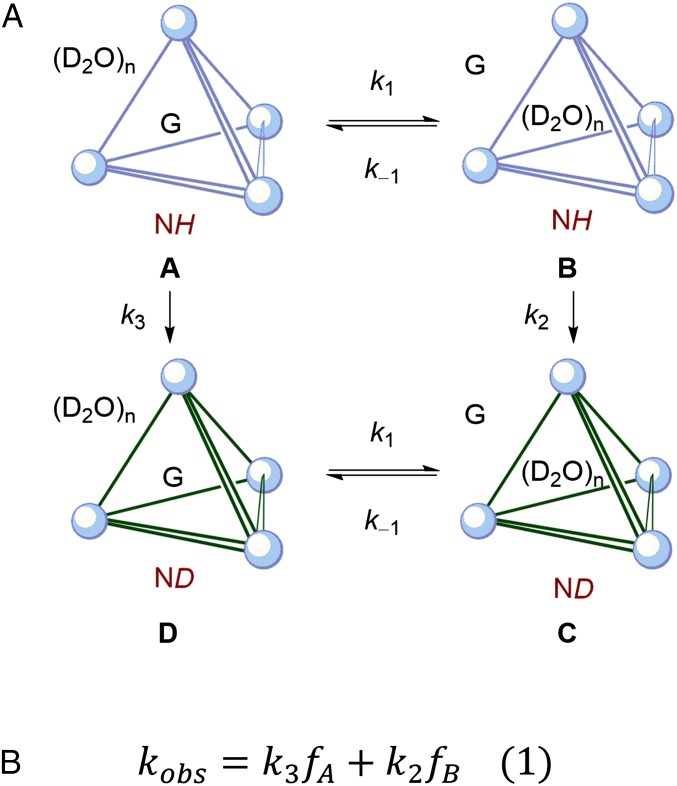
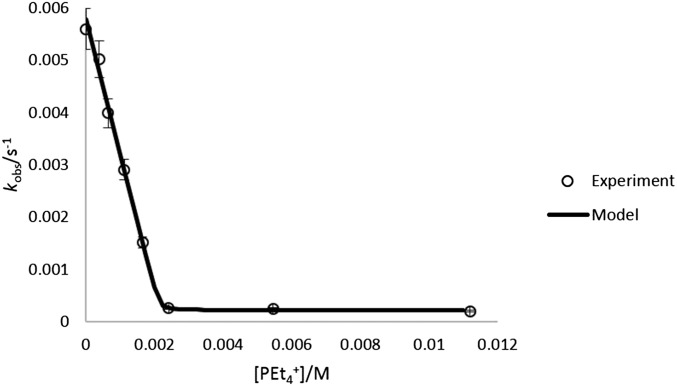
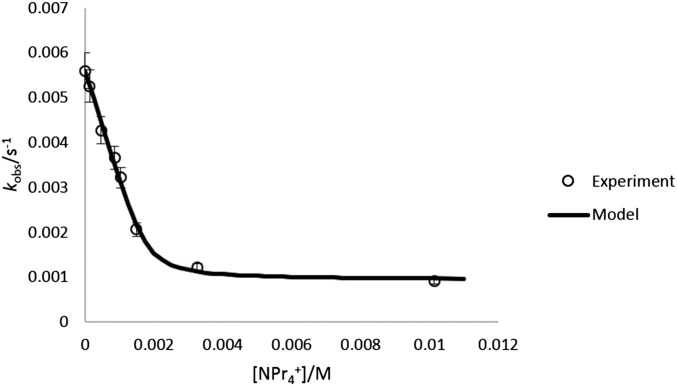
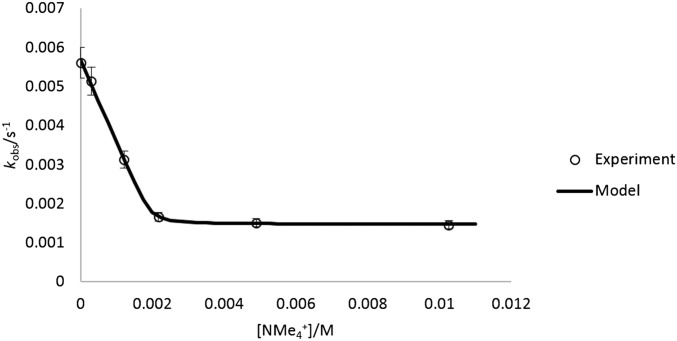
To probe the influence of guest encapsulation on the mechanism of amide HDX, 1 was treated with increasing concentrations of guest, and proton exchange rates were measured (pD 8.40). In all cases, the rate of amide HDX decreased with increasing guest concentration until one equivalent of guest had been added (Fig. 4). Increasing the concentration of added guest past one equivalent did not cause as large a decrease in kobs, suggesting that a saturation limit in guest had been obtained, which binds the interior of 1 and strongly inhibits proton exchange. In this manner, rate constants for HDX could be fit to kobs = k3fA + k2fB, where fA and fB are the mole fractions of species A and B, which may be characterized by the absence or presence of encapsulated water (Fig. 5). This fitting enabled k3 to be evaluated for G = NMe4+, NPr4+, and PEt4+ (see the Supporting Information). Based on this analysis, HDX proceeds up to 30× faster in B than A. We propose that encapsulated solvent provides a contribution to this difference (8–10 solvent molecules are expected to be encapsulated, based on thermogravimetric analysis) (42). It is also possible that HDX in the presence of excess G occurs simultaneously with guest exchange, as relevant guest self-exchange processes likely involve ligand distortion and occur 10–1,000× faster than the observed rates of HDX (43). Eyring analysis of HDX (G = PEt4+) revealed a large negative entropy of activation that is similar to the negative standard entropy of guest exchange and consistent with either hypothesis. The mechanism in Fig. 5 also provides a conceptual link between host–guest-influenced HDX and the Linderstrøm–Lang model of structurally hindered proton exchange by proteins (12, 13). Taken collectively, these data support a proton exchange mechanism involving encapsulated water, which can be modified by the presence of a competing guest.
Fig. 4.
Effect of increasing guest equivalents on kobs of amide HDX with G = PEt4+, NMe4+ (pD 8.40; lines represent fits of experimental points to Eq. 1. Error bars are omitted for clarity; see the Supporting Information).
Fig. 5.
(A) General guest-dependent mechanism for amide H/D exchange at constant pH and (B) rate law describing this process, where fA and fB are the fractions of host existing as A and B, respectively, and guest self-exchange is faster than k2, k3 (42).
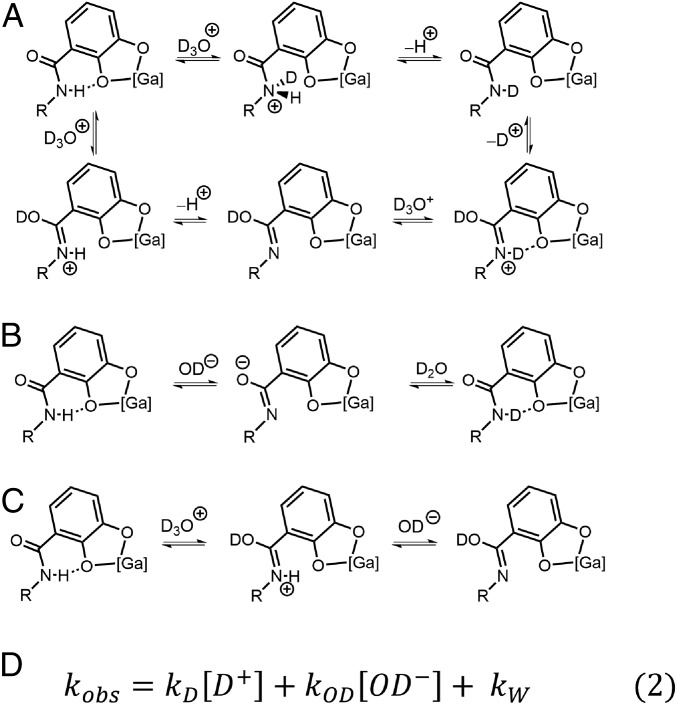
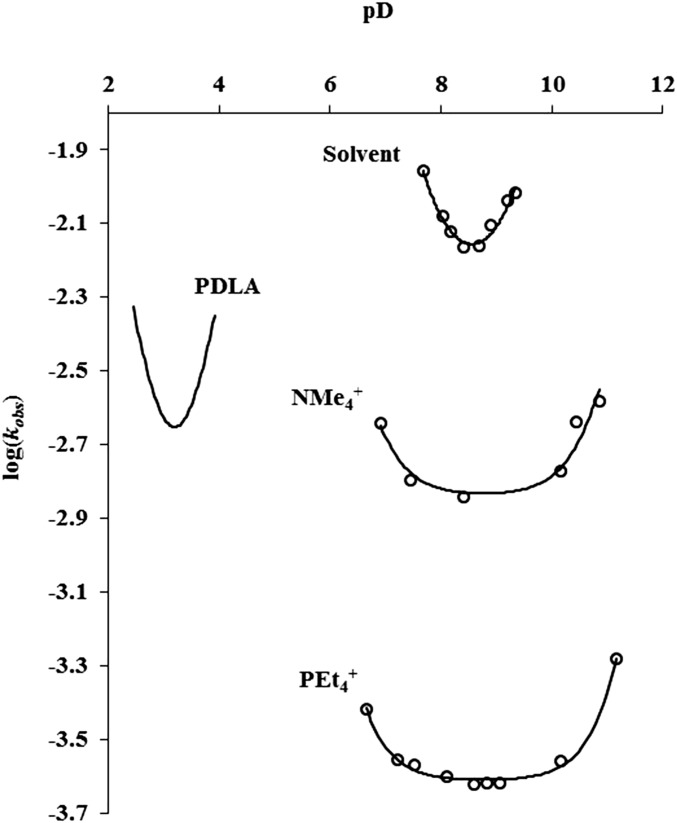
To assess the extent to which acid- and base-mediated proton exchange processes are accelerated or retarded in 1, the effects of bulk solution pD on HDX rates were investigated. Mechanistic studies have revealed that amide HDX proceeds by acid-, base-, and to a lesser extent, water-mediated mechanisms, of which the dominant mechanism depends on bulk solution pH or pD. Experimental rate constants obtained at various pD values were fit to the logarithmic form of Eq. 2 (in Fig. 7D), from which rate constants for exchange catalyzed by acid (kD), base (kOD), and water (kW) were obtained. These rate constants for host containing NMe4+, PEt4+, and solvent are tabulated in Table 1. The influences of the host are seen in the 105-fold acceleration of kD in solvent-filled host, and in the deceleration of kOD by 108-fold in PEt4+-filled host. For comparison, rate constants for a representative alanine polypeptide are included (44). The contributions of the various mechanisms are evident in the parabola-like relationship observed between log(kobs) of amide HDX and pD (Fig. 6).
Fig. 7.
Acid- (A), base- (B), and water- (C) mediated amide H/D exchange mechanisms, which can be described by (D) Eq. 2.
Table 1.
Rates of amide H/D exchange in 1
| Rate constant | PEt4+⊂1 | NMe4+⊂1 | (D2O)n⊂1 | PDLA* |
| logkD M−1⋅s−1 | 2.80 ± 0.03 | 3.81 ± 0.14 | 5.41 ± 0.04 | 0.0171 |
| logkOD M−1⋅s−1 | 0.16 ± 0.02 | 1.15 ± 0.09 | 3.16 ± 0.04 | 8.48 |
| logkW s−1 | −3.62 ± 0.01 | −2.84 ± 0.03 | −2.25 ± 0.02 | −3.04 |
Rate constants (298 K) for PDLA were calculated using Arrhenius parameters from ref. 44.
Fig. 6.
Influence of bulk solution pD on kobs of amide HDX. Varying the presence and type of guest affords order-of-magnitude changes in kobs, whose minimum pD values are shifted up to 6 pD units relative to poly-DL-alanine (PDLA). Lines represent fits of experimental kobs values to Eq. 2 (Fig. 7D; see Table 1 for rate constants).
It is apparent that the rate minimum for HDX in 1 occurs at pD ∼9, considerably higher than the minimum at pH ∼3 for polypeptides in aqueous solution. This minimum is where the dominant mechanism of amide HDX switches from acid to base catalysis. The shift of the minimum for 1 corresponds to a 200,000-fold increase in kD and decreases in kOD [compare (D2O)n⊂1; Table 1] relative to a representative polypeptide. These dramatic differences can be understood in terms of the mechanisms for acid and base catalysis, shown in Fig. 7 (45). Regardless of whether the acid-catalyzed exchange proceeds via N protonation (more likely in a polyanionic host) or via the imidic acid, the transition state bears a positive charge. In contrast, the transition state for the base-catalyzed exchange bears a negative charge. The positive charge is stabilized and the negative charge is destabilized by electrostatics, resulting from the polyanionic nature of 1. Similar behavior was seen in amide HDX in anionic (and cationic) micelles (9).
In the presence of a guest (NMe4+ or PEt4+), the pD-dependent curves in Fig. 5 are lowered and flattened. The data in Table 1 show that this is because both kD and kOD are decreased 40- to 100-fold for NMe4+ and 400- to 1,000-fold for PEt4+. This effect is not simply electrostatic, as it operates on both kD and kOD. Instead, when the interior of 1 is blocked by G, both acid- and base-mediated exchanges are retarded. We attribute this difference to a steric effect, whereby the guest interferes with the HDX, but we cannot be more specific as to how this operates.
It is surprising that kW is hardly affected by the presence of G. The range of variation is only about 10-fold, and kW is nearly the same as for a representative polypeptide. It is unusual for kW to be detectable above the errors in measuring the contributions of kD and kOD. What makes detection possible in the presence of G is that both of those contributions are retarded relative to the water-mediated pathway, leading to curves that are flattened near the pD minimum, as can be seen in Fig. 6.
Evidence for the water-mediated pathway has historically been tenuous, which has limited the establishment of a firm mechanism. Water-mediated HDX is highly unlikely to proceed simply through NH removal by D2O, as this process would have a maximum rate constant of 10−5 s−1, based on an amide pKa of ∼15 and an encounter-controlled reverse rate constant of 1010 M−1⋅s−1. We instead propose the mechanism shown in Fig. 7C. Ordinarily this path too is unlikely, with a rate constant of 10−6 s−1, based on a pKa of about −1 for the O-protonated amide, a pKW of ∼15 in D2O, and an encounter-controlled rate constant of 1010 M−1⋅s−1 for deprotonation by OD−. However, here the negative charge on the host can raise the pKa of the O-protonated amide up to 4 units (46), and retard encounter only slightly, leading to an estimate consistent with the observed kW. However, we cannot explain why the steric effect that retards kD and kOD is not also operative on kW.
In summary, the observed rates of amide HDX in biologically inspired, anionic K12Ga4L6 supramolecular assemblies 1 were correlated to the affinities of alkylammonium and phosphonium cations for the host interior. Guest titration experiments and modeling suggest that amide HDX proceeds through two different pathways contingent on the presence or absence of a guest, justifying this correlation. The rates of HDX catalyzed by acid and base are shifted by a factor of up to 200 million relative to those of an alanine polypeptide, an effect that we attribute to the polyanionic nature of 1. Collectively, these experiments suggest that amide HDX occurs through encapsulation of water and that the unique electrostatics of the host dramatically alters reactivity of this transient hydrate. These results improve both our understanding of water in molecular recognition, and contributions of proton transfer to multistep acid catalysis in supramolecular cavities. Knowledge of these properties may enable the more judicious development of both synthetic and biomolecular microenvironments. More broadly, these insights are envisioned to link the fundamental processes of hydron transfer and enzyme-mimic structure, thereby providing a significant step toward the development of catalysts that rival the utility of biological enzymes.
Experimental
General Procedures, Instrumentation, and Materials.
Unless otherwise noted, reactions and manipulations were performed using standard Schlenk techniques or in an oxygen-free wet box. NMR spectra were obtained on Bruker AV-500 or AV-600 spectrometers. Chemical shifts are reported as δ in ppm relative to residual protonated solvent resonances. Unless otherwise noted, chemicals were obtained from commercial suppliers and used without further purification. The synthesis of 1 and complexes with NMe4+, NPr4+, NEt4+, and PEt4+ have been described (33, 43). All solvents were degassed under N2 for 20 min before use. Buffered solutions were made by titrating a K3PO4 solution with DCl until the desired pD was reached. Reported pD and [OD–] values are corrected for the glass electrode artifact (i.e., pD = pHread + 0.40) (47) and for solvent dissociation constants (48). Least-squares fits were obtained using Origin (OriginLab) or Microsoft Excel. Rate constants were fit to log(kobs) = log(kD[D+] + kOD[OD–] + kW). Association constants (Ka) for the encapsulation of NMe4+ [log(Ka/M−1) = 2.51(1)] and NPr4+ [log(Ka/M−1) = 3.12(5)] were measured by NMR, UV-vis, and isothermal titration calorimetry (42). The association constants for PEt4+ and NEt4+ have been reported previously (42, 43).
General Procedure for Amide HDX Kinetic Experiments.
In a typical experiment, host 1 (5 mg, 1.4 μmol) was treated with 0–5 equivalents of NMe4Cl, NPr4Br, NEt4Cl, or PEt4I in H2O. After 10 min, water was removed in vacuo, affording a yellow solid. This solid was then dissolved in 500 µL 100 mM deuterated phosphate buffer, after which the resulting solution was transferred to a standard NMR tube. The tube was then inserted in a preheated NMR probe [25.0(1) °C] and the reaction was followed within 3 min of its preparation with single-scan 1H spectra acquired every 20–60 s. Decay of the integral of an amide NH resonance was assessed against either the aromatic resonances of 1 or an internal standard of sodium 3-trimethylsilylpropanesulfonate. First-order rate constants were obtained using the equation I = (Io − baseline)exp(−kobs) + baseline, where Io is the initial NH integral (44). In the case of solvent-filled host, only the final ∼20% conversion was observed and kobs was obtained using an adjustable baseline parameter. Initial rate kinetics were used to obtain kobs where the reaction was slower, such as in the presence of PEt4+. To reliably fit kobs in the cases of slow HDX, the baseline parameter was constrained to zero. Large differences in KCl concentration, i.e., 20 and 570 mM, afforded no measurable change in HDX rate [krel = 1.0(1)], demonstrating that this salt does not influence the HDX rates. Guest exchange of PEt4+ for NEt4+ was likewise unaffected by moderate changes in bulk solution acidity [pDcorr 6.47 and 10.45; krel = 1.1(1)]. Triplicate measurements of HDX [solvent-filled host, 25.0(1) °C, pD 8.40] had an SE in kobs of 7%, which is reflected in the fits of the next section.
Fits of Eq. 1 to kobs with Variable [G].
The plots below represent experimental kobs of amide HDX at variable [G], presented with fitted kobs values using Eq. 1 (see the main text). Fitted kobs were obtained using least-squares fitting of log(kobs) to the experimental log(kobs) data. Parameters K, [H]T, k2, and k3 were adjustable and [H] was calculated by solving the quadratic (Eq. S4) derived below, thereby enabling calculation of fA and fB from K, [H]T, and [G]T. The quadratic (Eq. S4) is obtained from Eqs. S1 and S2, where [H]T is the sum of [HG] and [H], and [G]T is the sum of [HG] and [G]. Notably, fitted K values in the fittings below are larger than independently determined encapsulation constants (see General Procedures, Instrumentation, and Materials). We attribute this difference to (i) steric effects resulting from external association and guest exchange processes, and (ii) the possible existence of a partially solvent-filled host that exhibits a slow HDX. Nonetheless, k2 and [H]T are in agreement with experimentally determined values. Close agreement between kW and k3 was also observed.
| [S1] |
| [S2] |
| [S3] |
| [S4] |
Fig. S1.
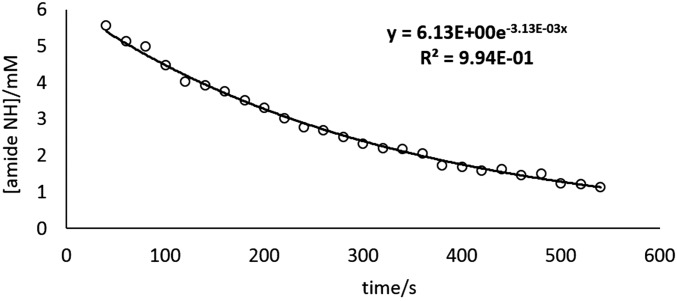
Representative time vs. concentration plot. Typical concentration of amide NH vs. time plot used to obtain kobs for HDX (guest = NMe4+), where the baseline parameter is constrained to zero. Similar results are obtained when the baseline parameter is adjustable (Io = 6.35 mM, baseline = 0.31 mM, k = 3.61 × 10−3 s−1).
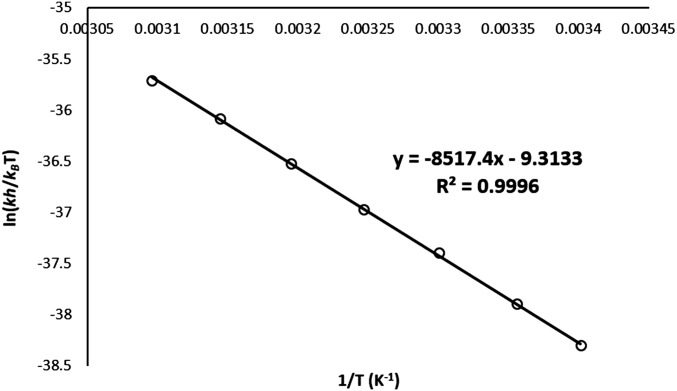
Fig. S2.
Eyring plot representing the dependence of amide HDX kobs on temperature [guest = PEt4+; pD 8.40, ∆H‡: 71(1) kJ·mol−1; ∆S‡: −77(2) J·mol−1⋅K−1].
Fig. S3.
Fit of amide HDX kobs to Eq. 1 (model) with variable concentrations of PEt4+ (k2 = 5.8 × 10−3 s−1, k3 = 2.2 × 10−4 s−1, logK = 5.7, pD 8.40; [H]T = 2.2 mM).
Fig. S4.
Fit of amide HDX kobs to Eq. 1 (model) with variable concentrations of NPr4+ (k2 = 5.6 × 10−3 s−1, k3 = 9.3 × 10−4 s−1, logK = 4.2, pD 8.40; [H]T = 1.8 mM).
Fig. S5.
Fit of amide HDX kobs to Eq. 1 (model) with variable concentrations of NMe4+ (k2 = 5.7 × 10−3 s−1, k3 = 1.5 × 10−3 s−1, logK = 4.9, pD 8.40; [H]T = 2.0 mM).
Acknowledgments
The authors are grateful to Drs. Derek Dalton, Kaking Yan, Chen Zhao, Tiffany Pham, David Tatum, and Michael Nippe, as well as Cynthia Hong, Rebecca Triano, and David Kaphan for helpful discussions. This research was supported by the Director, Office of Science, Office of Basic Energy Sciences, and the Division of Chemical Sciences, Geosciences, and Biosciences of the US Department of Energy at Lawrence Berkeley National Laboratory (DE-AC02-05CH11231).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515639112/-/DCSupplemental.
References
- 1.Buch-Pedersen MJ, Pedersen BP, Veierskov B, Nissen P, Palmgren MG. Protons and how they are transported by proton pumps. Pflugers Arch. 2009;457(3):573–579. doi: 10.1007/s00424-008-0503-8. [DOI] [PubMed] [Google Scholar]
- 2.Nagle JF, Tristram-Nagle S. Hydrogen bonded chain mechanisms for proton conduction and proton pumping. J Membr Biol. 1983;74(1):1–14. doi: 10.1007/BF01870590. [DOI] [PubMed] [Google Scholar]
- 3.Liang Z-X, Klinman JP. Structural bases of hydrogen tunneling in enzymes: Progress and puzzles. Curr Opin Struct Biol. 2004;14(6):648–655. doi: 10.1016/j.sbi.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Fillingame RH. H+ transport and coupling by the F0 sector of the ATP synthase: Insights into the molecular mechanism of function. J Bioenerg Biomembr. 1992;24(5):485–491. doi: 10.1007/BF00762366. [DOI] [PubMed] [Google Scholar]
- 5.Akeson M, Deamer DW. Proton conductance by the gramicidin water wire. Model for proton conductance in the F1F0 ATPases? Biophys J. 1991;60(1):101–109. doi: 10.1016/S0006-3495(91)82034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinman JP, Kohen A. Hydrogen tunneling links protein dynamics to enzyme catalysis. Annu Rev Biochem. 2013;82:471–496. doi: 10.1146/annurev-biochem-051710-133623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knapp MJ, Klinman JP. Environmentally coupled hydrogen tunneling. Linking catalysis to dynamics. Eur J Biochem. 2002;269(13):3113–3121. doi: 10.1046/j.1432-1033.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 8.Anderson JS, Hernández G, Lemaster DM. A billion-fold range in acidity for the solvent-exposed amides of Pyrococcus furiosus rubredoxin. Biochemistry. 2008;47(23):6178–6188. doi: 10.1021/bi800284y. [DOI] [PubMed] [Google Scholar]
- 9.Perrin CL, Chen J-H, Ohta BK. Amide proton exchange in micelles. J Am Chem Soc. 1999;121(11):2448–2455. [Google Scholar]
- 10.Leichtling BH, Klotz IM. Catalysis of hydrogen-deuterium exchange in polypeptides. Biochemistry (Mosc) 1966;5(12):4026–4037. [Google Scholar]
- 11.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Englander SW, Mayne L, Bai Y, Sosnick TR. Hydrogen exchange: The modern legacy of Linderstrøm-Lang. Protein Sci. 1997;6(5):1101–1109. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinner JJ, Lim WK, Bédard S, Black BE, Englander SW. Protein dynamics viewed by hydrogen exchange. Protein Sci. 2012;21(7):996–1005. doi: 10.1002/pro.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariga K, et al. Challenges and breakthroughs in recent research on self-assembly. Sci Technol Adv Mater. 2008;9(1):014109. doi: 10.1088/1468-6996/9/1/014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahsar KR, Schwartz DK, Medlin JW. Control of metal catalyst selectivity through specific noncovalent molecular interactions. J Am Chem Soc. 2014;136(1):520–526. doi: 10.1021/ja411973p. [DOI] [PubMed] [Google Scholar]
- 16.Soberats B, Sanna E, Martorell G, Rotger C, Costa A. Programmed enzyme-mimic hydrolysis of a choline carbonate by a metal-free 2-aminobenzimidazole-based cavitand. Org Lett. 2014;16(3):840–843. doi: 10.1021/ol403612e. [DOI] [PubMed] [Google Scholar]
- 17.Oshovsky GV, Reinhoudt DN, Verboom W. Supramolecular chemistry in water. Angew Chem Int Ed Engl. 2007;46(14):2366–2393. doi: 10.1002/anie.200602815. [DOI] [PubMed] [Google Scholar]
- 18.Breslow R. Artificial enzymes. Science. 1982;218(4572):532–537. doi: 10.1126/science.7123255. [DOI] [PubMed] [Google Scholar]
- 19.Breslow R. Biomimetic chemistry and artificial enzymes: Catalysis by design. Acc Chem Res. 1995;28(3):146–153. [Google Scholar]
- 20.Murase T, Nishijima Y, Fujita M. Cage-catalyzed Knoevenagel condensation under neutral conditions in water. J Am Chem Soc. 2012;134(1):162–164. doi: 10.1021/ja210068f. [DOI] [PubMed] [Google Scholar]
- 21.Habicher T, Diederich F, Gramlich V. Catalytic dendrophanes as enzyme mimics: Synthesis, binding properties, micropolarity effect, and catalytic activity of dendritic thiazolio-cyclophanes. Helv Chim Acta. 1999;82(7):1066–1095. [Google Scholar]
- 22.Houk KN, Leach AG, Kim SP, Zhang X. Binding affinities of host-guest, protein-ligand, and protein-transition-state complexes. Angew Chem Int Ed Engl. 2003;42(40):4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 23.Leung DH, Bergman RG, Raymond KN. Enthalpy-entropy compensation reveals solvent reorganization as a driving force for supramolecular encapsulation in water. J Am Chem Soc. 2008;130(9):2798–2805. doi: 10.1021/ja075975z. [DOI] [PubMed] [Google Scholar]
- 24.Wang L-Y, Yang Y, Liu K, Li B-L, Zhang Y. A new “opened-cube” (H2O)10 cluster and undulated water chain in porous metal−organic frameworks. Cryst Growth Des. 2008;8(11):3902–3904. [Google Scholar]
- 25.Lakshminarayanan PS, Suresh E, Ghosh P. A hybrid water-chloride structure with discrete undecameric water moieties self-assembled in a heptaprotonated octaamino cryptand. Angew Chem Int Ed Engl. 2006;45(23):3807–3811. doi: 10.1002/anie.200600254. [DOI] [PubMed] [Google Scholar]
- 26.Hazra A, Kanoo P, Mohapatra S, Mostafa G, Maji TK. A flexible supramolecular host with a crowned chair octameric water cluster and highly selective adsorption properties. CrystEngComm. 2010;12(10):2775. [Google Scholar]
- 27.Li Y, Jiang L, Feng X-L, Lu T-B. Anion dependent water clusters encapsulated inside a cryptand cavity. Cryst Growth Des. 2008;8(10):3689–3694. [Google Scholar]
- 28.Mandal S, et al. An unusual (H(2)O)(20) discrete water cluster in the supramolecular host of a charge transfer platinum(ii) complex: Cytotoxicity and DNA cleavage activities. Dalton Trans. 2010;39(40):9514–9522. doi: 10.1039/c0dt00527d. [DOI] [PubMed] [Google Scholar]
- 29.Kurotobi K, Murata Y. A single molecule of water encapsulated in fullerene C₆₀. Science. 2011;333(6042):613–616. doi: 10.1126/science.1206376. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Lin H, Mu B, Tian A, Liu G. Encapsulation of discrete (H(2)O)(12) clusters in a 3D three-fold interpenetrating metal-organic framework host with (3,4)-connected topology. Dalton Trans. 2010;39(27):6187–6189. doi: 10.1039/c0dt00302f. [DOI] [PubMed] [Google Scholar]
- 31.Yoshizawa M, et al. Endohedral clusterization of ten water molecules into a “molecular ice”within the hydrophobic pocket of a self-assembled cage. J Am Chem Soc. 2005;127(9):2798–2799. doi: 10.1021/ja043953w. [DOI] [PubMed] [Google Scholar]
- 32.Massera C, Melegari M, Ugozzoli F, Dalcanale E. Formation of tetrameric water clusters driven by a cavitand template. Chem Commun (Camb) 2010;46(1):88–90. doi: 10.1039/b917931c. [DOI] [PubMed] [Google Scholar]
- 33.Caulder DL, Powers RE, Parac TN, Raymond KN. The self-assembly of a predesigned tetrahedral M4L6 supramolecular cluster. Angew Chem Int Ed. 1998;37(13-14):1840–1843. [Google Scholar]
- 34.Zhao C, et al. Chiral amide directed assembly of a diastereo- and enantiopure supramolecular host and its application to enantioselective catalysis of neutral substrates. J Am Chem Soc. 2013;135(50):18802–18805. doi: 10.1021/ja411631v. [DOI] [PubMed] [Google Scholar]
- 35.Janser I, Albrecht M, Hunger K, Burk S, Rissanen K. Formation of triple-stranded dinuclear helicates with dicatecholimine ligands: The influence of steric hindrance at the spacer. Eur J Inorg Chem. 2006;2006(1):244–251. [Google Scholar]
- 36.Zhao C, Toste FD, Raymond KN, Bergman RG. Nucleophilic substitution catalyzed by a supramolecular cavity proceeds with retention of absolute stereochemistry. J Am Chem Soc. 2014;136(41):14409–14412. doi: 10.1021/ja508799p. [DOI] [PubMed] [Google Scholar]
- 37.Hart-Cooper WM, et al. The effect of host structure on the selectivity and mechanism of supramolecular catalysis of Prins cyclizations. Chem Sci (Camb) 2015;6(2):1383–1393. doi: 10.1039/c4sc02735c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart-Cooper WM, Clary KN, Toste FD, Bergman RG, Raymond KN. Selective monoterpene-like cyclization reactions achieved by water exclusion from reactive intermediates in a supramolecular catalyst. J Am Chem Soc. 2012;134(43):17873–17876. doi: 10.1021/ja308254k. [DOI] [PubMed] [Google Scholar]
- 39.Pluth MD, Bergman RG, Raymond KN. Proton-mediated chemistry and catalysis in a self-assembled supramolecular host. Acc Chem Res. 2009;42(10):1650–1659. doi: 10.1021/ar900118t. [DOI] [PubMed] [Google Scholar]
- 40.Fiedler D, van Halbeek H, Bergman RG, Raymond KN. Supramolecular catalysis of unimolecular rearrangements: Substrate scope and mechanistic insights. J Am Chem Soc. 2006;128(31):10240–10252. doi: 10.1021/ja062329b. [DOI] [PubMed] [Google Scholar]
- 41.Hastings CJ, Pluth MD, Bergman RG, Raymond KN. Enzymelike catalysis of the Nazarov cyclization by supramolecular encapsulation. J Am Chem Soc. 2010;132(20):6938–6940. doi: 10.1021/ja102633e. [DOI] [PubMed] [Google Scholar]
- 42.Sgarlata C, et al. External and internal guest binding of a highly charged supramolecular host in water: Deconvoluting the very different thermodynamics. J Am Chem Soc. 2010;132(3):1005–1009. doi: 10.1021/ja9056739. [DOI] [PubMed] [Google Scholar]
- 43.Davis AV, et al. Guest exchange dynamics in an M4L6 tetrahedral host. J Am Chem Soc. 2006;128(4):1324–1333. doi: 10.1021/ja056556+. [DOI] [PubMed] [Google Scholar]
- 44.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrin CL. Proton exchange in amides: Surprises from simple systems. Acc Chem Res. 1989;22(8):268–275. [Google Scholar]
- 46.Pluth MD, Bergman RG, Raymond KN. Making amines strong bases: Thermodynamic stabilization of protonated guests in a highly-charged supramolecular host1. J Am Chem Soc. 2007;129(37):11459–11467. doi: 10.1021/ja072654e. [DOI] [PubMed] [Google Scholar]
- 47.Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. J Phys Chem. 1960;64(1):188–190. [Google Scholar]
- 48.Covington AK, Robinson RA, Bates RG. The ionization constant of deuterium oxide from 5 to 50°. J Phys Chem. 1966;70(12):3820–3824. [Google Scholar]