Significance
Transcription factors typically bind to more sites than are functionally affected upon transcription factor inactivation. What, then, determines whether transcription factor binding impacts gene expression? Here we address this question by investigating Rho, the essential transcription termination factor that associates with most newly transcribed RNAs in bacteria, but promotes transcription termination only in a fraction of these transcripts. We uncover a novel RNA element that sequesters Rho in an inactive complex, thereby advancing transcription without affecting Rho binding. Our results indicate that the site of action of transcription factors is defined not only by sequences that mediate their recruitment but also by sequences that antagonize their activity.
Keywords: Rho-dependent termination, antitermination, Salmonella, MgtC, transcriptional polarity
Abstract
The transcription termination factor Rho associates with most nascent bacterial RNAs as they emerge from RNA polymerase. However, pharmacological inhibition of Rho derepresses only a small fraction of these transcripts. What, then, determines the specificity of Rho-dependent transcription termination? We now report the identification of a Rho-antagonizing RNA element (RARE) that hinders Rho-dependent transcription termination. We establish that RARE traps Rho in an inactive complex but does not prevent Rho binding to its recruitment sites. Although translating ribosomes normally block Rho access to an mRNA, inefficient translation of an open reading frame in the leader region of the Salmonella mgtCBR operon actually enables transcription of its associated coding region by favoring an RNA conformation that sequesters RARE. The discovery of an RNA element that inactivates Rho signifies that the specificity of nucleic-acid binding proteins is defined not only by the sequences that recruit these proteins but also by sequences that antagonize their activity.
Transcription factors recognize sequence and structural elements in DNA and RNA to turn genes on or off. The transcription termination factor Rho is responsible for the majority of factor-dependent termination events in enteric bacterial species (1). Rho prevents the production of dysfunctional, and potentially dangerous, RNAs (2). For instance, Rho implements transcriptional polarity, a process whereby compromised translation of a promoter-proximal gene reduces transcription of downstream genes in an operon (3–5).
Rho is a hexameric helicase that binds RNA and translocates in the 5′-to-3′ direction using the energy derived from ATP hydrolysis. Initially, RNA is anchored to Rho primary binding sites on the surface of the hexamer. Rho recruitment sites tend to be rich in Cs and Us and free of strong secondary structures (6–8). Interaction with a recruitment site causes the Rho hexamer to open briefly, allowing the RNA 3′ region to pass through the center, where secondary binding sites are located (9, 10). Further contact between the RNA and the secondary binding sites stimulates Rho's ATPase activity (11). Using the energy derived from ATP hydrolysis, Rho translocates along an RNA until it reaches a paused RNA polymerase (RNAP) and promotes transcription termination (12).
The specificity of Rho-dependent transcription termination has remained enigmatic because Rho associates with many newly transcribed RNAs in Escherichia coli (13), but only a portion of these messages is affected when bacteria are treated with bicyclomycin (BCM) (1), a Rho-specific inhibitor (14). In other words, binding of Rho to a particular RNA is not sufficient for subsequent transcription termination.
Rho-dependent terminators are typically found at the end of operons; however, they can also be present in the leader region of mRNAs, where they perform regulatory functions (15–17). This appears to be the case for the 296-nt-long leader region of the mgtCBR transcript from Salmonella enterica serovar Typhimurium, which controls transcription elongation into its associated coding region (18, 19) but lacks sequences that resemble an intrinsic terminator [i.e., a G + C-rich stem-loop followed by a stretch of Us (12)]. The mgtCBR operon specifies the virulence protein MgtC (20), the Mg2+ transporter MgtB (21), and the regulatory peptide MgtR (22).
We now report the identification of a Rho-antagonizing RNA element (RARE) in the mgtCBR leader that prevents transcription termination by factor Rho. We establish that RARE traps Rho in a nonfunctional state (as opposed to hindering Rho binding to its recruitment sites), and we define the sequences and positions governing RARE activity. In contrast to transcriptional polarity, wherein a translating ribosome protects RNA from Rho invasion (12), translation of a short open reading frame (ORF) in the mgtCBR leader actually compromised RARE action, thereby preventing transcriptional readthrough into the associated coding region. Our findings indicate that the arrangement of genetic elements governing Rho activity, rather than simply the coupling of transcription and translation, determines how translation of an upstream ORF influences downstream transcription. Furthermore, they imply that nucleic-acid binding proteins manifest specificity both by sequences that recruit these proteins and by sequences that hinder their function.
Results
The mgtCBR Leader Harbors a Rho-Dependent Transcription Terminator.
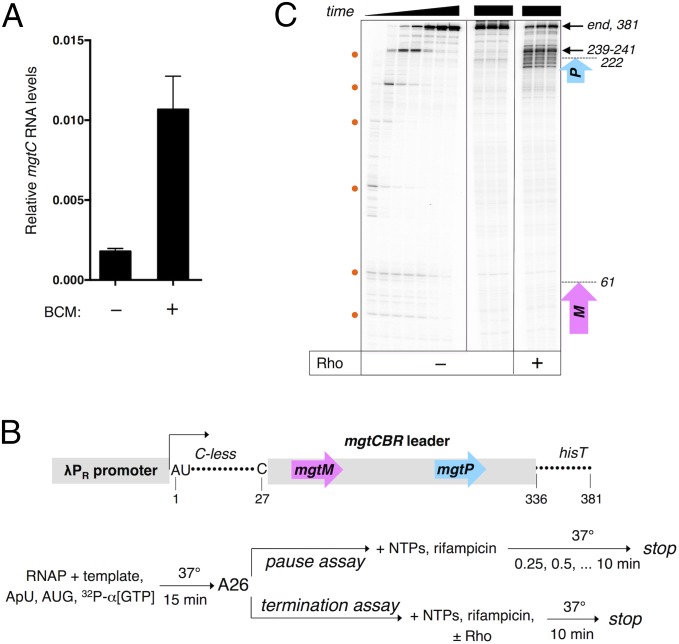
We determined that the mgtCBR leader includes a Rho-dependent terminator because the mRNA levels of the mgtC coding region increased sixfold when bacteria were treated with the Rho-specific inhibitor BCM (Fig. 1A). To identify the site of transcription termination in the mgtCBR leader, we carried out single-round in vitro transcription assays with purified RNAP and a DNA template that contained the λPR promoter and a 26-nt long C-less region followed by the sequence corresponding to the mgtCBR leader (Fig. 1B). On this template, transcription elongation complexes halted by the omission of CTP can resume transcription upon addition of all four NTPs.
Fig. 1.
The mgtCBR transcript harbors a Rho-dependent transcription terminator within its leader. (A) The Rho-specific inhibitor BCM derepresses mgtC transcription in vivo. mRNA levels of the mgtC coding region produced by wild-type Salmonella (14028s) in the presence or absence of BCM as determined by qRT-PCR. Cells were harvested after 3 h of growth in N-minimal medium with 10 μM Mg2+ followed by a 15-min treatment with BCM. Shown are the mean and SD from at least three independent experiments. (B) Schematic of the linear DNA template and experimental strategy used in the pause and termination assays. (C) A representative 6% denaturing gel of pause and termination assays performed as described in (B). Pause assay is shown (Left) and positions of pause sites are indicated by the orange dots. Termination assay performed in triplicate is shown in the next three panels. Positions of the pause sites and termination products mapped in the presence of chain-terminating NTPs are indicated on the Right (Materials and Methods). The purple and blue arrows show the positions of the mgtM and mgtP ORFs located in the mgtCBR leader.
We detected bands that faded away over time (Fig. 1C), which correspond to RNA products of RNAPs pausing at several sites until they reach the end of the template. In the absence of Rho, the majority of the RNAPs reached the end of the template (Fig. 1C), supporting the notion that the mgtCBR leader lacks an intrinsic transcription terminator. Addition of Rho to the in vitro transcription reaction gave rise to prematurely terminated RNA products (Fig. 1C). The 3′ ends of these termination products were distributed from positions 130–290 relative to the transcription start site, with the strongest termination site located at positions 239–241 (Fig. 1C). This site corresponds to the most prominent pause site in the mgtCBR leader (Fig. 1C), consistent with Rho preferentially inducing RNA release from paused elongation complexes (12). Together, these results demonstrate that the mgtCBR leader contains a Rho-dependent transcription terminator.
The Conformation of the mgtCBR Leader Dictates Rho-Dependent Termination.
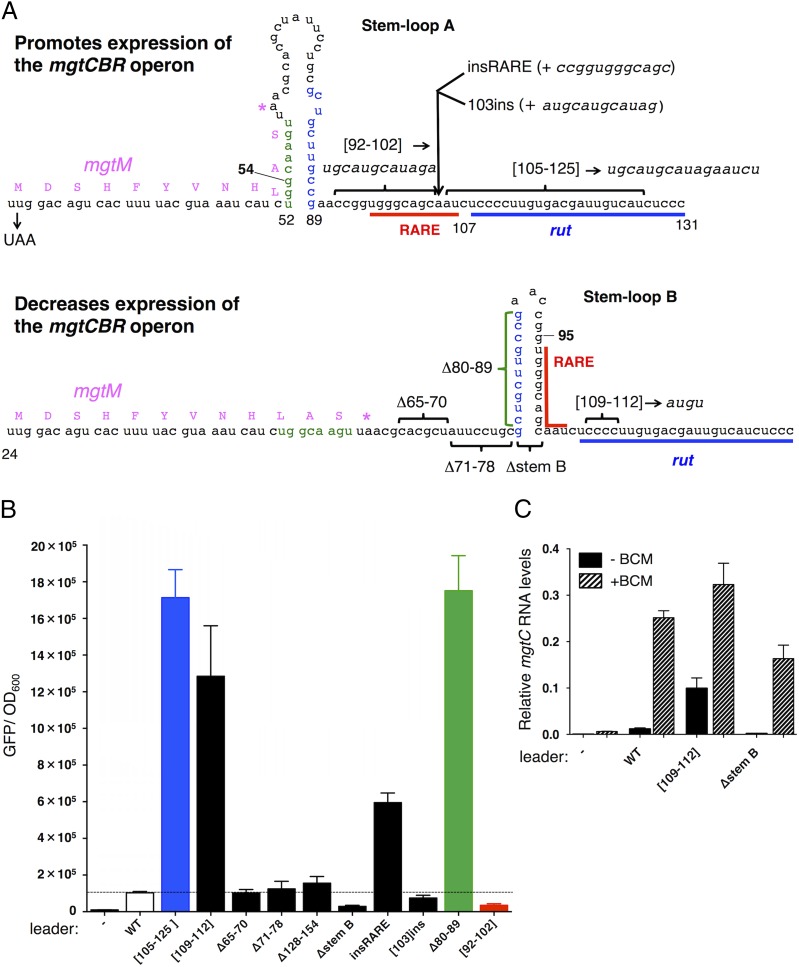
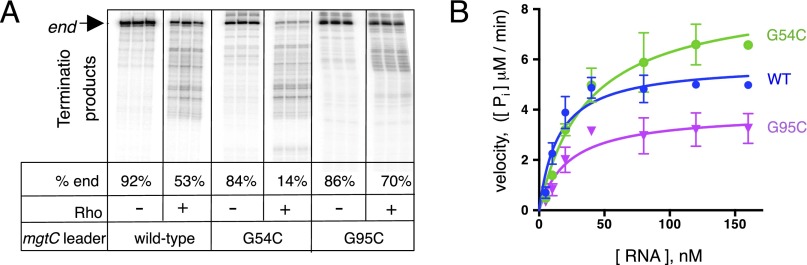
The mgtCBR leader harbors two short ORFs, mgtM and mgtP, within or immediately adjacent to sequences with the ability to adopt alternative stem-loop structures (Figs. 1B and 2A) (18, 19). Mutations favoring one of these structures (i.e., stem-loop A) advance transcription into the coding region, whereas those furthering the alternative structure (i.e., stem-loop B) promote transcription termination within the mgtCBR leader (Fig. 2A) (18). We investigated the ability of Rho to terminate transcription in mgtCBR leader variants genetically locked in the stem-loop A or B conformations. The G54C substitution, which hinders formation of stem-loop A (Fig. 2A) (18), increased Rho-dependent termination in vitro (Fig. S1A). By contrast, the G95C substitution, which prevents formation of stem-loop B (Fig. 2A), reduced the fraction of prematurely terminated products (Fig. S1A). We concluded that stem-loops A and B exert their regulatory effects directly by influencing Rho’s ability to terminate transcription in the absence of any additional factors.
Fig. 2.
RARE prevents Rho-dependent termination in the mgtCBR leader RNA. (A) Schematic of two alternative conformations that the 5′ end of the mgtCBR leader RNA can adopt (18). Positions of insertions are indicated by arrows, deletions and substitutions by brackets, and mutations that selectively destabilize stem-loops A and B by numbers in bold face. The calculated ∆G values for stem-loops A and B are −14.8 kcal/mole and −17.8 kcal/mole, respectively. (B) Fluorescence levels exhibited by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion between the wild-type mgtCBR promoter and leader, or mgtCBR leader variants, to a promotorless gfp gene. (−) corresponds to wild-type Salmonella (14028s) harboring the plasmid vector pFPV25. Shown are the mean and SD from at least three independent experiments. (C) gfp mRNA levels produced by wild-type Salmonella (14028s) harboring a plasmid with a transcriptional fusion of the wild-type mgtCBR promoter and leader, or mgtCBR leader variants with the Cs at positions 109–112 substituted by AUGU, or deleted for nucleotides 79–103 (∆stemB), to a promotorless gfp gene. mRNA levels were determined by qRT-PCR. Cells were harvested after 3 h growth in N-minimal medium with 10 μM Mg2+ followed by a 15-min treatment with BCM. (−) corresponds to wild-type Salmonella (14028s) harboring the plasmid vector pFPV25. Shown are the mean and SD from at least three independent experiments.
Fig. S1.
Conformation of the mgCBR leader mRNA controls Rho recruitment. (A) A representative 6% denaturing gel of in vitro transcription termination assay with templates containing the wild-type mgtCBR leader or variants with the G54C or G95C substitutions, which hinder formation of stem-loops A and B, respectively. Assays were performed in triplicate as described in Materials and Methods. Only the relevant portion of the gel is shown. The percentage of the full-length transcripts (end) was determined from at least three independent experiments. (B) Stimulation of Rho’s ATPase activity by the wild-type mgtCBR leader RNA or variants with the G54C or G95C substitutions. Kinetics of Rho ATPase activity were measured by monitoring the accumulation of free phosphate with increasing concentrations of RNA. Each data point is an average of at least three independent experiments (see Materials and Methods for details).
A Rho Recruitment Site Is Necessary for Transcription Termination in the mgtCBR Leader.
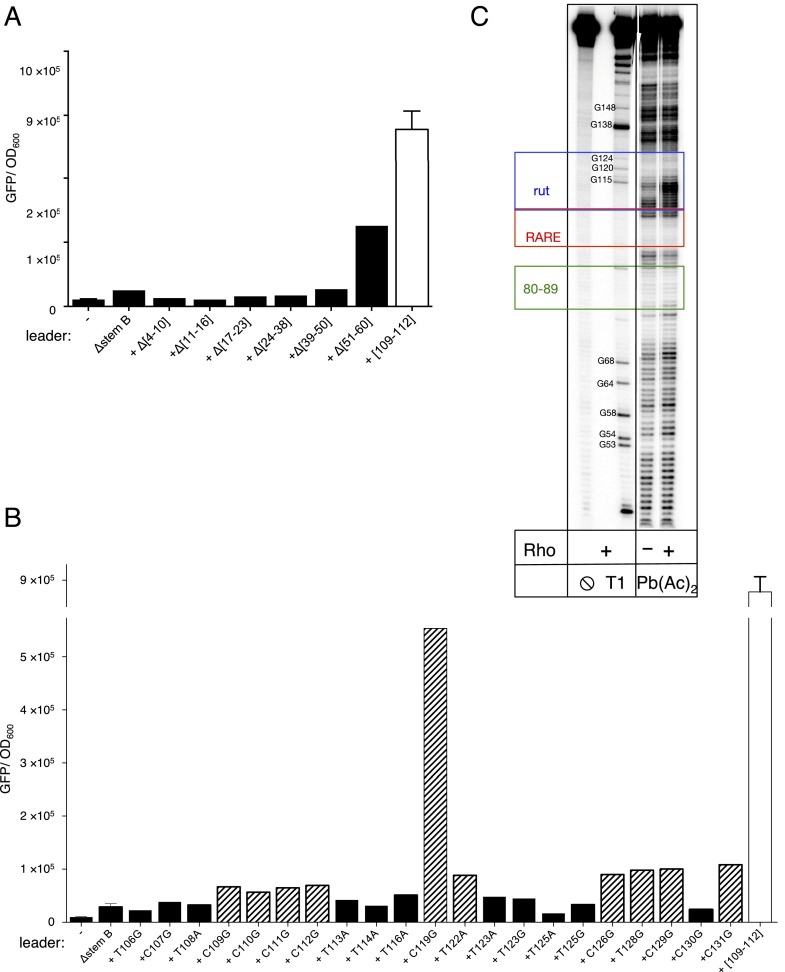
Transcription termination by Rho occurs in several steps, beginning with Rho binding to a rut (Rho utilization) site in a transcript (2, 23, 24). We identified a potential rut site immediately downstream of stem-loop B (Fig. 2A). Substitution of nucleotides in this region (positions 105–125) dramatically increased transcription of the associated coding region in vivo (Fig. 2B, [105–125]), and this was also the case when only four C residues were substituted by AUGU (Fig. 2B, [109–112]), indicating that the identified region functions as an authentic rut site. By contrast, deletion of nucleotides upstream and downstream of this site had no effect on transcription elongation into the coding region (Fig. 2B, ∆65–70, ∆71–78, ∆128–154, and Fig. S2A).
Fig. S2.
Sequences of the mgtCBR leader regulating Rho-dependent termination. (A) Fluorescence levels exhibited by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion between the mgtCBR promoter and mutant leader variants to a promotorless gfp gene after 4 h of growth in N-minimal medium with 10 μM Mg2+. (−) corresponds to wild-type Salmonella harboring the plasmid vector pFPV25. The data demonstrate that the first 60 nt of the mgtCBR leader do not contribute to Rho-dependent termination. Note that all substitutions were made in a derivative of the mgtCBR leader deleted for stem-loop B (Fig. 2A, Δstem B) to exclude the possibility of mutations generating sequence combinations that can adopt a secondary structure alternative to stem-loop B. Shown are the mean and SD from at least three independent experiments. (B) Fluorescence levels exhibited by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion of the mgtCBR promoter and mutant leader variants to a promotorless gfp gene after 4 h of growth in N-minimal medium with 10 μM Mg2+. The data show the contribution of individual pyrimidines at positions 106–131 of the mgtCBR leader to Rho activity. To exclude the possibility of mutations generating sequence combinations that can adopt a secondary structure alternative to stem-loop B, all substitutions were made in a derivative of the mgtCBR leader deleted for stem-loop B (Fig. 2A, Δstem B). The fluorescence levels exhibited by the wild-type Salmonella (14028s) harboring a mutant variant of the reporter with substitutions in the putative Rho-binding site ([109–112]) combined with Δstem B are shown at the right of the figure for comparison purposes. Mutants that caused a more than twofold increase in fluorescence compared with the parental construct are highlighted with a striped pattern. (C) Rho induces conformational changes in the region of the mgtCBR leader corresponding to the identified rut site. Wild-type mgtCBR leader RNA was treated with lead acetate [Pb(Ac)2] in the presence or absence of Rho. RNA–Rho complexes were prepared as for enzymatic digestion (Materials and Methods) and treated with 20 mM of freshly diluted Pb(Ac)2 for 1 min. The reaction was stopped by addition of 20 V of 10 mM EDTA. Proteins were removed by phenol extraction, and RNA fragments were separated at the 6% (vol/vol) denaturing gel. Pb2+ induces highly specific cleavage at positions of tight Mg2+-binding sites as well as single-stranded regions, loops, and bulges. Addition of Rho caused distinct changes in the cleavage pattern, with the most prominent difference observed at positions 108–114, a region critical for Rho function (Fig. 2B). Rho increased reactivity in this region, which is in contrast to canonical footprinting assays where a protein protects its binding site on RNA or DNA from cleavage. Such an unusual pattern suggests that the cleavage by Pb2+ is more sensitive to conformational changes in the mgtCBR leader RNA upon Rho binding than to RNA–protein contacts, which is consistent with previous reports on other RNA–protein complexes treated by lead acetate (7). A limited protection induced by Rho was observed around position 160. This region is located far downstream from the region crucial for Rho function on the mgtCBR leader RNA, as identified by mutagenesis (Fig. 3, compare [105–125] and [128–154]) and might correspond to auxiliary interactions with the Rho primary binding site on the surface of the hexamer, or interactions with the secondary binding site inside the hexameric ring.
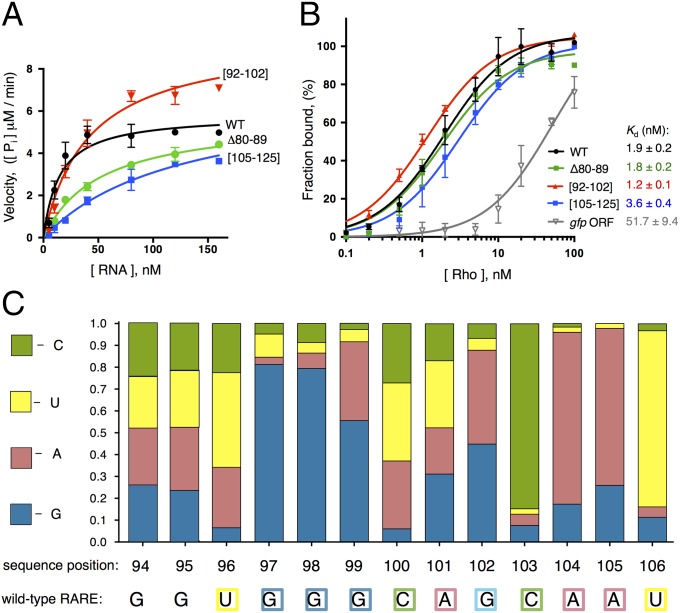
The increased transcription resulting from nucleotide substitutions in the rut site is due to diminished Rho-dependent termination. This is because BCM addition to cells harboring a leader mutated in the Cs at positions 109–112 increased the mRNA levels of the associated coding region only threefold, significantly less than the 16-fold increase BCM promoted in the isogenic strain with the wild-type leader (Fig. 2C). Moreover, the [105–125] RNA variant stimulated Rho's ATPase activity much less than the wild-type leader RNA (Fig. 3A), indicating that this region is required for Rho function (Rho’s ATPase activity reflects both Rho loading and subsequent translocation on its RNA substrate).
Fig. 3.
RARE traps Rho in a catalytically inactive complex. (A) Stimulation of Rho’s ATPase activity by the wild-type mgtCBR leader RNA or three variants (Fig. 2A). ATPase activity was measured by monitoring the accumulation of free phosphate with increasing concentrations of RNA (15). Each data point is an average of at least three independent experiments. (B) RARE does not affect Rho binding to the mgtCBR leader RNA. A 32P-labeled fragment of the wild-type mgtCBR leader RNA, its mutant variants or control RNA corresponding to the 3′ region of the gfp ORF was incubated with increasing concentrations of Rho, and protein-bound RNA fractions were separated by filtering. The amount of filter-bound 32P-RNA was plotted against the concentration of Rho, and an apparent Kd was determined by fitting data to a hyperbolic equation. Each data point is an average of at least three independent experiments. (C) Relative activity of RARE variants with single nucleotide substitutions at each of the positions. All mutants were derived from an mgtCBR leader variant lacking nucleotides 80–89 to lock RARE in a single-stranded form. Fluorescence values produced by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion between the wild-type mgtCBR promoter and the Δ80–89 mgtCBR leader variant with single nucleotide substitutions at each position of RARE to a promotorless gfp gene were summed up and set as 1. The colored segments of each bar represent relative activity of RARE containing G, A, U, or C at a given position. The wild-type RARE sequence is indicated below. Bacteria were grown as described in the legend to Fig. 2B.
To evaluate the contribution of individual nucleotides within and around the identified rut site on transcription termination, we used a variant of the mgtCBR leader locked in a conformation favoring Rho recruitment (Fig. 2 A and B, ∆stem B). This allowed us to refine the borders of the rut site to positions 108–131 (Fig. S2B). With the exception of the C119G substitution, single nucleotide substitutions of the Cs or Us in the 108–131 region displayed only a modest increase in expression of the associated coding region (Fig. S2B). These results are consistent with the premise that Rho recruitment relies on multiple nonspecific and sequence-specific contacts in RNA spread out over a large segment of a transcript (2, 23, 24). Structural probing of an RNA fragment corresponding to the wild-type mgtCBR leader with lead acetate revealed small but distinct changes in sensitivity within the identified rut region upon addition of Rho (Fig. S2C). Cumulatively, these data identified a region of the mgtCBR leader mRNA critical for Rho-dependent transcription termination.
RARE Inhibits Rho-Dependent Termination When Not Sequestered in a Hairpin.
Leader RNAs that harbor Rho-dependent transcription terminators often have the ability to adopt mutually exclusive conformations, one favoring and one hindering the ability of Rho to terminate transcription. For example, a rut site is sequestered within a stem in the mgtA leader conformer hindering Rho-dependent termination, but single stranded and available to Rho in the conformer that favors Rho-dependent termination (15). By contrast, the rut site identified in the mgtCBR leader is single stranded in both the stem-loop A and B conformations (Fig. 2A), which prevent and stimulate Rho activity, respectively (Fig. S1). What, then, determines Rho recruitment and transcription termination in the mgtCBR leader?
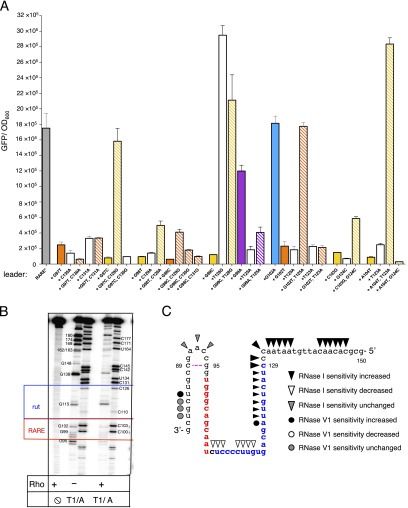
We hypothesized that nucleotides 92–102 regulate Rho-dependent transcription termination because these nucleotides are single stranded when the leader forms stem-loop A but double stranded when the leader folds into stem-loop B (Fig. 2A). We designated this region RARE for Rho-antagonizing RNA element. If stem-loop B promotes transcription termination by sequestering RARE, then rendering RARE single-stranded should overcome the inhibitory effect of stem-loop B on transcriptional readthrough. As hypothesized, expression of the associated coding region increased sixfold when a second copy of RARE was inserted after stem-loop B (Fig. 2 A and B, insRARE), but not when a random sequence of the same length was inserted at the identical position (Fig. 2 A and B, [103]ins).
Several lines of evidence provide additional support to the notion that a single-stranded RARE hinders termination by Rho within the mgtCBR leader. First, removing the left arm of stem-loop B (i.e., positions 80–89, Fig. 2A), which rendered RARE single stranded, increased expression of the coding region 17-fold (Fig. 2B, Δ80–89). We ascribe the increased expression of the latter mutant to impaired Rho function because the corresponding RNA stimulated Rho’s ATPase activity significantly less than did the wild-type RNA (Fig. 3A). This means that RARE exerts its antitermination effect specifically by impairing Rho function and not by modifying RNAP or recruiting a cellular cofactor. Second, replacement of RARE by a scrambled sequence promoted Rho-dependent termination in vivo (Fig. 2B, [92–102]), and, as expected, the corresponding RNA stimulated Rho’s ATPase activity more than the wild-type leader RNA in vitro (Fig. 3A). These results indicate that the removal of RARE from the mgtCBR leader makes the resulting RNA a better substrate for Rho. This result is despite the fact that the mutation prevented formation of stem-loop B. And third, removal of stem-loop B (i.e., positions 79–103, Fig. 2A) reduced transcription of the associated coding region to near background levels (Fig. 2B, ∆stem B), like the mutant with the scrambled RARE. The silencing effect resulting from removal of stem-loop B is due to Rho activity because addition of BCM to cells harboring this variant increased the mRNA levels of the associated coding region 40-fold (Fig. 2C, ∆stem B), significantly more than the increase observed in the isogenic strain with the wild-type mgtCBR leader (16-fold, Fig. 2C) or in the strain with a mutation in the rut site (threefold, Fig. 2C, [109–112]). We conclude that single-stranded RARE directly prevents Rho from terminating transcription in the mgtCBR leader, thereby allowing elongation into the associated coding region.
Rho Binds Normally to mgtCBR Leader Variants with RARE Single Stranded or with a Mutant rut.
The selectivity of Rho-dependent termination has been ascribed to the initial binding of Rho to its RNA substrate (2). However, we have now established that RARE exerts its regulatory effect after the initial binding of Rho, arguing against this notion. That is, Rho bound with a high affinity to the leader variant with RARE single stranded, similar to that exhibited toward the wild-type mgtCBR leader RNA (Fig. 3B, apparent Kd = 1.8 ± 0.2 nM, Δ80–89 versus 1.9 ± 0.2 nM, WT), and only slightly better to an RNA lacking RARE (Fig. 3B, apparent Kd = 1.2 ± 0.1 nM, [92–102]). By contrast, Rho bound with ∼30-fold lower affinity to a control RNA segment of the same length originating from the gfp coding sequence (Fig. 3B, apparent Kd = 51.7 ± 9.4 nM, gfp ORF).
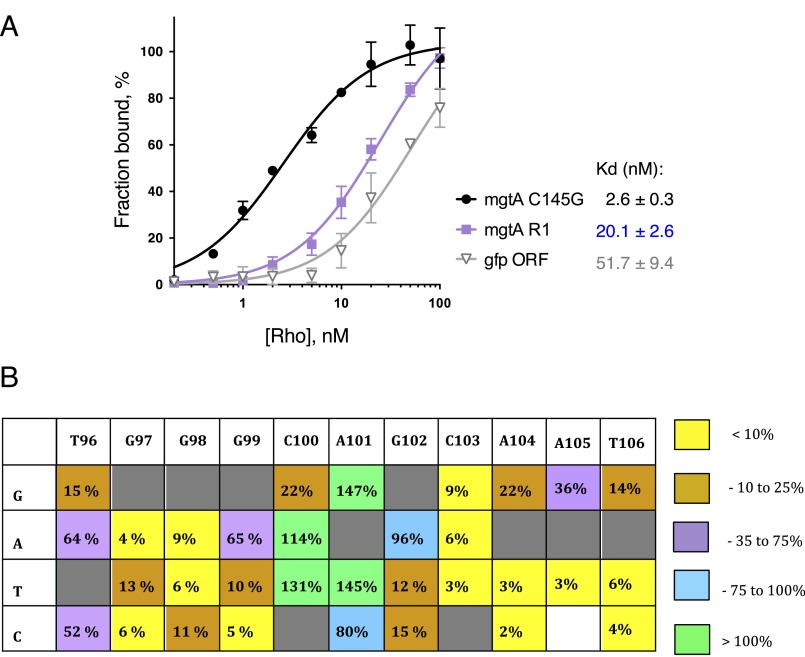
Surprisingly, Rho’s affinity for the RNA with mutations in the rut was only twofold lower than that of the wild-type mgtCBR leader RNA (Fig. 3B, apparent Kd = 3.6 ± 0.4 nM, [105–125] versus 1.9 ± 0.2 nM, WT) even though this mutation led to dramatic derepression in vivo (Fig. 2B, [105–125]). The modest decrease in affinity displayed by the RNA with mutations in a rut in the mgtCBR leader is in contrast to the effect of mutation of a rut site in the mgtA leader region (15, 25), which decreased affinity for Rho eightfold [Fig. S3A, apparent Kd = 2.6 ± 0.3 nM for the C145G mgtA variant promoting an RNA conformation favorable for Rho binding (15) versus 20.1 ± 2.6 nM for the R1 variant with substitutions in a rut site (15)]. Nevertheless, extensive deletion analysis revealed that nucleotides in the [105–125] region were the only ones required for Rho-dependent termination within the 5′ half of the mgtCBR leader (Fig. 2B, compare [105–125] and [109–112] to Δ65–70, Δ71–80, ΔstemB, Δ128–154, and Fig. S2B, compare [109–112] to Δ4–10, Δ11–16, Δ17–23, Δ24–38, Δ39–50, and Δ51–60), suggesting that this region serves as a genuine rut site. Cumulatively, our findings revealed two distinctive features of Rho interaction with the mgtCBR RNA. First, RARE exerts its inhibitory activity after the initial Rho binding takes place and in the absence of any additional cellular factors. And second, the role of the identified rut site is to stimulate Rho’s activity after binding rather than simply to tether the RNA to Rho.
Fig. S3.
Mutation in rut disrupts Rho binding to the mgtA leader RNA, sequences important for RARE activity in the mgtCBR leader. (A) Rho binding to the mgtA leader RNA variants (3′ segment 243 nt long) or control RNA. The C145G mgtA variant promotes an RNA conformation favorable for Rho binding (8). The R1 mgtA variant has substitutions in rut (8). A 32P-labeled fragment of mgtA leader RNA variants or control RNA corresponding to a 3′ region of gfp ORF was incubated with increasing concentrations of Rho, and protein-bound RNA fractions were separated by filtering. The amount of filter-bound 32P-RNA was plotted against the concentration of Rho, and an apparent Kd was determined by fitting data to a hyperbolic equation (see Materials and Methods for details). Each data point is an average of at least three independent experiments. (B) Relative values of fluorescence produced by wild-type Salmonella harboring a plasmid that contains a transcriptional fusion between the wild-type mgtCBR promoter and an mgtCBR leader variant lacking nucleotides 80–89 so that RARE is locked single stranded (or having single nucleotide derivatives in RARE), to a promotorless gfp gene. Bacteria were grown as described in the legend to Fig. 4B. Fluorescence produced by wild-type Salmonella strains harboring the RNA variant lacking nucleotides 80–89 was set as 100%. Colors reflect the severity of the effect caused by the mutations (see key to the Right of the table). Data correspond to the average from at least three independent experiments.
The Sequences and Positions Required for RARE Activity.
To define the RARE residues hindering Rho-dependent transcription termination, we mutated each of the 13 nucleotides within the 94–106 region to each of the other three. We used as parental template a leader variant deleted for nucleotides 80–89 so that RARE would always be single stranded, and thus available to inhibit Rho (Fig. 2A). Then, we measured the fluorescence produced by wild-type Salmonella harboring a plasmid with a transcriptional fusion between the wild-type mgtCBR promoter and the mgtCBR leader variant lacking nucleotides 80–89, and a promotorless gfp gene.
We established that G97, G98, C103, A104, and U106 are the most important RARE residues because substitutions at these positions to any other nucleotide severely decreased fluorescence (Fig. 3C and Fig. S3A). Mutations at positions 99 and 102 had different effects depending on the substituting nucleotide. For example, the G102A mutant retained the parental behavior, whereas substitution of G102 to U or C decreased fluorescence six- to eightfold (Fig. 3C and Fig. S3A). This result suggests that G102 is involved in wobble base pairing with a U residue that may also establish a canonical base pair with A, but not with U or C. Mutations at positions 100 and 101 actually increased fluorescence, suppressing transcription termination even more efficiently than the parental RARE (Fig. S3A). Finally, substitutions at positions 94 and 95 did not have detectable effects on fluorescence (Fig. 3C).
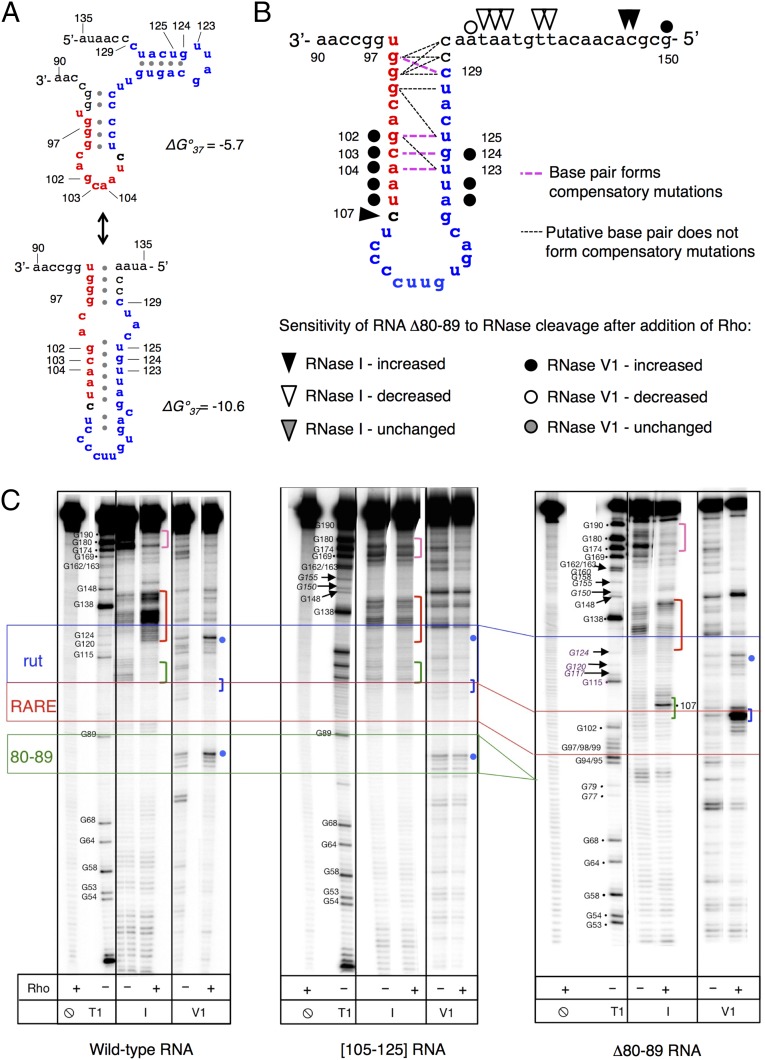
How do the identified RARE nucleotides affect termination by Rho? In silico secondary structure predictions raised the possibility of stem-loops forming between RARE residues and the adjacent rut nucleotides (Fig. 4A). Analysis of compensatory mutations within rut (Fig. S4A) that restored activity of RARE with single-nucleotide substitutions provided evidence of limited base-pairing interaction between these regions (e.g., G102–U125, C103–G124, and A104–U123; Fig. 4B and Fig. S4A). However, no interactions between Gs at positions 97–99 and the CU-rich motif at 128–131, which would form the base of a hypothetical stem-loop, were observed (Fig. 4 A and B). The only putative base pair identified by a compensatory mutation analysis (C97–C129) is incompatible with the in silico model (Fig. 4 A and B), and the remaining interactions appear insufficient to form a stable secondary structure (Fig. 4B).
Fig. 4.
RARE promotes the formation of an unusual complex between the mgtCBR leader RNA and Rho. (A) Putative secondary structures formed by RARE (red) and rut (blue) in the mgtCBR leader. Structures were generated using the Mfold web server for nucleic acid folding and hybridization prediction (46). (B) Graphic representation of the proposed structure formed by RARE (red) and the rut (blue) in the mgtCBR leader based on RARE mutagenesis (Fig. 3C), compensatory mutation analysis (Fig. S4A), and the structural probing shown below. Mutant combinations that restored fluorescence to the levels displayed by the Δ80–89 variant are shown in bold pink dashed lines; mutant combinations that were tested but did not restore Δ80–89 function are shown in thin black dashed lines. See text for details. (C) Enzymatic probing of the wild-type mgtCBR leader RNA and the [105–125] and Δ80–89 variants. The 5′ 32P-labeled fragment of the mgtCBR leader RNA was treated with different RNases in the presence or absence of Rho. T1 is for RNase T1 (cuts at G residues, used as a molecular marker), I is for RNase I, and V1 is for RNase V1. Positions of interest are marked with colored dots and brackets (see text). Colored lines show borders of relevant elements: blue for rut, red for RARE, and green for the left arm of stem-loop B (80–89). Only the relevant lanes from a representative gel are shown. The experiment was performed at least three times for each sample.
Fig. S4.
RARE interacts with rut in the mgtCBR leader. (A) Primary data for compensatory analysis of mutations in the mgtCBR leader that restore RARE function graphically shown in Fig. 4B. Fluorescence produced by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion of the mgtCBR promoter and mutant leader variants to a promotorless gfp gene. The Δ80–89 leader variant, which locks RARE in single-stranded conformation, was used to derive the single- and double-nucleotide substitution variants. Experiments were performed as described in the legend to Fig. 2B. Shown are the mean and SD from at least three independent experiments. (B) Rho protects G residues within RARE in the Δ80–89 mgtCBR leader RNA from cleavage by the G-specific RNase T1. Cleavage by RNase A (cuts after C and U residues) is used as molecular marker. (C) Graphic representation of the structural probing of the wild-type mgtCBR RNA shown in Fig. 4C.
RARE Promotes a Singular Conformation of the Rho–RNA Complex.
How does RARE prevent Rho from terminating transcription? To address this question, we examined the interaction between Rho and mgtCBR leader RNA substrates using enzymatic probing. We determined that the cleavage pattern that Rho promoted in the leader RNA with RARE single stranded (Δ80–89) differed from the patterns observed in the wild-type and the rut mutant variant [105–125] RNAs (Fig. 4C). For instance, Rho protected the wild-type mgtCBR leader RNA at positions 105–114 from cleavage by RNase I, an enzyme that cleaves unpaired residues (Fig. 4C, green bracket), but not in the rut mutant variant RNA (Fig. 4C, green bracket) even when added at saturating levels. This protected region corresponds to nucleotides required for Rho to terminate transcription (Fig. 2B). By contrast, Rho promoted an RNase I hypersensitive site in the Δ80–89 RNA at position 107 (Fig. 4C, green bracket), which is located between RARE and rut (Fig. 2A). In addition, Rho induced the appearance of RNase I hypersensitive sites at positions 130–145 in the wild-type RNA but not in the Δ80–89 RNA (Fig. 4C, red brackets). These results suggest that Rho promotes a conformation in the mgtCBR leader RNA with RARE single stranded that does not stimulate Rho’s ATPase activity (Fig. 3A, Δ80–89).
RARE appears to contact both Rho and rut in the mgtCBR leader (Fig. 4B) for two reasons. First, Rho partially protected G residues within RARE in the Δ80–89 RNA from the G-specific RNase T1 (Fig. S4B). Second, the digestion patterns of Rho bound to the Δ80–89 RNA promoted by the RNase V1 and RNase I suggested the presence of a stem-loop structure. That is, there was an RNase V1-sensitive site at positions 103–105 (Fig. 4C, blue bracket) followed by RNase I-sensitive site at position 107 (Fig. 4C, green bracket) (RNase V1 cleaves double-stranded or stacked residues). The location of this hairpin is in excellent agreement with the model suggested by genetic analysis (Fig. 4B). We believe that this is a bona fide stem-loop because its RNase digestion pattern is similar to the one displayed by stem-loop B, whose presence was verified by lead acetate (Fig. S2C), in-line probing (18) and genetic analysis (18). In the same RNase probing assay, stem-loop B is characterized by RNase V1-sensitive site at positions 82–85, (Fig. 4C, blue dot, and Fig. S4C) followed by a site sensitive to RNase I at positions 90–92 (Fig. 4C and Fig. S4C) in the wild-type leader RNA.
Crucially, the stem-loop pattern in the Δ80–89 RNA appears only in the presence of Rho, indicating that the contacts between RARE and rut are unstable in the absence of Rho.
mgtM Translation Favors Rho-Dependent Termination in the mgtCBR Leader by Sequestering RARE Within a Stem.
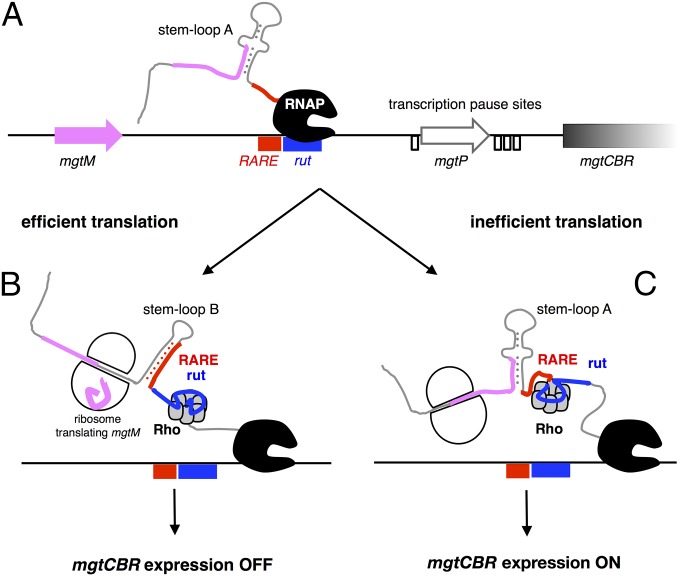
What determines whether RARE is single stranded and allows expression of downstream genes, or sequestered within a stem and leads to premature termination within the mgtCBR leader? On the one hand, there are intrinsic properties of the mgtCBR leader RNA: stem-loop B is more stable than stem-loop A (∆GA ∼ −14.8 kcal/mole versus ∆GB ∼ −17.8 kcal/mole), but stem-loop A has a kinetic advantage over stem-loop B because it emerges from the elongating RNAP before stem-loop B (Fig. 5A). On the other hand, a ribosome translating mgtM will unfold stem-loop A because the last four mgtM codons are embedded in stem-loop A (Fig. 2A). This will favor formation of stem-loop B, which will sequester RARE, resulting in transcription termination within the mgtCBR leader (Fig. 5B). By contrast, if mgtM translation is inefficient, stem-loop A will prevail, RARE will be single stranded, Rho will be trapped, and transcription will continue into the associated coding region (Fig. 5C).
Fig. 5.
Model of transcription termination control by the mgtCBR leader. (A) Schematic of the DNA region corresponding to the mgtCBR leader. Small ORFs are indicated by pink and white arrows; the position of RARE and rut are indicated by rectangles, and transcription pause sites (Fig. 2C) by open boxes. The leader RNA emerges from the transcribing RNAP and folds into stem-loop A. (B) When translation is efficient, the ribosome translating mgtM unwinds stem-loop A, allowing folding of stem-loop B, which sequesters RARE. Rho loads onto the RNA and translocates toward a paused RNAP. Rho triggers transcription termination, thereby turning mgtCBR expression OFF. (C) When translation is inefficient, single-stranded RARE traps Rho in an inactive state. Transcription continues, turning mgtCBR expression ON.
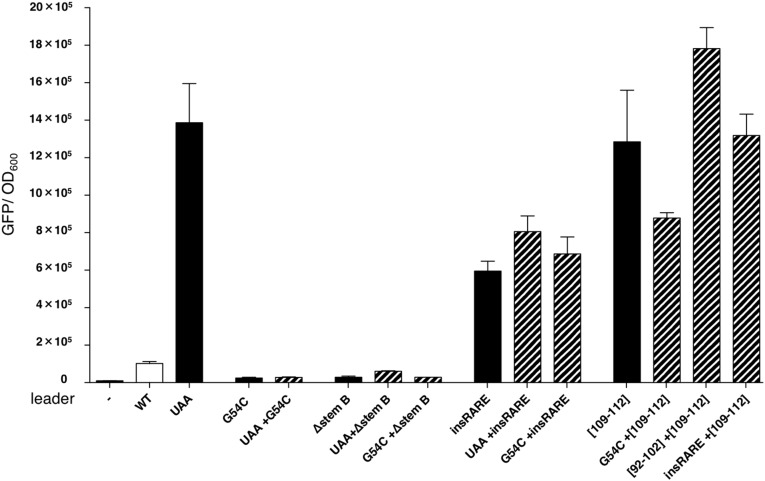
According to the model described above, mutations that disrupt individual steps in the pathway should display 3′-to-5′ hierarchy when combined. In other words, mutations abolishing formation of stem-loop A should be dominant over those compromising mgtM translation; mutations that preclude sequestration of RARE should be dominant over those impacting RNA folding and mgtM translation; and mutations disrupting the Rho-binding site should be dominant over all of the above. We have now established that all these predictions are fulfilled.
First, a mutation that abolished mgtM translation due to replacement of its start codon by a stop codon (Fig. 2A, UAA) caused a 15-fold derepression of the associated coding region (Fig. 6, UAA) but had no effect when combined with the substitution G54C (Fig. 6, UAA + G54C), which weakens stem-loop A (Fig. 2A, G54C) (18), or when stem-loop B was deleted (Fig. 6, UAA + ∆stem B), thereby eliminating RARE (Fig. 2A, ∆stem B). These results support the notion that a ribosome translating mgtM exerts its action through conformational changes in the mgtCBR leader RNA. Second, mutations that disrupt mgtM translation or favor formation of stem-loop B had little or no effect when RARE was single stranded (Fig. 6, UAA + insRARE, G54C + insRARE). These findings indicate that formation of stem-loop B and mgtM translation promote Rho-dependent termination in the mgtCBR leader by sequestering RARE. And third, substitutions in rut were dominant over mutations that promote formation of stem-loop B, lack RARE, or render RARE single stranded (Fig. 6, G54C + [109–112], [−RARE] + [109–112], insRARE + [109–112]). Thus, the ribosome translating mgtM disrupts stem-loop A, allowing sequestration of RARE in stem-loop B, leading to Rho-dependent termination within the mgtCBR leader.
Fig. 6.
mgtM translation promotes transcription termination by sequestering RARE. Epistasis analysis of mutations affecting Rho-dependent termination in the mgtCBR leader. Fluorescence levels exhibited by wild-type Salmonella (14028s) harboring a plasmid that contains a transcriptional fusion of the wild-type mgtCBR promoter and leader, or mgtCBR leader variants, to a promotorless gfp gene. The experiment was performed as described in the legend to Fig. 1B. Single mutations are shown with solid bars and double mutations are shown with dashed bars. (−) corresponds to wild-type Salmonella (14028s) harboring the plasmid vector pFPV25. Data correspond to the average from three independent experiments.
Discussion
We have now uncovered an RNA element that sequesters termination factor Rho in an inactive complex, thereby preventing transcription termination without affecting Rho binding to its target. Rho had been known to bind preferentially to C-rich regions of RNA that are free of strong secondary structures and translating ribosomes (12). However, Rho promotes termination only at a subset of the transcripts it binds (13). The identification of RARE, an RNA motif that induces formation of a catalytically incompetent complex with Rho, indicates that Rho's specificity is defined not only by sequences that mediate its recruitment but also by sequences that antagonize its activity.
Why Inefficient Translation of an Upstream ORF May Not Result in Transcriptional Polarity.
Translation is thought to be a universal defense mechanism against Rho-dependent transcription termination because translating ribosomes protect an elongating RNAP from Rho invasion (5, 26). This is why disrupting the coupling of transcription with translation gives rise to transcriptional polarity, a process whereby compromised translation of a promoter-proximal ORF impairs transcription of downstream genes in the same transcription unit (12). For example, nonsense mutations in the first or second genes of the gal operon silence expression of the second or/and the third genes, respectively (27). Similarly, significant repression of tryptophanase expression due to constitutive Rho-dependent termination was observed when the start codon of the short ORF tnaC in the leader region of the tryptophanase tnaAB operon was replaced by a stop codon (28). By contrast, replacement of mgtM’s start codon with a stop codon derepressed the associated coding region (18).
Our data suggest that it is the particular arrangement of rut, RARE, and short ORF in a leader region that determines whether translation of the short ORF actually favors transcription elongation into the associated coding region instead of resulting in polarity. Rho-dependent transcription termination in the mgtCBR leader is actually stimulated by translation of the mgtM ORF (Fig. 6) because a ribosome translating mgtM favors a conformation in the mgtCBR RNA that sequesters RARE and allows Rho recruitment, thereby silencing the mgtCBR coding region (Fig. 5B). And in the case of the tnaAB leader, excess tryptophan induces ribosome stalling within tnaC in a manner uniquely dependent on the nascent TnaC peptide (29). A ribosome stalled at a Trp codon in the middle of tnaC occludes the rut site, thereby suppressing Rho-dependent termination.
RARE Inhibits Rho Function.
Rho-dependent termination in the mgtCBR leader is modulated by RARE, an RNA element that hinders Rho function (Fig. 2A). RARE inhibits termination only when not sequestered in stem-loop B (Fig. 2B, Δ80–89 and insRARE). RARE does not affect Rho binding to the mgtCBR leader RNA (Fig. 3B), but it prevents formation of a catalytically competent complex (Fig. 3A).
The mgtCBR leader RNA with RARE single stranded adopts a conformation when bound to Rho that is distinct from that of the wild-type leader RNA (Fig. 4C). In the presence of Rho, RARE makes limited contact with the Rho-binding site: Δ80–89 RNA displays an RNase V1-sensitive site at positions 103–105 (Fig. 4C, blue bracket) followed by an RNase I-sensitive site at position 107 (Fig. 4C, green bracket), a pattern suggesting the presence of a hairpin. This pattern is in contrast to that displayed by the wild-type RNA in the presence of Rho: RNase V1-sensitive sites at positions 82–85, (Fig. 3C, blue dot) followed by RNase I- and lead acetate-sensitive sites at positions 90–92 (Fig. 4C and Fig. S2C). The former cleavage pattern was observed only in the presence of Rho, suggesting that base pairing between RARE and rut is unstable in the absence of Rho.
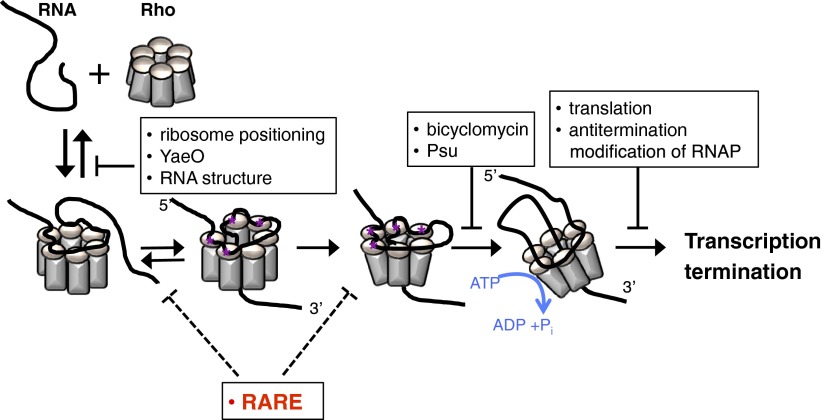
What is the fate of the complex formed by Rho and the mgtCBR leader RNA with single-stranded RARE? Kinetic and structural studies of Rho recruitment indicate that a Rho–RNA complex undergoes several isomerization events before reaching a translocation-competent state (Fig. S5) (30, 31). RARE might stabilize one of the intermediates on the recruitment pathway; for example, RARE might prevent the Rho ring from opening or closing, and in this way hinder Rho movement toward a paused RNAP (Fig. S5). Alternatively, changes in conformation of rut induced by RARE in the presence of Rho might affect Rho translocation at later steps. The idea that rut can participate in Rho-dependent termination postrecruitment is supported by the behavior of elongation factor NusG, which is particularly important for terminators with poor rut sites and affects RNA release in the process of Rho-dependent termination (5).
Fig. S5.
How RARE may inhibit Rho function. Schematic of proposed Rho recruitment pathway (9, 10), with modifications. A complex forms upon initial transient interaction between RNA (black) and a positively charged surface of the Rho hexamer (gray). Sequence-specific contacts (purple asterisks) between RNA and Rho primary binding site lead to ring opening and RNA threading through the central hole of the hexamer. RNA establishes contacts with Rho secondary binding site, leading to ring closure. Using the energy derived from ATP hydrolysis, Rho begins to translocate in 5′-to-3′ direction until it catches up with RNAP. RARE induces arrest of Rho recruitment process after the initial binding but before formation of the translocation-competent complex. The steps of the pathway targeted by other Rho inhibitors are shown for comparison purposes.
RARE differs from RNA or DNA signals that mediate antitermination by recruiting protein cofactors or modifying elongating RNAP (see ref. 32 for a review). This is because inhibition of Rho by RARE does not require cellular cofactors (Fig. S1A) or active transcription (Fig. 3A and Fig. S1B).
Sequence Elements Providing Specificity to Rho.
Rho exerts a widespread effect in transcription (2, 33). Once Rho is recruited to a particular site on a nascent RNA, it is thought to translocate toward the transcribing RNAP. A translating ribosome or antitermination factors can shield the transcribing RNAP from interaction with Rho (32, 33). Although these mechanisms contribute to Rho-dependent termination, independent lines of evidence indicate that they are not sufficient to explain Rho specificity.
First, genome-wide distribution of Rho-dependent termination revealed that Rho affects about 20% of nascent transcripts (1), whereas ChIP-chip data determined that Rho is associated with almost all nascent RNAs (13). In other words, only a fraction of transcripts that recruit Rho are subjected to Rho-dependent termination. Second, in vitro studies indicate that Rho can use RNA substrates that are very different from canonical Rho targets (34). Third, kinetic analysis of Rho's ATPase activity revealed that the process of hexamer ring opening, followed by RNA threading and ring closing, is a rate-limiting step in Rho recruitment (30). Fourth, structural studies demonstrated that RNA interactions with the Rho secondary binding site bring about major structural rearrangements within the protein ring (9, 35). And fifth, a study of transcription termination potency of various Rho mutants in vivo suggested that closure of the Rho hexamer around the substrate RNA serves as a regulatory point in Rho recruitment (31).
We propose that RARE represents a distinct class of RNA signal: one that suppresses Rho activity by inducing formation of a translocation-incompetent complex. This mode of action differs from those adopted by other Rho inhibitors (Fig. S5). For example, the YaeO protein prevents Rho from interacting with its RNA substrate (36), whereas processive antiterminators prevent RNA release by modifying elongating RNAP (32). Although both the Psu protein from bacteriophage P4 (37) and the antibiotic BCM (14) inhibit Rho’s translocation, we believe that RARE operates by a different mechanism. The small size and location of RARE upstream of rut suggest that it likely contacts Rho nearby Rho’s primary binding sites, whereas Psu binds at the opposite site of the Rho ring (37), and BCM binds deep inside the channel where it interferes with ATP binding (14, 38). Arrest of Rho-dependent termination by RARE is also distinct from regulation of Rho recruitment by alternative RNA structures that entrap rut (15) because the mgtCBR leader rut remained single stranded in either RNA conformation in the absence of Rho (Fig. 2A) and because RARE did not prevent Rho binding to its recruitment site (Fig. 3B). Moreover, once RARE was single stranded (Fig. 2A, insRARE), conformational changes in RNA had no effect on Rho-dependent termination (Fig. 6, UAA + insRARE, G54C + insRARE). Therefore, formation of alternative structures in the mgtCBR leader RNA serves merely as a means of modulating the availability of RARE. The proposed mechanism expressly allows RARE to determine which of the transcripts bound by Rho will be terminated.
Finally, our findings open the possibility of other RNA elements with similar properties to RARE existing in the bacterial transcriptome and modulating Rho specificity. The availability of RARE and RARE-like elements may be controlled by mechanisms other than formation of an RNA hairpin, including ribosome positioning, protein binding, a riboswitch, or a small RNA. Such elements may also affect the site of Rho-dependent termination when protecting the chromosome from R loops and double-strand breaks (39–41) and the suppression of pervasive antisense transcription (5).
Materials and Methods
Bacterial Strains, Plasmids, Oligodeoxynucleotides, Proteins, and Reagents.
Bacterial strains and plasmids used in this study are listed in Dataset S1. Primers used in this study are listed in Dataset S2. E. coli strain DH5α was used as the host for preparation of plasmid DNA. Ampicillin was used at 50 µg/mL and kanamycin at 20 µg/mL. For in vivo experiments, wild-type S. enterica serovar Typhimurium 14028s (42) was used as the host strain. Bacteria were grown at 37 °C in LB broth or in N-minimal medium (pH 7.4) (43) supplemented with 0.1% casamino acids, 38 mM glycerol, and 10 µM of MgCl2. One milliliter of the overnight culture was washed in the N-minimal medium without Mg2+ and resuspended in 1 mL of the same media. The suspended bacteria were inoculated 1/100 volume in N-minimal medium with 10 μM of MgCl2 and ampicillin and grown for 4 h unless indicated otherwise. Fluorescence and OD600 of the cultures were measured in a Victor3 plate reader from Perkin-Elmer. For the experiment involving BCM treatment, cells were harvested after 3 h of growth in N-minimal medium with 10 μM Mg2+ followed by a 15-min treatment with BCM added to the final concentration of 20 µg/mL. PCR reagents were from Invitrogen, [γ32P]-ATP and [α32P]-GTP, from Perkin-Elmer, and other chemicals from Sigma or Fisher. Plasmid DNAs and PCR products were purified using spin kits from Qiagen and Promega. RNAP was purified as described (44). Rho was purified as described in SI Materials and Methods.
Construction of Plasmids.
Plasmid pAS69, carrying the mgtCBR leader sequence downstream of the λPR promoter and a C-less initial transcribed sequence, was synthesized in vitro by GenScript USA based on vector pUC57; the full sequence is available upon request. Cloning was performed using modification enzymes from NEB as indicated in the plasmid description. Site-directed mutagenesis was performed using the Quikchange II kit from Agilent with plasmid pAS69 (used to generate templates for in vitro transcription) and pGFP303 (used for in vivo fluorescence measurements) and their derivative as templates and indicated oligonucleotides. See Dataset S1 for details.
Templates for in Vitro Transcription.
Templates for pause and termination assays were generated by PCR amplification using primers 12230 and 13192, and plasmids pAS69 (wild-type), pAS75 (G54C), and pAS77 (G95C). Templates for RNA synthesis were generated by PCR amplification using primers 13347 and 13016, and plasmids pAS69 (wild type), pAS75 (G54C), pAS77 (G95C), pAS155 ([109–112]), pAS237 (Δ[80–89]), and pAS241 ([105–125]). Primers 14787/14788 were used for pYS1010-C145G (mgtA C145G) or pYS10116 (mgtA R1) and primers 15187/15188, for pFVP25 (gfp ORF).
Single-Round Pause Assay.
Halted elongation complexes (designated A26 in Fig. 2B) were prepared with 50 nM of E. coli RNAP (core enzyme to σ70 ratio is 1:4) and 40 nM of linear DNA template prepared as described above, in Rho buffer [40 mM Tris⋅HCl pH 7.9, 50 mM KCl, 2 mM MgCl2, 0.1 mM DTT, 3% (vol/vol) glycerol] supplemented with ApU (RiboMed) at 100 μM, ATP and UTP at 5 μM, GTP at 1 μM, and 5 mCi of [α32P]-GTP (3000 Ci/mmol) during a 15-min incubation at 37 °C. Transcription was restarted by the addition of CTP, ATP, GTP, and UTP to 50 μM, and rifapentin to 25 µg/mL. Samples were removed at 15, 30, 45, 60, 120, 300, and 600 s and quenched by the addition of an equal volume of STOP buffer [95% (vol/vol) deionized formamide, 50 mM EDTA, 45 mM Tris-borate; pH 8.3, 0.1% bromophenol blue, 0.1% xylene cyanol]. For RNA sequencing, 10-μL aliquots of halted complexes were chased by 25 μM RNA sequencing mixtures (a single 3'OMeNTP at 1/10 to the corresponding rNTP) for 10 min and quenched by the addition of an equal volume of STOP buffer.
Samples were heated at 90 °C for 2 min and separated by electrophoresis in 6% denaturing acrylamide (19:1) gels (7 M urea, 0.5× TBE). RNA products were visualized and quantified using a PhosphorImager Storm 820 System (GE Healthcare), ImageQuant Software, and Microsoft Excel.
Rho-Dependent Termination Assay.
Halted elongation complexes were prepared as described above. Transcription was restarted by the addition of GTP, CTP, ATP, and UTP to 1 mM, rifapentin to 25 µg/mL, and Rho to 20 nM where indicated. Reactions were carried out at 37 °C for 15 min and stopped as described above.
ATPase Assay.
Rho ATPase activity was determined with the EnzCheck Phosphate Assay kit from Invitrogen as described (15) using RNA fragments corresponding to the mgtCBR leader or variants prepared as described in SI Materials and Methods.
Filter-Binding Assay.
Determination of equilibrium Kd values for binding of RNAs to Rho was carried out by the nitrocellulose binding method (45). The 5′-end radiolabeled RNAs prepared as described in SI Materials and Methods were diluted to 0.01 nM in binding buffer [50 mM KCl, 1 mM MgCl2, 20 mM Hepes pH 7.9, 3% (vol/vol) glycerol, 0.1 mM EDTA], heated to 80 °C for 3 min, and renatured at room temperature. To prevent nonspecific binding, yeast tRNA was added to 0.06 mg/mL. Samples were incubated with a series of dilutions of Rho prepared in binding buffer supplemented with 10% (vol/vol) glycerol and 4 mg/mL BSA, or storage buffer for 10 min at room temperature; the samples were filtered through 0.45-μm nitrocellulose filters (HAWP, Millipore) under vacuum; and the filters were washed with 5 mL of the binding buffer, air dried, and quantified using PhosphorImager and ImageQuant Software (GE Healthcare). To calculate the apparent equilibrium dissociation constants, the data were fit to a hyperbolic equation, B = Bmax × [Rho]/([Rho] + Kd), where B is a percentage of RNA bound, Bmax is the maximum binding at infinite concentration of Rho, and Kd is the dissociation constant. The fitting was performed by nonlinear regression algorithm using Prism 6 (GraphPad Software). For each RNA–Rho combination, Kd measurements were independently repeated two to three times, and averages were calculated.
RNA Probing.
The 5′-end radiolabeled RNAs prepared as described in SI Materials and Methods were diluted to 20 nM in Rho buffer, heated to 80 °C for 3 min, and renatured at room temperature. MgCl2 was added to 5 mM and ATPγS [adenosine 5′-O(3-thiotriphosphate) from Calbiochem] to 2 mM. To prevent nonspecific binding, yeast tRNA was added to 0.05 mg/mL. Samples were incubated with 100 nM of Rho or equivalent volume of storage buffer at 37 °C for 3 min. For limited enzymatic digestion, 8-μL reaction aliquots were mixed with 2 µL of RNase T1 diluted to 0.3 units/µL, or RNaseI diluted to 0.3 units/µL, or RNase V1 diluted to 0.005 units/µL. Reactions were quenched by addition of 140 µL of 10 mM EDTA, and RNA was purified by phenol-chloroform extraction followed by ethanol precipitation. Pellets were resuspended in formamide loading buffer, loaded onto 6% (wt/vol) gel, and analyzed as described above.
Quantitative RT-PCR.
Total RNA was isolated using RNeasy Kit (Qiagen) according to the manufacturer’s instructions. The purified RNA was quantified using a NanoDrop machine (NanoDrop Technologies). cDNA was synthesized using High Capacity RNA-to cDNA Master Mix (Applied Biosystems). The mRNA levels of the gfp and mgtC genes were measured by quantification of cDNA using Fast SYBR Green PCR Master Mix (Applied Biosystems) and appropriate primers (mgtC coding: 7530/7531; gfp: 13725/13726) and monitored using a Fast ABI7500 machine (Applied Biosystems). Data were normalized to the levels of 16S ribosomal RNA amplified with primers 6970 and 6971.
SI Materials and Methods
Rho Overexpression and Purification.
An overnight culture of E. coli strain XJb (λDE3) harboring plasmid pAS85 (with the rho ORF amplified from S. enterica serovar Typhimurium 14028s DNA and cloned between the NdeI and HindIII sites of pET28) was diluted 1/100 into fresh LB media supplemented with kanamycin and grown with agitation at 37 °C until OD600 ∼0.6). To induce expression of the recombinant protein, IPTG was added to 0.2 mM and the temperature was lowered to 30 °C. After 6 h, arabinose was added to 0.06% to induce the expression of endolysin and cells were grown for another hour. Cells were collected by centrifugation and frozen at −80 °C. Pellet was resuspended in lysis buffer [25 mM Tris⋅HCl pH 7.9, 500 mM KCl, 10% (vol/vol) glycerol, and 0.1 mM DTT] supplemented with Complete EDTA-free Protease Inhibitors Mixture from Roche and lysozyme. Cells were lysed by ultrasonication, followed by centrifugation (two times, 30 min at 29,500 × g, 4 °C), and the cleared lysate was loaded onto a Ni–Sepharose (GE Healthcare) gravity column preequilibrated with lysis buffer. The column was washed with 10 volumes of lysis buffer, 10 volumes of HepA buffer [50 mM Tris⋅HCl pH 6.9, 5% (vol/vol) glycerol, 1 mM β-mercaptoethanol], and 10 volumes of HepA supplemented with 50 mM imidazole. The protein was eluted with HepA + 200 mM imidazole, diluted twice, and loaded onto a HiTrap Heparin HP column (GE Healthcare). Bound proteins were eluted by NaCl gradient and Rho eluted as a single peak at 42 mSi. Fractions containing Rho were pulled and dialyzed against Rho storage buffer [50% (vol/vol) glycerol, 10 mM Tris⋅HCl pH 7.9, 100 mM KCl, 0.1 mM EDTA, and 0.1 mM DTT] and stored at −20 °C.
RNA Preparation.
The first 203 nt of the mgtCBR leader RNAs were used in the ATPase assay because longer fragments, including the full-length leader, displayed low activity in the ATPase assay, presumably due to strong secondary structures located after mgtP interfering with Rho translocation. RNA fragments corresponding to the first 203 nt of the wild-type mgtCBR leader or mutant derivatives were synthesized using 12 μg of linear DNA templates containing T7 promoter prepared as described above, 600 units of T7 RNAP from NEB, and 2 mM NTPs in 500 μL of transcription buffer (40 mM Tris⋅HCl pH 7.9, 10 mM NaCl, 6 mM MgCl2, 2 mM spermidine, and 10 mM DTT) supplemented with 100 units of SUPERas-In RNase inhibitor from Ambion and 0.3 units of inorganic pyrophosphatase from Affymetrix. After 2 h at 37 °C, 10 units of DNase I was added and reaction mixture was incubated for another 0.5 h. RNA was recovered by phenol-chloroform extraction followed by isopropanol precipitation. The pellet was resuspended in 100 μL of 1× DNase I buffer containing 10 units of DNase I and 2 units of RNasin RNase inhibitor for the second treatment, RNA was recovered as described above. The pellet was resuspended in STOP buffer and loaded onto 6% (vol/vol) denaturing gel, as described above. RNA fragments were visualized by UV shadowing, excised, crushed, and soaked in gel-extraction buffer (0.3 mM Na-acetate pH 5.2, 25 mM Tris-acetate pH 7.5, EDTA 1 mM) in the presence of 50 μL of phenol to prevent degradation and incubated overnight with nutation at room temperature. The gel slurry was separated with a MWCO 100 spin filtration device from Millipore and water-soluble fraction was subjected to chloroform extraction and ethanol precipitation. RNA was resuspended in 100 μL H20 and stored at −80 °C while not in use.
For RNA probing and filter binding assays, RNAs were labeled with 32P as follows: 100 pmol of RNAs were dephosphorylated using APex phosphatase from Epicentre and RNAs were recovered by phenol-chloroform extraction and precipitated with ethanol in the presence of GlycoBlue (Ambion). RNAs were 5′ end-labeled with [γ32P]-ATP and T4 polynucleotide kinase from New England Biolabs and purified by PAGE as described above.
Supplementary Material
Acknowledgments
We thank Max Gottesman for providing bicyclomycin; Hubert Salvail for technical advice regarding the RNA probing experiments; Ronald Breaker for useful discussions; and Irina Artsimovitch, Paul Babitzke, Max Gottesman, Robert Landick, and Jeffrey Roberts for comments on the manuscript. This work was supported, in part, by NIH Grant AI49561 (to E.A.G., who is an investigator of the Howard Hughes Medical Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515383112/-/DCSupplemental.
References
- 1.Peters JM, et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106(36):15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudvillain M, Figueroa-Bossi N, Bossi L. Terminator still moving forward: Expanding roles for Rho factor. Curr Opin Microbiol. 2013;16(2):118–124. doi: 10.1016/j.mib.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Yanofsky C, Ito J. Nonsense codons and polarity in the tryptophan operon. J Mol Biol. 1966;21(2):313–334. doi: 10.1016/0022-2836(66)90102-1. [DOI] [PubMed] [Google Scholar]
- 4.Newton WA, Beckwith JR, Zipser D, Brenner S. Nonsense mutants and polarity in the lac operon of Escherichia coli. J Mol Biol. 1965;14(1):290–296. doi: 10.1016/s0022-2836(65)80250-9. [DOI] [PubMed] [Google Scholar]
- 5.Peters JM, et al. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26(23):2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CY, Richardson JP. Sequence elements essential for rho-dependent transcription termination at lambda tR1. J Biol Chem. 1987;262(23):11292–11299. [PubMed] [Google Scholar]
- 7.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell. 2003;114(1):135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 8.Morgan WD, Bear DG, Litchman BL, von Hippel PH. RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res. 1985;13(10):3739–3754. doi: 10.1093/nar/13.10.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127(3):553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 10.Burgess BR, Richardson JP. RNA passes through the hole of the protein hexamer in the complex with the Escherichia coli Rho factor. J Biol Chem. 2001;276(6):4182–4189. doi: 10.1074/jbc.M007066200. [DOI] [PubMed] [Google Scholar]
- 11.Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577(2):251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 12.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol. 2011;412(5):793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33(1):97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohn H, Widger W. The molecular basis for the mode of action of bicyclomycin. Curr Drug Targets Infect Disord. 2005;5(3):273–295. doi: 10.2174/1568005054880136. [DOI] [PubMed] [Google Scholar]
- 15.Hollands K, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109(14):5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166(1):217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EJ, Groisman EA. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature. 2012;486(7402):271–275. doi: 10.1038/nature11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee EJ, Choi J, Groisman EA. Control of a Salmonella virulence operon by proline-charged tRNA(Pro) Proc Natl Acad Sci USA. 2014;111(8):3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee EJ, Pontes MH, Groisman EA. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell. 2013;154(1):146–156. doi: 10.1016/j.cell.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J Bacteriol. 1989;171(9):4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alix E, Blanc-Potard AB. MgtC: A key player in intramacrophage survival. Trends Microbiol. 2007;15(6):252–256. doi: 10.1016/j.tim.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Hart CM, Roberts JW. Deletion analysis of the lambda tR1 termination region. Effect of sequences near the transcript release sites, and the minimum length of rho-dependent transcripts. J Mol Biol. 1994;237(3):255–265. doi: 10.1006/jmbi.1994.1229. [DOI] [PubMed] [Google Scholar]
- 24.Graham JE. Sequence-specific Rho-RNA interactions in transcription termination. Nucleic Acids Res. 2004;32(10):3093–3100. doi: 10.1093/nar/gkh630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groisman EA, et al. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet. 2013;47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudvillain M, Nollmann M, Margeat E. Keeping up to speed with the transcription termination factor Rho motor. Transcription. 2010;1(2):70–75. doi: 10.4161/trns.1.2.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adhya SL, Shapiro JA. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics. 1969;62(2):231–247. doi: 10.1093/genetics/62.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart V, Yanofsky C. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1986;167(1):383–386. doi: 10.1128/jb.167.1.383-386.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gollnick P, Yanofsky C. tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J Bacteriol. 1990;172(6):3100–3107. doi: 10.1128/jb.172.6.3100-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DE, Patel SS. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J Biol Chem. 2001;276(17):13902–13910. doi: 10.1074/jbc.M011043200. [DOI] [PubMed] [Google Scholar]
- 31.Shashni R, Qayyum MZ, Vishalini V, Dey D, Sen R. Redundancy of primary RNA-binding functions of the bacterial transcription terminator Rho. Nucleic Acids Res. 2014;42(15):9677–9690. doi: 10.1093/nar/gku690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9(5):319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washburn RS, Gottesman ME. Regulation of transcription elongation and termination. Biomolecules. 2015;5(2):1063–1078. doi: 10.3390/biom5021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz A, Walmacq C, Rahmouni AR, Boudvillain M. Noncanonical interactions in the management of RNA structural blocks by the transcription termination rho helicase. Biochemistry. 2007;46(33):9366–9379. doi: 10.1021/bi700493m. [DOI] [PubMed] [Google Scholar]
- 35.Thomsen ND, Berger JM. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell. 2009;139(3):523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutiérrez P, et al. Solution structure of YaeO, a Rho-specific inhibitor of transcription termination. J Biol Chem. 2007;282(32):23348–23353. doi: 10.1074/jbc.M702010200. [DOI] [PubMed] [Google Scholar]
- 37.Ranjan A, Sharma S, Banerjee R, Sen U, Sen R. Structural and mechanistic basis of anti-termination of Rho-dependent transcription termination by bacteriophage P4 capsid protein Psu. Nucleic Acids Res. 2013;41(14):6839–6856. doi: 10.1093/nar/gkt336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skordalakes E, Brogan AP, Park BS, Kohn H, Berger JM. Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin. Structure. 2005;13(1):99–109. doi: 10.1016/j.str.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146(4):533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci USA. 2011;108(2):792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leela JK, Syeda AH, Anupama K, Gowrishankar J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(1):258–263. doi: 10.1073/pnas.1213123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243(4894 Pt 1):1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 43.Snavely MD, Miller CG, Maguire ME. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266(2):815–823. [PubMed] [Google Scholar]
- 44.Belogurov GA, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26(1):117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carey J, Cameron V, de Haseth PL, Uhlenbeck OC. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- 46.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.