Significance
Inactivating pancreatic endoplasmic reticulum kinase (PERK) mutations cause pancreatic degeneration and diabetes in patients with Wolcott-Rallison syndrome. Pancreatic injury is also observed in mice upon PERK genetic ablation or treatment with PERK inhibitors. This toxicity (the mechanisms of which are poorly understood) impedes the clinical development of PERK inhibitors, which show promise against cancers and neurodegenerative diseases. Here we demonstrate that activation of type 1 interferon signaling occurs upon PERK ablation and is responsible for pancreatic injury and the loss of exocrine and endocrine tissues and functions. Neutralization of interferon signaling protects the pancreas from deleterious effects of PERK inhibitors. Temporally targeting the interferon pathway may help with the treatment of patients with Wolcott-Rallison syndrome and the use of PERK inhibitors against other diseases.
Keywords: PERK, pancreas, interferon, diabetes, Wolcott-Rallison syndrome
Abstract
The great preclinical promise of the pancreatic endoplasmic reticulum kinase (PERK) inhibitors in neurodegenerative disorders and cancers is marred by pancreatic injury and diabetic syndrome observed in PERK knockout mice and humans lacking PERK function and suffering from Wolcott-Rallison syndrome. PERK mediates many of the unfolded protein response (UPR)-induced events, including degradation of the type 1 interferon (IFN) receptor IFNAR1 in vitro. Here we report that whole-body or pancreas-specific Perk ablation in mice leads to an increase in IFNAR1 protein levels and signaling in pancreatic tissues. Concurrent IFNAR1 deletion attenuated the loss of PERK-deficient exocrine and endocrine pancreatic tissues and prevented the development of diabetes. Experiments using pancreas-specific Perk knockouts, bone marrow transplantation, and cultured pancreatic islets demonstrated that stabilization of IFNAR1 and the ensuing increased IFN signaling in pancreatic tissues represents a major driver of injury triggered by Perk loss. Neutralization of IFNAR1 prevented pancreatic toxicity of PERK inhibitor, indicating that blocking the IFN pathway can mitigate human genetic disorders associated with PERK deficiency and help the clinical use of PERK inhibitors.
Tumor microenvironment-associated deficit in oxygen and nutrients activate numerous pathways that aid cancer and tumor stroma cells by increasing their ability to survive, withstand anticancer therapies, and ultimately select for more aggressive and viable clones capable of metastasizing (1). Activation of the unfolded protein response (UPR) plays a central role in these processes (2). Three branches of this response include stimulation of activating transcription factor-6 and activation of two kinases, inositol requiring enzyme 1α/β and the eukaryotic translation initiation factor 2-alpha kinase 3 [also termed double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase, or pancreatic endoplasmic reticulum kinase (PERK)]. The latter kinase contributes to phosphorylation of the eukaryotic translation initiation factor 2-alpha and controls the rate of global translation and noncanonical induction of specific proteins that help cope with stress (reviewed in ref. 2).
Among three main UPR pathways, signaling through PERK has received the most attention for its central role in cancer (3–6). Genetic studies have demonstrated that PERK is essential in supporting tumor growth and progression via diverse mechanisms, including stimulation of angiogenesis (7–12), potential effects on antitumor immunity (13, 14), and direct increase in cancer cell viability by altering its metabolic status (15), promoting survival autophagy (16–18), and induction of prosurvival microRNAs (19). Accordingly, development of novel, potent, and selective PERK inhibitors as a means to treat cancers has been proposed (20, 21). Several PERK inhibitors have shown promising results in various preclinical tumor models (22–24). Furthermore, some of these inhibitors can protect against the prion-mediated neurogenerative disorders (25).
Regrettably, PERK knockout and small-molecule inhibitors also showed serious toxic effects primarily affecting the pancreas (22, 25–27). Importantly, PERK has been indeed shown to play a key role in the maintenance of normal pancreatic exocrine, and especially endocrine, function (28–31). Failure of the insulin-producing pancreatic function is characteristic for Wolcott-Rallison syndrome, caused by inactivating mutations of PERK in humans (28). Pancreatic inflammation, loss of pancreatic tissue (including the β cells), and development of insulin-dependent diabetic syndrome was also described in mice either constitutively lacking Perk or undergoing inducible Perk ablation (32–38).
Intriguingly, we have recently identified an important role of PERK in the hypoxia- or virus replication-induced UPR-mediated ubiquitination and down-regulation of the IFNAR1 chain of type 1 IFN receptor (39–42). IFNs play important antiviral, antitumor, and immunomodulatory functions (43), yet can elicit and mediate pathologic scenarios (44). IFN has long been linked to pancreatic dysfunction in humans via elevated IFN expression in pancreatic tissues of patients with type 1 diabetes mellitus (45, 46) and induction of pancreatitis (47–49) and diabetogenic effects (50, 51) by pharmaceutical IFN used for treatment of tumors or viral infections. In addition, experiments in mouse models demonstrated that transgenic expression of IFN in β cells leads to diabetes (52), and that development of diabetes in the nonobese diabetic mice depends on production of IFN (53), as well as functional status of IFNAR1 (54).
Given that maintenance of threshold IFNAR1 levels is essential for the antiproliferative, proapoptotic, and immunopathological effects of IFN (55, 56), and that stabilization of IFNAR1 exacerbates acute and chronic inflammation in the pancreas (57), we proposed to test the role of IFN in the pancreatic toxicity of PERK inactivation. Work described here reveals that IFN is induced in the pancreas of mice lacking Perk ubiquitously or specifically in the pancreas. Knockout of IFNAR1 alleviated pancreatic tissue damage and endocrine dysfunction induced by Perk ablation. Conversely, an accelerated development of diabetic syndrome can be generated in mice lacking Perk, yet expressing the mutant Ifnar1 allele, whose protein product is insensitive to all known inducers of ubiquitination and degradation. Furthermore, either knockout of Ifnar1 or the use of neutralizing anti-IFNAR1 antibodies attenuated the pancreatic toxicities of PERK inhibitor in vitro and in vivo. These results indicate that IFN signaling plays a central role in mediating the pancreatic toxicity of PERK inactivation and suggests that modulating IFN responses may help treat the patients with Wolcott-Rallison and broaden the use of PERK inhibitors for therapeutic purposes.
Results
Activation of IFN Signaling upon PERK Inactivation Contributes to Apoptosis in Pancreatic Islets in Vitro.
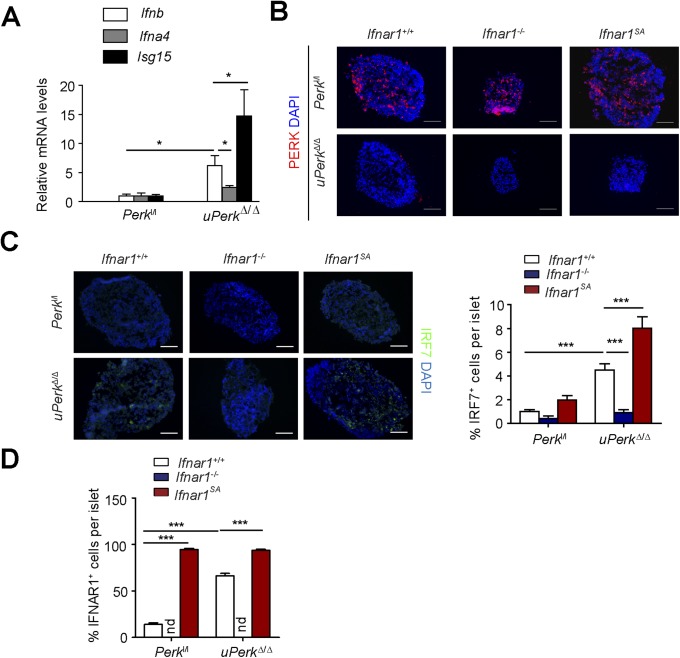
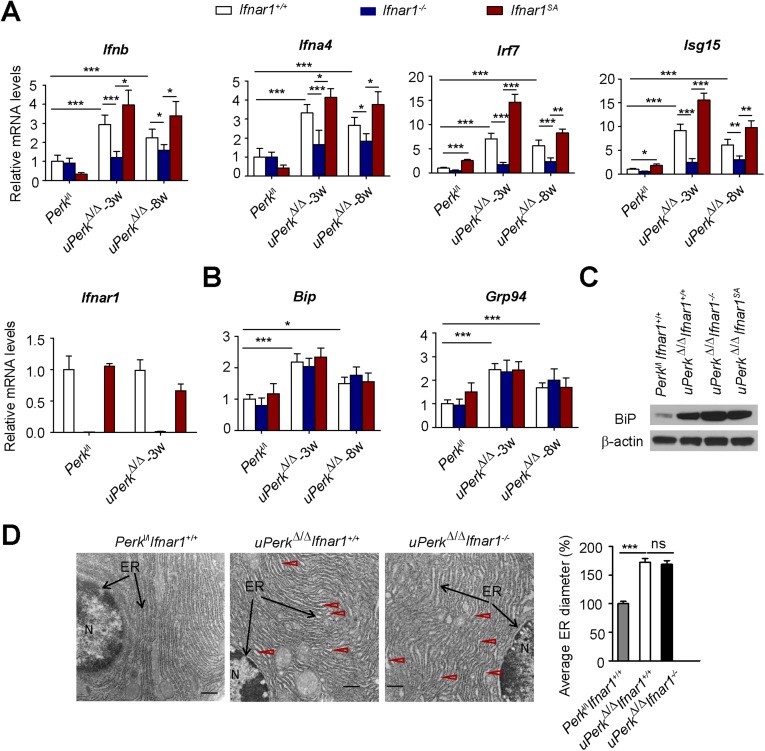
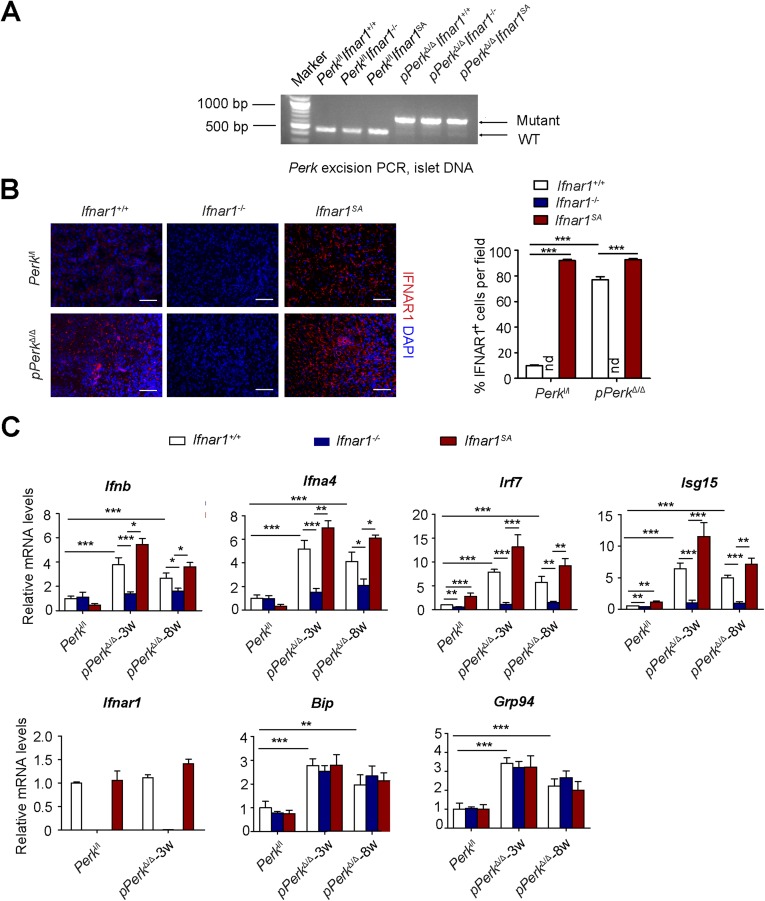
We previously demonstrated that acute excision of PERK in Perkl/l;Ubc9-CreERT mice (ubiquitous Perk deletion, uPerkΔ/Δ) resulted in a rapid decline in β-ell number and in development of a diabetic syndrome (32). Analysis of mRNA from purified islets from these mice revealed a significant induction of Ifnb and Ifna4 and IFN-stimulated gene Isg15 mRNA (Fig. S1A) associated with PERK excision. This induction may reflect stimulation of the IFN pathway in the pancreatic gland cells or/and infiltration of degenerating pancreas by immune cells that highly express these genes. To directly assess IFN signaling in these islet cells, we purified and cultured pancreatic islets from Perkl/l;Ubc9-CreERT mice before 4-hydroxytamoxifen-mediated Cre excision in vitro. This treatment efficiently decreased levels of PERK in cultured islets (Fig. S1B). Analysis of mRNA from these islets demonstrated that ablation of Perk resulted in a moderate induction of UPR-stimulated binding immunoglobulin protein (Bip) and Grp94 (Fig. 1A). Remarkably, Perk knockout robustly increased expression of IFN ligands (Ifna4 and Ifnb) and IFN-stimulated gene interferon regulatory factor 7 (Irf7) (Fig. 1A). Furthermore, levels of IRF7 protein were increased in islet cells after 4-hydroxytamoxifen treatment (Fig. S1C). These results suggest that Perk ablation leads to activation of IFN signaling in the pancreatic islets in vitro.
Fig. S1.
PERK suppresses IFN signaling in pancreatic islets. (A) mRNA levels of indicated genes in islets freshly isolated from Perkl/l; Ubc9-CreERT mice before (Perkl/l; values taken as 1.0) or after (uPerkΔ/Δ) tamoxifen treatment. Asterisk denotes P < 0.05. (B) Immunofluorescence analysis of PERK in the indicated genotypes of 4-ydroxytamoxifen-treated in vitro cultured islets. (Scale bars, 100 μm.) (C) Immunofluorescence analysis of IRF7 in the indicated genotypes of in vitro cultured islets. (Scale bars, 100 μm.) (D) Quantitation of results of immunofluorescent analysis of IFNAR1 protein levels in the cultured islets from indicated mice (from Fig. 1B). At least 10 islets observed in three independent experiments are shown. ***P < 0.001.
Fig. 1.
Activation of IFN signaling upon PERK inactivation contributes to apoptosis in pancreatic islets in vitro. (A) Expression of mRNA of indicated genes from in vitro cultured pancreatic islets from indicated mice was assessed by quantitative PCR. Asterisks here and thereafter indicate statistical significance: *P < 0.05, **P < 0.01, ***P < 0.001. (B) Immunofluorescent analysis of IFNAR1 protein levels in the cultured islets from indicated mice. (Scale bars, 100 μm.) (C) Analysis of cell death in the indicated cultured islets was carried out using the TUNEL labeling counterstained with antibody against insulin and DAPI. (Right) Quantitation of results from at least 10 islets observed in three independent experiments. (Scale bars, 50 μm.) (D) Analysis of cell death in the indicated islets cultured in presence of anti-IFNAR1 neutralizing or control antibody and treated with vehicle or inhibitors of CK1 (D4476, 50 µM), p38 kinase (SB203580, 10 µM), or PERK (GSK2606414, 1 µM) for 2 d. (Scale bars, 50 μm.) (E) Quantitation of results from D. Average data from at least 10 islets observed in three independent experiments are shown.
In cultured fibroblasts and HeLa cells, UPR signaling accelerated ligand-independent ubiquitination and degradation of IFNAR1 (58) in a manner that required activities of PERK (39, 41, 42), p38 kinase (40), and casein kinase 1α (59). Accordingly, ablation of Perk in cultured islets in vitro led to a robust increase in the IFNAR1 levels (Fig. 1B and Fig. S1D). The specificity of this signal was ensured by comparison with a negative control represented by the islets from Perkl/l mice lacking Ifnar1 and positive control [islets from Perkl/l mice that harbor the knocked-in Ifnar1SA alleles encoding the IFNAR1S526A mutant protein, which is insensitive to ubiquitination induced via PERK, as well as other PERK-independent pathways (57)]. Given the data from biochemical studies implicating PERK in the regulation of IFNAR1 ubiquitination and turnover (39, 42), and the fact that we did not observe a concurrent increase in Ifnar1 mRNA levels (Fig. 1A), these data are indicative of posttranscriptional mechanisms that increase IFNAR1 protein (e.g., stabilization) in Perk-deficient pancreatic islets in vitro.
Importantly, ablation of both Perk and Ifnar1 attenuated IRF7 mRNA and protein induction (Fig. 1A and Fig. S1C), consistent with reduced IFN-dependent signaling. Conversely, even a greater induction of IRF7 was seen in islets whose cells expressed an ubiquitination-deficient IFNAR1SA (Fig. S1 A and C). Collectively, these results suggest that ablation of Perk may induce IFN signaling by concurrent induction of IFN expression and partial stabilization of IFNAR1.
IFN is well-known for its proapoptotic and antiproliferative effects (43). Given that ablation of Perk triggers islets cell death (32, 38), we next sought to examine the role of IFN and IFNAR1 in the islet cells. The increase in TUNEL-positive cells in Perk-deficient cultures was attenuated after Ifnar1 loss, yet further enlarged in PerkΔ/Δ, Ifnar1SA islets (Fig. 1C). Furthermore, cell death induced by treatment of islets from wild-type mice (but not from mice lacking Ifnar1) with the PERK inhibitor, GSK2606414, could be attenuated by culturing the islets in the presence of an IFNAR1-blocking antibody (Fig. 1 D and E). Importantly, use of this antibody or knockout of IFNAR1 also attenuated cell death induced by inhibitors of either p38 kinase (SB203580) or CK1 (D4476, Fig. 1 D and E), which are kinases known to function downstream of PERK in stimulating IFNAR1 ubiquitination and degradation (40–42, 59). Collectively, these results suggest that PERK inactivation triggers the induction of IFN and stabilization of IFNAR1, leading to the activation of the cell death pathways in pancreatic islets in vitro.
Perk Antagonizes IFN Signaling, Thereby Preventing Pancreatic Exocrine and Endocrine Tissue Injury and Dysfunction.
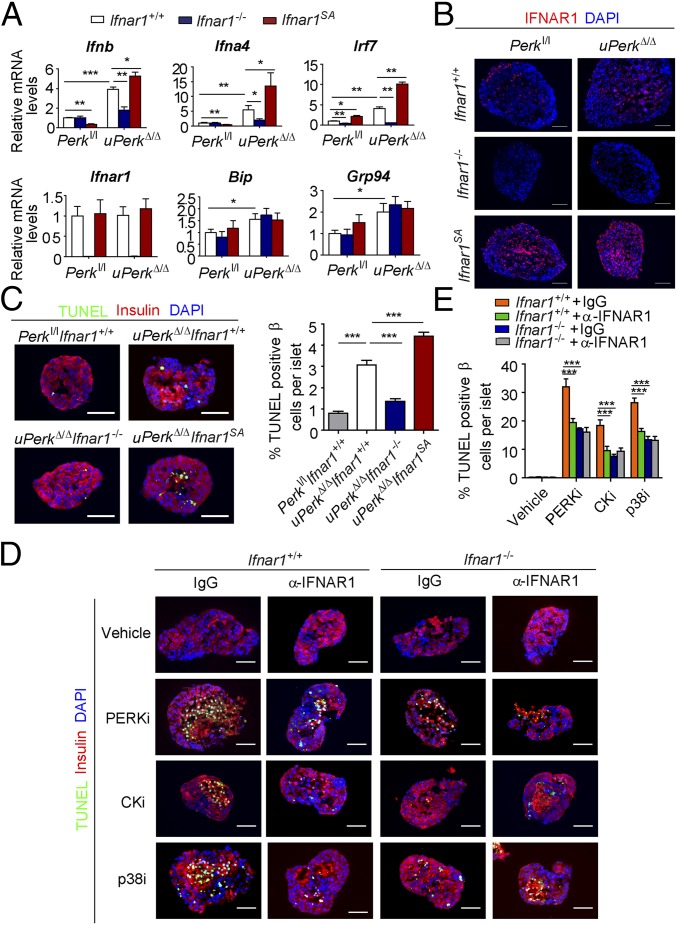
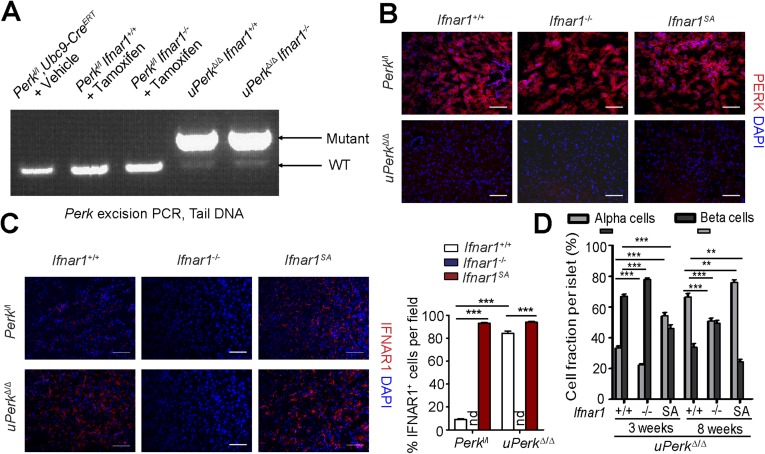
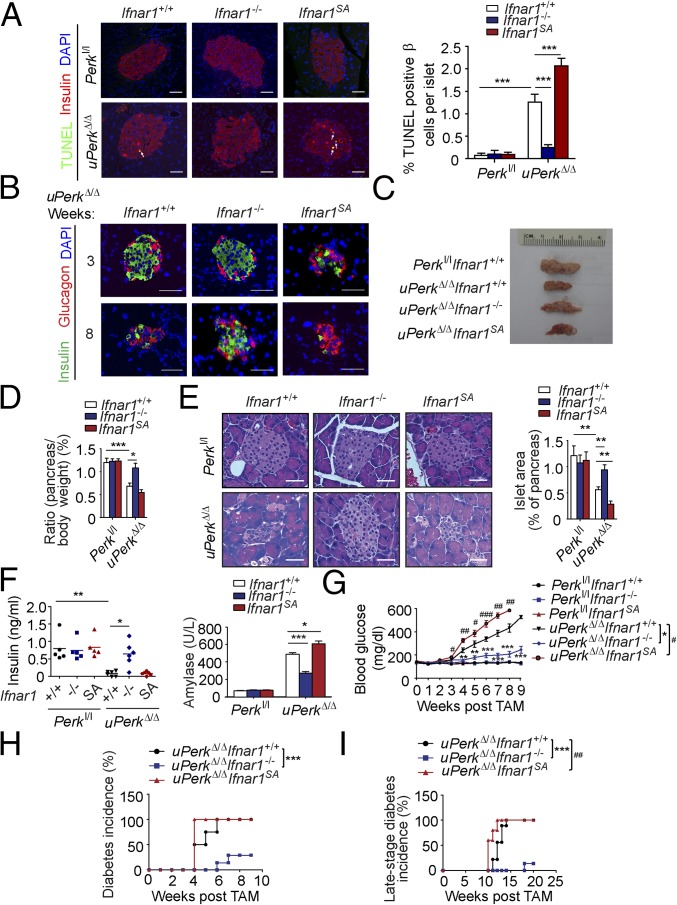
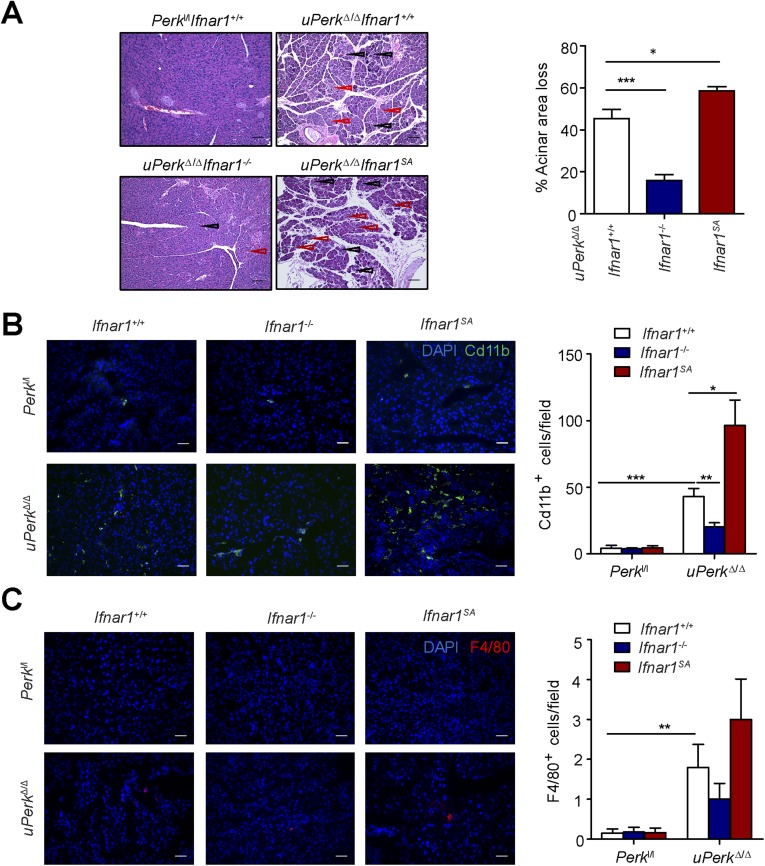
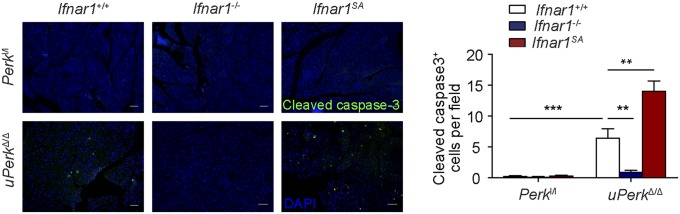
We sought to determine the role of IFN signaling in pancreatic dysfunction caused by ubiquitous Perk inactivation (uPerkΔ/Δ) in vivo. Consistent with previous work (32), tamoxifen treatment of Perkl/l;Ubc9-CreERT mice led to efficient Perk ablation (Fig. S2 A and B) and signs of pancreatic injury, including increased β-cell death, loss in β-cell mass, reduced pancreatic mass, decreased insulin production, increased amylase levels, signs of acinar cells loss, and the development of a fully manifested diabetic syndrome within 5 wk (Fig. 2 and Fig. S3A). Analysis of pancreatic tissues from these uPerkΔ/Δ mice revealed a robust increase in the levels of IFNAR1 protein (Fig. S2C). Pancreatic tissues from wild-type, Ifnar1-deficient, and Ifnar1SA mice exhibited a similar efficacy of Perk excision (Fig. S2 A and B). Furthermore, IFNAR1 status did not affect the severity of UPR, judging by the similar induction of UPR-stimulated genes and proteins (e.g., BiP; Fig. S4 B and C) or similarly extended endoplasmic reticulum analyzed by electron microscopy (Fig. S4D). However, ablation of Ifnar1 significantly decreased the expression of IFN (Ifnb and Ifna4) and practically abrogated the expression of IFN-stimulated genes (Irf7 and Isg15), whereas the latter levels were superinduced in Ifnar1SA mice (Fig. S4A). Importantly, these changes were mirrored by changes in the frequency of pancreatic cell death after Perk excision. Concurrent ablation of Ifnar1 attenuated cell death in the Perk-deficient pancreata, whereas a significantly greater level of cell death was seen in Ifnar1SA tissues (Fig. 2A and Fig. S5). These results collectively suggest that PERK functions to partially suppress the IFN signaling in the normal pancreas. Furthermore, these data further implicate IFN signaling in pancreatic cell death caused by Perk inactivation.
Fig. S2.
PERK induces down-regulation of IFNAR1 protein in the pancreas. (A) PCR analysis of Perk excision in genomic DNA obtained from tails of mice of the indicated genotypes. (B) Immunofluorescence analysis of PERK in the pancreas frozen sections of the indicated genotypes. (Scale bars, 100 μm.) (C) Immunofluorescence analysis of IFNAR1 in the pancreas frozen sections of the indicated genotypes. (Scale bars, 100 μm.) (D) Quantitation of percentage of glucagon-positive alpha cells and insulin-positive beta cells per islet from indicated mice (from Fig. 2B). Fourteen to 22 islets observed in three independent experiments is shown. **P < 0.01; ***P < 0.001.
Fig. 2.
Ubiquitous inactivation of PERK in vivo up-regulates IFNAR1 and its signaling, leading to pancreatic exocrine and endocrine tissue injury and dysfunction. (A) Analysis of TUNEL-positive β cells in pancreatic tissues from indicated mice was carried out 3 wk after tamoxifen treatment. Arrows indicate TUNEL (green), insulin (red), and DAPI triple-positive cells. (Right) Quantitation of results from 14 to 22 islets. (Scale bars, 50 µm.) (B) A representative islet immunostaining for insulin (green) and glucagon (red) was performed on pancreatic tissues from indicated mice killed at the indicated time after tamoxifen treatment. (Scale bars, 50 µm.) (C) Gross appearance of whole pancreata harvested from uPerkΔ/Δ and control mice 12 wk after tamoxifen. (D) Quantification of specific weight of pancreata from indicated mice (n = 3–4 for each group). (E) H&E staining of the islets of the indicated mice 12 wk after tamoxifen treatment. (Right) Quantification of the islet area from three to four mice. (Scale bars, 50 µm.) (F) Analysis of serum insulin (left) and amylase (right) levels from the indicated mice 12 wk after tamoxifen treatment. (G) Blood glucose was measured every week after tamoxifen treatment (n = 5–8 mice in each case; data shown as mean ± SEM). (G–I) #P = uPerkΔ/ΔIfnar1SA vs. uPerkΔ/ΔIfnar1+/+. #P < 0.05; ##P < 0.01; ###P < 0.001. *P = uPerkΔ/ΔIfnar1−/− vs. uPerkΔ/ΔIfnar1+/+. *P < 0.05; **P < 0.01; ***P < 0.001. (H) Diabetes incidence (blood glucose >250 mg/dL for two consecutive measurements) was analyzed from the indicated mice in F. (I) Late-stage diabetes (blood glucose >600 mg/dL for three consecutive measurements) incidence was analyzed from the indicated mice in F.
Fig. S3.
PERK suppresses IFN signaling in the pancreas to prevent pancreatic damage and immune cell infiltration. (A) H&E staining of the pancreas of the indicated mice 12 wk after tamoxifen treatment. Black and red arrows point the regions of acinar atrophy and leukocyte infiltration, respectively. (Scale bars, 100 µm.) (Right) Quantification of acinar area loss from three to four mice. (B and C) Immunofluorescent analysis of myeloid cell (B) and macrophage (C) infiltration into pancreata from uPerkΔ/Δ and control mice (harvested 3 wk after tamoxifen treatment) was carried out, respectively, on frozen sections using anti-CD11b antibody (B) or anti-F4/80 antibody (C). (Right) Quantitation of results from 10 fields observed in tissues from three mice. (Scale bars, 100 µm.) *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S4.
Perk ablation-induced IFN signaling did not affect UPR signaling or endoplasmic reticulum status in the pancreas. Expression of IFN and IFN-stimulated genes (A) and UPR-induced genes (B) was analyzed by real-time PCR. Expression of UPR-induced BiP protein (C) was analyzed by immunoblot. Levels of β-actin are shown as a loading control. (D) Electron microscopy pictures of pancreatic acinar cells from pancreatic tissues from mice of the indicated genotypes. The nucleus is marked as “N,” and black arrows point to the endoplasmic reticulum. Red arrows points to the dilated endoplasmic reticulum filled with electron-dense material. (Scale bars, 0.5 μm.) Quantification data were analyzed from at least 100 randomly chosen endoplasmic reticulum tubules from mice of the indicated genotypes. *P < 0.05; **P < 0.01; ***P < 0.001; ns indicates not significant (P > 0.05).
Fig. S5.
Immunofluorescence analysis of cleaved caspase-3 in the pancreatic tissues paraffin sections obtained from mice of indicated genotypes. (Right) Quantification of cleaved caspase-3 positive cells from more than 10 fields of three mice in each indicated genotypes. (Scale bars, 100 μm.) **P < 0.01; ***P < 0.001.
Loss of IFNAR1 (Ifnar1−/−) attenuated, whereas stabilization of IFNAR1 (Ifnar1SA) dramatically exacerbated, Perk excision-induced alterations in the islet structure, size, and β-cell numbers (Fig. 2 B and E and Fig. S2D); in overall size and weight of wet pancreata (Fig. 2 C and D); and in underlying histopathologic changes (Fig. 2E and Fig. S3A). These changes included atrophic alterations in the acinar cells whose death manifested itself in increased serum amylase levels attenuated in Ifnar1-null mice and aggravated in Ifnar1SA animals (Fig. 2F). Profound loss of islets (Fig. 2E) and a decrease in serum insulin levels (Fig. 2F) followed the same trend. Finally, stabilization of IFNAR1 in Ifnar1SA animals accelerated development of Perk deficiency-induced diabetes, which was dramatically delayed and moderated in the Ifnar1 knockout animals (Fig. 2 F–I). In all, these data strongly suggest that effects of IFN are largely responsible for pancreatic toxicity that occurs upon inactivation of Perk.
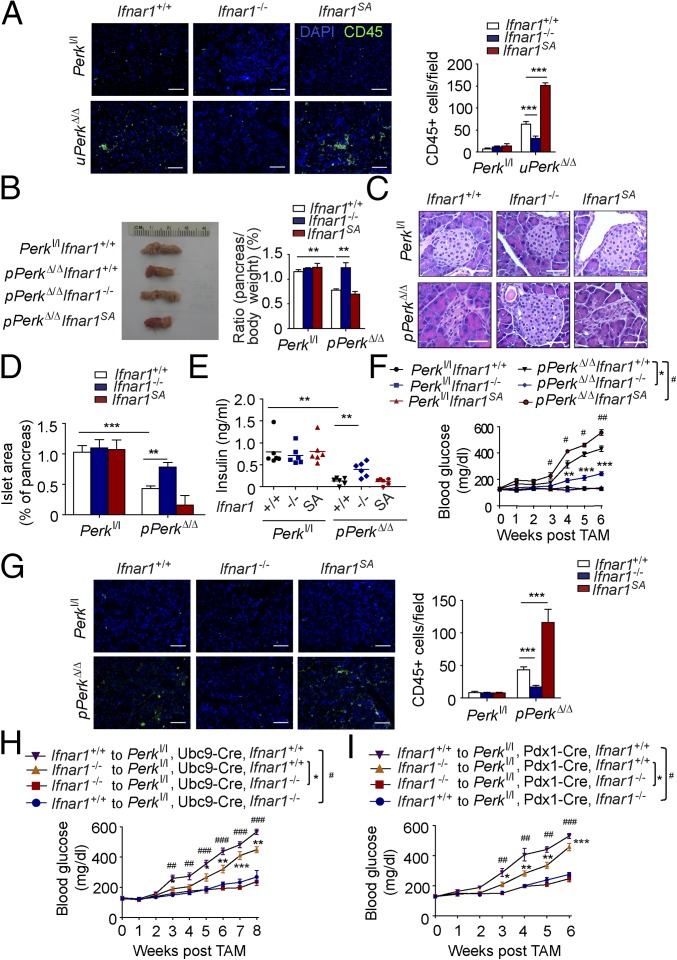
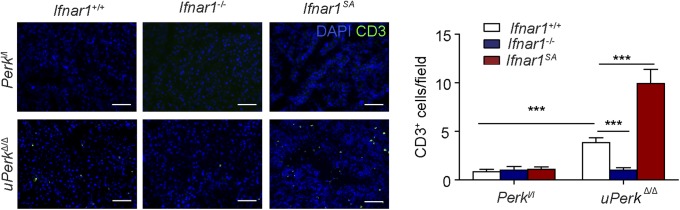
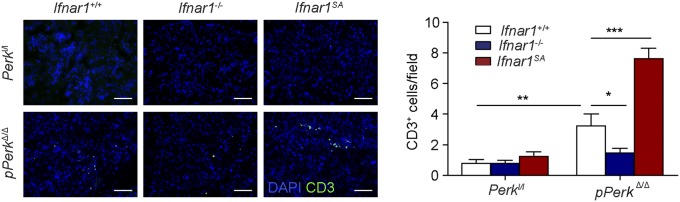
Ubiquitous ablation of Perk in these studies could use IFN signaling to elicit detrimental changes in the pancreas directly or via Perk-dependent alterations in the immune cells. Indeed, the severity of IFNAR1-dependent changes in pancreatic injury and dysfunction (Fig. 2) paralleled the extent of pancreatic tissue infiltration with leukocytes (Fig. 3A), including myeloid cells (but not macrophages, Fig. S3 B and C) and T lymphocytes (Fig. S6). Thus, we have examined the role of IFN signaling under conditions in which Perk ablation occurs via inducing CreERT, which is expressed under the Pdx1 promoter specifically in the pancreas (60). In Perkl/l, Pdx1-CreERT; Ifnar1+/+ mice, treatment with tamoxifen (resulting in pPerkΔ/Δ genotype) also triggered efficient Perk excision in the pancreas (Fig. S7A) and was associated with the β-cell loss and the development of a diabetic syndrome (ref. 32 and Fig. 3 B–F).
Fig. 3.
Pancreatic PERK suppresses IFN signaling in the pancreas to prevent pancreatic tissue injury and dysfunction. (A) Immunofluorescent analysis of leukocyte infiltration into pancreata from uPerkΔ/Δ and control mice (harvested 3 wk after tamoxifen treatment) was carried out on frozen sections using anti-CD45 antibody. (Right) Quantitation of results from 10 fields observed in tissues from three mice. (Scale bars, 100 µm.) (B) The picture of whole pancreata harvested from pPerkΔ/Δ and control mice killed 9 wk after tamoxifen treatment. (Right) Quantification of results from three to four mice. (C) H&E staining of the islets of pPerkΔ/Δ and control mice 9 wk after tamoxifen treatment. (D, Right) Quantification of the islet area from three to four mice. (Scale bars, 50 µm.) (E) Analysis of serum levels of insulin in pPerkΔ/Δ and control mice 9 wk after tamoxifen treatment. (F) Blood glucose was measured every week after tamoxifen treatment (n = 5–8 mice in each case; data shown as mean ± SEM). #P = uPerkΔ/ΔIfnar1SA vs. uPerkΔ/ΔIfnar1+/+. #P < 0.05; ##P < 0.01; ###P < 0.001. *P = uPerkΔ/ΔIfnar1−/− vs. uPerkΔ/ΔIfnar1+/+. *P < 0.05; **P < 0.01; ***P < 0.001. (G) Immunofluorescent analysis of pancreata from pPerkΔ/Δ and control mice (harvested 3 wk after tamoxifen treatment) was carried out on frozen sections using anti-CD45 antibody. (Right) Quantification. (Scale bars, 100 µm.) (H) Weekly blood glucose levels in Perkl/l; Ubc9-CreERT mice (harboring indicated Ifnar1 status) that received bone marrow from Ifnar1−/− or Ifnar1+/+ mice, and 8 wk later being treated with tamoxifen (n = 5–8 mice in each case; data shown as mean ± SEM). (I) Weekly blood glucose levels in Perkl/l; Pdx1-CreERT mice (harboring indicated Ifnar1 status) that received bone marrow from Ifnar1−/− or Ifnar1+/+ mice, and 8 wk later being treated with tamoxifen (n = 5–8 mice in each case; data shown as mean ± SEM).
Fig. S6.
Immunofluorescence analysis of CD3-positive cells in the frozen sections from pancreata of indicated genotypes. (Right) Quantification of data from more than 10 fields of three mice in each indicated genotype. (Scale bars, 100 μm.) ***P < 0.001.
Fig. S7.
Pancreatic Perk suppresses IFN signaling. (A) PCR analysis of Perk excision in genomic DNA obtained from pancreatic islets from mice of the indicated genotypes. (B) Immunofluorescence analysis of IFNAR1 in the frozen sections of pancreatic tissues obtained from mice of indicated genotypes. (Scale bars, 100 μm.) (C) Real-time PCR analysis of mRNA levels of genes downstream of IFN and UPR signaling in pancreas of mice of the indicated genotypes harvested at 3 or 8 weeks after tamoxifen treatment. *P < 0.05; **P < 0.01; ***P < 0.001.
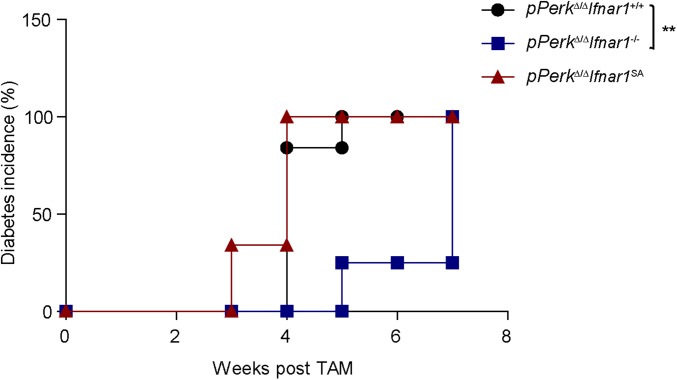
Intriguingly, pancreas-specific Perk ablation in pPerkΔ/Δ mice also increased levels of pancreatic IFNAR1 (Fig. S7B), indicating that this increase is unlikely to result from recruitment of Perk-deficient leukocytes that highly express IFNAR1, as might be argued in uPerkΔ/Δ mice wherein PERK was excised ubiquitously. Similar to the results obtained in the uPerkΔ/Δ mice (Fig. 2), pancreas-specific pPerkΔ/Δ mice exhibited elevated IFN signaling that reflected the extent of expression and stability of IFNAR1 (Fig. S7C); the latter did not markedly alter UPR signaling (Fig. S7C). Importantly, experiments using Perkl/l, Pdx1-CreERT Ifnar1-null and Ifnar1SA mice clearly demonstrated that IFN signaling plays a key role in the Perk deficiency-induced loss of total pancreatic tissue (Fig. 3B), islets mass (Fig. 3 C and D), insulin levels (Fig. 3E), and development of diabetes (Fig. 3F and Fig. S8). These results in pPerkΔ/Δ mice support a model in which Perk functions to moderate IFN signaling in the pancreatic tissues to prevent IFN-dependent pancreatic injury and functional deficiency.
Fig. S8.
Diabetes incidence (blood glucose >250 mg/dL for two consecutive measurements) observed in the indicated mice. **P < 0.01.
Notably, pancreas-specific deletion of Perk still elicited immune infiltration of pancreas that was modulated by the IFNAR1 status (Fig. 3G and Fig. S9). Given that null or SA alleles of Ifnar1 are ubiquitous, it is plausible that Perk status in the pancreas signals to IFN pathway in the immune system, rather than in the pancreatic cells themselves. To test this possibility, we transferred bone marrow from Ifnar1+/+ or Ifnar1−/− mice into lethally irradiated Perkl/l; Ubc9-CreERT or Perkl/l; Pdx1-CreERT recipients before administering tamoxifen to excise Perk. Although chimeras receiving bone marrow from Ifnar1−/− mice exhibited only a modest delay in diabetes development (most likely associated with an additional immune role of IFN), the status of IFNAR1 in peripheral tissues was the major determinant of pancreatic toxicity because the Ifnar1-deficient recipients lacking Perk in all peripheral tissues (Fig. 3H), or specifically in pancreas (Fig. 3I), displayed a dramatic suppression of diabetic phenotype. In all, these results suggest that Perk negatively regulates IFN signaling within the peripheral tissues (including pancreas) to prevent the injury and dysfunction of the pancreas.
Fig. S9.
Immunofluorescence analysis of CD3-positive cells in the pancreas frozen sections of indicated genotypes. (Right) Quantification of data from more than 10 fields of three mice in each of indicated genotypes. (Scale bars, 100 μm.) *P < 0.05; **P < 0.01; ***P < 0.001.
Pharmacologic Inactivation of IFN Signaling Protects Pancreas from Toxic Effects of PERK Inhibitor.
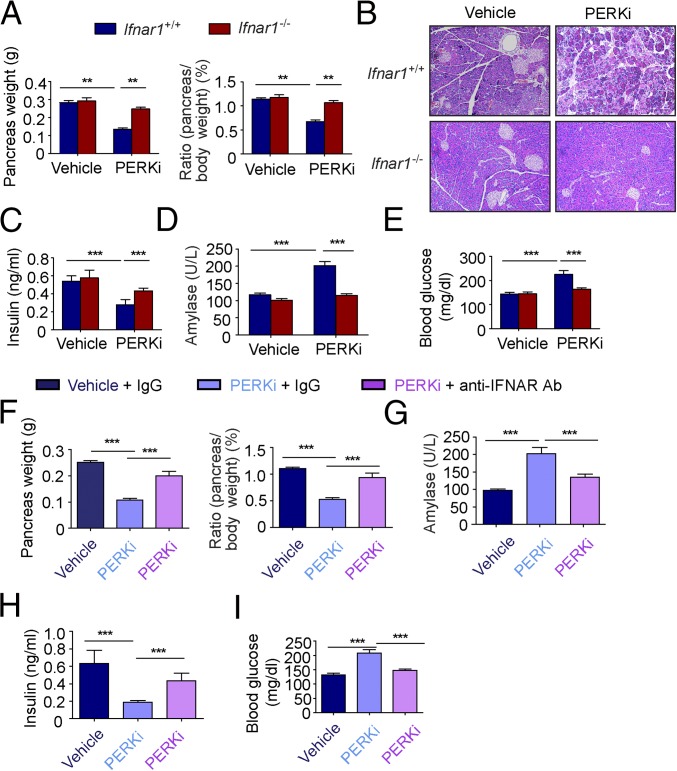
Given that IFNAR1 signaling contributes to apoptosis induced by GSK2606414 in cultured pancreatic islets (Fig. 1D), we aimed to determine whether knockout or antibody-based blocking of IFNAR1 can protect animals from pancreatic toxicity generated after treatment with this PERK inhibitor in vivo. Consistent with previously published reports (22), extended treatment with PERK inhibitor markedly decreased pancreas weight in wild-type mice (Fig. 4A) and caused noticeable degenerative changes in pancreatic islets and acinar cells (Fig. 4B), as well as decreased insulin levels (Fig. 4C) and increase in serum levels of amylase (Fig. 4D) and glucose (Fig. 4E). In addition, upon glucose challenge, wild-type mice treated with PERK inhibitor exhibited defects in stimulated insulin secretion and glucose tolerance (Fig. S10). Importantly, all these detrimental changes in pancreatic morphology and function were alleviated by Ifnar1 knockout (Fig. 4 A–E and Fig. S10).
Fig. 4.
Pharmacologic inactivation of IFN signaling protects pancreas from toxic effects of PERK inhibitor. (A) Analysis of absolute (Left) and specific (Right) weight of the pancreatic glands from wild-type or Ifnar1−/− mice treated with vehicle or PERK inhibitor GSK2606414 (150 mg/kg) for 14 d. (B) Representative H&E staining of the pancreas from the mice treated as in A. (Scale bars, 100 µm.) (C) Analysis of serum insulin levels from the mice treated as in A. (D) Analysis of serum amylase levels from the mice treated as in A. (E) Analysis of blood glucose levels from the mice treated as in A. (F) Analysis of absolute (Left) and specific (Right) weight of the pancreatic glands from wild-type mice treated with vehicle or PERK inhibitor GSK2606414 for 14 d combined with anti-IFNAR1 antibody or its isotype control treatment (every 5 d, 1 mg i.p. injection per mouse). (G) Analysis of serum amylase levels from the mice treated as in F. (H) Analysis of serum insulin levels from the mice treated as in F. (I) Analysis of blood glucose levels from the mice treated as in F. **P < 0.01; ***P < 0.001.
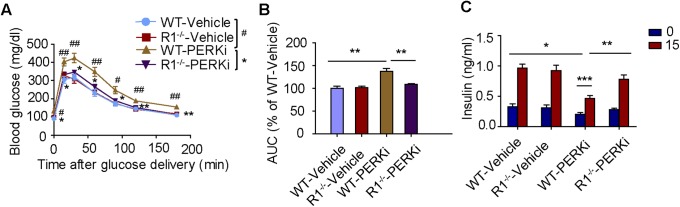
Fig. S10.
PERK inhibitor elicits pancreatic toxicity in an IFNAR1-dependent manner. GSK2606414-treated wild-type mice are more glucose intolerant and secrete less stimulated insulin compared with the same treated Ifnar1−/− mice. (A) Glucose tolerance test of vehicle or GSK2606414-treated wild-type or Ifnar1−/−mice (n = 5 for each). #P < 0.05; ##P < 0.01 compared to R1−/−-PERKi. Areas under curve were quantified in B. Blood was also sampled for secreted insulin at 0 and 15 min after glucose delivery during glucose tolerance test (C). *P < 0.05; **P < 0.01; ***P < 0.001.
Furthermore, administration of anti-IFNAR1 neutralizing antibody [previously shown to alleviate development of diabetes in NOD mice (54)] at least partially rescued GSK2606414-induced changes in pancreatic mass (Fig. 4F), amylase (Fig. 4G), insulin (Fig. 4H), and glucose levels and tolerance (Fig. 4I). These data indicate that IFN signaling plays an important role in pancreatic dysfunction caused by PERK inhibitors and provides a proof of principle for blocking the IFN pathway to reduce pancreatotoxic effects of PERK inactivation.
Discussion
Genetic (28–31) or pharmacologic (22, 25–27) inactivation of Perk in mice and humans contributes to degenerative changes in the pancreas and its ensuing exocrine and endocrine disorders, including development of diabetes mellitus. Here we demonstrate that either knockout of IFNAR1 or its blockade, using specific antibody, elicits a profound rescue effect on the viability and number of pancreatic exocrine and endocrine cells. The results provide strong support for a model in which Perk normally functions to restrict IFN signaling in the normal pancreas. Furthermore, pancreatic injury and dysfunction triggered by Perk inactivation reflect increased IFN signaling and are largely (but not exclusively) mediated by IFN effects.
Comparison of in vivo data from uPerkΔ/Δ mice (Fig. 2) with in vitro data obtained in cultured islets (Fig. 1) suggests that toxic effects of IFN are at least in part mediated by direct IFN action on the β cells. Importantly, IFN signaling is induced and can cause damage even if Perk is specifically inactivated in the pancreas itself (pPerkΔ/Δ; Fig. 3). Additional experiments using bone marrow transplantation indicate that pancreatotoxicity is largely caused by the IFN action on the peripheral tissues. Although the role of IFN in eliciting the immunopathologic injury to the pancreas via additional changes in the immune system cannot be ruled out, our current data strongly suggest that Perk-deficient pancreatic parenchymal cells expressing IFNAR1 are directly sensitive to the toxic effects of IFN.
PERK was shown to mediate the UPR-induced IFNAR1 ubiquitination and degradation in vitro (42, 58, 59).The increase in IFNAR1 protein, but not mRNA levels after Perk ablation, has provided in vivo proof regarding the importance of PERK in regulating the IFNAR1 protein levels. Hyperactivation of the IFN pathway upon Perk deletion is likely a sum of IFN induction triggered by the UPR, as well as increased ability of this IFN to elicit the signaling via engaging highly expressed IFNAR1 (Figs. 1–3). Indeed, although complete IFNAR1 stabilization (Ifnar1SA mice) further exacerbated Perk deficiency-induced pancreatic toxicity, ablation of IFNAR1 alleviated detrimental effects to pancreatic cells and tissues in vitro and in vivo (Figs. 1–4).
The ability of anti-IFN therapy to antagonize pancreatic toxicity caused by Perk loss or inhibition is of significant practical importance. Current medical efforts are focused on the development of antibody-based drugs (sifalimumab, rontalizumab, etc.) for neutralizing IFNAR1 in patients with diverse inflammatory/autoimmune syndromes (61, 62). Our data suggest that a similar strategy could be potentially envisioned for relieving IFN-mediated pancreatotoxicity in patients with Wolcott-Rallison or patients who receive PERK inhibitors for treating tumors (22–24) and prion-mediated neurogenerative disorders (25).
Materials and Methods
Animals.
All experiments with animals were carried out under the protocols 803995 and 804470 approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. All mice had water ad libitum and were fed regular chow. Ubc9-CreERT (gift from E. Brown, University of Pennsylvania) and Pdx1-CreERT mice (gift from X. Hua, University of Pennsylvania) were crossed with Perkl/l mice (kindly provided by D. Cavener, Penn State University) and either Ifnar1−/− mice (a kind gift of Dr. Dong-Er Zhang, University of California, San Diego) or Ifnar1S526A/S526A mice (Ifnar1SA, described in ref. 57) to generate future uPerkΔ/Δ or pPerkΔ/Δ littermates and their controls. Genotyping of mice using tail DNA or islet DNA was performed by PCR. Only male mice were used for the experiments. The method to induce Perk deletion is described in SI Materials and Methods.
For induction of pancreatic toxicity in mice by GSK2606414 treatment, IFNAR1 neutralizing antibody (63) treatment in mice, bone marrow transplantation assay, glucose tolerance test, and measurement of blood glucose, insulin, and amylase, see SI Materials and Methods.
Islet Culture and Treatments.
Islet isolation was described previously (32). For details regarding islet isolation and treatment, and TUNEL assay to assess β-cell death, see SI Materials and Methods.
Histopathology, Immunological, and Other Techniques.
For the immunostaining of frozen sections, H&E staining, the immunostaining of paraffin sections, TUNEL assay, and immunoblotting, see SI Materials and Methods. Detailed imaging protocols, Fuji software (64) and Illustrator image processing, data analyzing, and statistics were included in SI Materials and Methods. For details about electron microscopy, methods for RNA isolation, cDNA synthesis, quantitative PCR, and the sequences of the primers, see SI Materials and Methods.
Statistics.
Every shown quantified result is an average of at least three independent experiments carried out in either triplicate or quadruplicate and calculated as means ± SE. The P values were calculated using the two-tailed Student t test. Diabetes incidence was compared by using the log-rank test.
SI Materials and Methods
Animals.
All experiments with animals were carried out under protocols 803995 and 804470, approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. All mice had water ad libitum and were fed regular chow. Ubc9-CreERT (gift from E. Brown, University of Pennsylvania) and Pdx1- CreERT (gift from X. Hua, University of Pennsylvania) mice were crossed with Perkl/l mice (kindly provided by D. Cavener, Penn State University) and either Ifnar1−/− mice (a kind gift of Dr. Dong-Er Zhang, University of California, San Diego) or Ifnar1S526A/S526A mice (Ifnar1SA, described in ref. 57) to generate future uPerkΔ/Δ or pPerkΔ/Δ littermates and their controls. Genotyping of mice using tail DNA or islet DNA was performed by PCR. Only male mice of 8–10 wk of age were used for the experiments. To induce Perk deletion, tamoxifen (Sigma) was given once daily via oral gavage for 5 consecutive days at a dose of 0.2 mg/g of body weight/day. Time after tamoxifen treatment was determined after the last treatment.
To induce pancreatic toxicity in mice, 8–12-wk Ifnar1+/+ or Ifnar1−/− mice were treated with 150 mg/kg GSK2606414 (formulated in 0.5% hydroxypropyl methyl cellulose, 0.1% tween-80 in water at pH 4.0) or the same volume of vehicle (orally, twice daily) for 14 d. To investigate whether IFNAR1 neutralizing antibody could influence GSK2606414-induced pancreatic toxicity, low endotoxin functional formulation anti-IFNAR1 antibody (I-401, clone is MAR1-5A3; Leico) or isotype control (mouse IgG, I-536; Leinco) were injected (i.p., 1 mg per mouse) at the first day of GSK2606414 treatment and then every 5 d after the first injection. The half-life of MAR1-5A3 has been demonstrated as 5.2 d when a sufficient amount of antibody is administered (63).
For bone marrow transplantation assay, bone marrow harvesting and transfers were carried out as described elsewhere (1). Briefly, the bone marrows of 8–12-wk Perkl/l, Ifnar1−/− or Perkl/l, Ifnar1+/+ mice were transferred to irradiated Ubc9-CreERT, Perkl/l, Ifnar1+/+; Ubc9-CreERT, Perkl/l, Ifnar1−/− or Pdx1-CreERT, Perkl/l, Ifnar1+/+ and Pdx1-CreERT, Perkl/l, Ifnar1−/− mice. Sixty days after bone marrow transplantation, the recipient mice were treated with tamoxifen for 5 d to induce Perk excision, as described earlier.
Blood glucose was measured using a Freestyle meter (Accu-Chek Aviva; Roche). Insulin levels were measured by the RIA and Biomarkers Core, University of Pennsylvania. Blood amylase levels were determined by using QuantiChrom α-Amylase Assay Kit (Bioassay Systems; DAMY-100). Glucose tolerance test were performed by injection of glucose (i.p., at 2 g/kg of body weight).
Islet Culture and Treatments.
Islet isolation was described previously (32). Islets were isolated from mice by collagenase digestion and cultured with 10 mM glucose in RPMI medium 1640 (Sigma) for 1 d. Then the islets were treated with vehicle or 1 μm 4-OH tamoxifen (Sigma) for 4 d or 1 μM PERK inhibitor GSK2606414 (Chembest), 10 μM p38 inhibitor SB203580 (Calbiochem), 50 μM CK1 inhibitor D4476 (Calbiochem) with or without 10 µg/mL α-IFNAR1 neutralizing antibody, or isotype control for 2 d. TUNEL assay were then carried out to assess β-cell death.
Histopathology and Immune Techniques.
For the immunostaining of frozen sections, pancreata harvested from mice or islets were frozen in tissue-Tek optimum cutting temperature (O.C.T.) compound and cryosectioned using the Leica CM3050 S Cryostats, fixed in acetone, washed, and blocked with PBS containing 5% (vol/vol) goat or donkey serum. The sections were incubated for 1 h with primary antibodies detecting PERK (Santa Cruz, 1/200), IFNAR1 (Sino Biological; 2 μg/mL), cleaved caspase-3 (Cell Signaling #9661; 1/400), and IRF7 (Abcam ab62505; 5 μg/mL). Then sections were washed and incubated with Alexa Fluor 488/594 goat anti-rabbit antibodies (Invitrogen; 1:500) or Alexa Fluor 594 Donkey anti-goat antibodies (Invitrogen; 1:500) for 1 h, washed again, and coverslipped using mounting solution with DAPI (Prolong Gold). To detect leukocytes, lymphocytes, or myeloid cell infiltration, the pancreatic sections were respectively stained with anti-CD45-FITC, anti-CD3-FITC, or anti-CD11b-FITC for 1 h. To detect macrophage infiltration, the sections were stained with biotinylated anti-F4/80 antibodies for 1 h, washed twice, and then incubated with DyLight 594-conjugated streptavidin for another 1 h. The slides were then washed twice and coverslipped, using mounting solution with DAPI. All the antibodies to detect immune cell infiltration were from Biolegend.
For H&E staining, pancreata were fixed in 4% (vol/vol) buffered formalin overnight. H&E sections were prepared according to standard protocols. For the immunostaining of paraffin sections, pancreata were dehydrated in gradient ethanol after fixation in 4% (vol/vol) buffered formalin and then embedded in paraffin. Tissue sections (6 μm) were subjected to immunostaining by using primary antibodies against insulin (Invitrogen, 1/200) and glucagon (Abcam, 1/200). Secondary antibodies labeled with Alexa Fluor 488/594 were applied after the primary antibody incubation. The slides were then washed twice and coverslipped, using mounting solution with DAPI.
TUNEL was performed on pancreas paraffin sections using TUNEL enzyme (Roche) and TUNEL label (Roche) kits according to the manufacturer’s instructions. Sections were subsequently stained for insulin and appropriate secondary antibody, according to the description given earlier.
Microscopy was carried out on Olympus systems, with magnification ranging from ×10 to ×40, and equipped with proprietary software; pictures from individual experiments were recorded with identical settings and identically processed with the Fuji software (64) and Illustrator. A total of at least randomly selected 10 fields observed in tissues from three mice were scored per group for quantification of percentage of cells, which were positive for CD3, CD45, CD11b, F4/80, or cleaved caspase-3. For TUNEL assay on pancreas sections, 14–22 islets observed from three mice were scored per group for quantification of TUNEL-positive β cells. At least 10 primary islets observed in three independent experiments were scored for IRF7-positve or TUNEL-positive β cells. All the quantifications were carried out in a double-blind manner.
Real-Time PCR.
Harvested islets or pancreas were flash-frozen and pulverized in liquid nitrogen, homogenized in TRIzol reagent, and extracted with chloroform. Reverse transcription was carried out using Revertaid first-strand cDNA synthesis kit (Thermo Scientific), and the cDNA was used for quantitative PCR carried out using Applied Biosystems 7500 Fast Real-Time PCR system, using the following primers: Ifnb (FW, 5′- GTCAGAGTGGAAATCCTAAG-3′, REV, 5′- ACAGCATCTGCTGGTTGAAG-3′), Ifna4 (FW, 5′-CCTGTGTGATGCAGGAACC-3′, REV, 5′- TCACCTCCCAGGCACAGA-3′), Ifnar1 (FW, 5′-CGACCAAGTGTGAATTCTCTTTAC-3′, REV, 5′- ATCAACCTCATTCCACGAAGAT-3′), Isg15 (FW, 5′-GGAACGAAAGGGGCCACAGCA-3′, REV, 5′-CCTCCATGGGCCTTCCCTCGA-3′), Irf7 (FW, 5′- CCACACCCCCATCTTCGA-3′, REV, 5′-CCTCCGAGCCCGAAACTC-3′), Bip (FW, 5′- ACCCTTACTCGGGCCAAATT-3′, REV, 5′- AGAGCGGAACAGGTCCATGT- 3′), Grp 94 (FW, 5′- CTGGGTCAAGCAGAAAGGAG-3′, REV, 5′- TCTCTGTTGCTTCCCGACTT -3′), and β-actin (FW, 5′- AGAGGGAAATCGTGCGTGAC-3′, REV, 5′- CAATAGTGATGACCTGGCCGT-3′).
Electron Microscopy.
Sample preparation was performed at the Biomedical Imaging Core of Abramson Cancer Center of the University of Pennsylvania, and the sectioned samples were imaged using a Jeol-1010 transmission electron microscope.
Immunoblotting.
First antibody detecting β-actin (Sigma) and BiP (Invitrogen) were used. Secondary antibodies conjugated to horseradish peroxidase were purchased from Millipore Bioscience Research Reagents. Immunoblotting procedures were described previously (4).
Statistics.
Every shown quantified result is an average of at least three independent experiments carried out in either triplicate or quadruplicate and calculated as means ± SE. The P values were calculated using the two-tailed Student t test. Diabetes incidence was compared by using the log-rank test.
Acknowledgments
We thank D. R. Cavener (Pennsylvania State University), D. E. Zhang (University of California, San Diego), E. Brown, and Xianxin Hua (University of Pennsylvania) for providing mouse strains, and the members of the C.K., J.A.D., S.Y.F., Roger A. Greenberg, and L. Busino laboratories for helpful discussion. This work was supported by NIH/National Cancer Institute Grant PO1 CA165997 (to J.A.D, C.K., and S.Y.F.), including help from the Scientific Cell/Tissue Morphology Core and its principal investigator, Dr. Qian-Chun Yu. Additional support from National Institutes of Health/National Cancer Institute Grant R01 CA092900 (to S.Y.F.) is greatly appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516362112/-/DCSupplemental.
References
- 1.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 2.Diehl JA, Fuchs SY, Koumenis C. The cell biology of the unfolded protein response. Gastroenterology. 2011;141:38–41. doi: 10.1053/j.gastro.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobrovnikova-Marjon E, et al. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29(27):3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6(1):55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 5.Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther. 2006;5(7):723–728. doi: 10.4161/cbt.5.7.2967. [DOI] [PubMed] [Google Scholar]
- 6.Bi M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24(19):3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res. 2012;72(20):5396–5406. doi: 10.1158/0008-5472.CAN-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamison S, Lin Y, Lin W. Pancreatic endoplasmic reticulum kinase activation promotes medulloblastoma cell migration and invasion through induction of vascular endothelial growth factor A. PLoS One. 2015;10(3):e0120252. doi: 10.1371/journal.pone.0120252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karali E, et al. VEGF Signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol Cell. 2014;54(4):559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Wouters BG, et al. Control of the hypoxic response through regulation of mRNA translation. Semin Cell Dev Biol. 2005;16(4-5):487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Blais JD, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol. 2006;26(24):9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blais JD, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24(17):7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins I, et al. Restoration of the immunogenicity of cisplatin-induced cancer cell death by endoplasmic reticulum stress. Oncogene. 2011;30(10):1147–1158. doi: 10.1038/onc.2010.500. [DOI] [PubMed] [Google Scholar]
- 14.Zitvogel L, et al. Immunogenic tumor cell death for optimal anticancer therapy: the calreticulin exposure pathway. Clin Cancer Res. 2010;2010;16(12):3100–3104. doi: 10.1158/1078-0432.CCR-09-2891. [DOI] [PubMed] [Google Scholar]
- 15.Hou X, et al. PERK silence inhibits glioma cell growth under low glucose stress by blockage of p-AKT and subsequent HK2's mitochondria translocation. Sci Rep. 2015;5:9065. doi: 10.1038/srep09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124(3):1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey S, Tameire F, Koumenis C. PERK-ing up autophagy during MYC-induced tumorigenesis. Autophagy. 2013;9(4):612–614. doi: 10.4161/auto.23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart LS, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122(12):4621–4634. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitnis NS, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48(3):353–364. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blais J, Bell JC. Novel therapeutic target: the PERKs of inhibiting the integrated stress response. Cell Cycle. 2006;5(24):2874–2877. doi: 10.4161/cc.5.24.3597. [DOI] [PubMed] [Google Scholar]
- 21.Maly DJ, Papa FR. Druggable sensors of the unfolded protein response. Nat Chem Biol. 2014;10(11):892–901. doi: 10.1038/nchembio.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73(6):1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 23.Axten JM, et al. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) J Med Chem. 2012;55(16):7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 24.Axten JM, et al. Discovery of GSK2656157: An Optimized PERK Inhibitor Selected for Preclinical Development. ACS Med Chem Lett. 2013;4(10):964–968. doi: 10.1021/ml400228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno JA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5(206):206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 26.Halliday M, et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015;6:e1672. doi: 10.1038/cddis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harding HP, Zyryanova AF, Ron D. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J Biol Chem. 2012;287(53):44338–44344. doi: 10.1074/jbc.M112.428987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julier C, Nicolino M. Wolcott-Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman RJ, Back SH, Song B, Han J, Hassler J. The unfolded protein response is required to maintain the integrity of the endoplasmic reticulum, prevent oxidative stress and preserve differentiation in β-cells. Diabetes Obes Metab. 2010;12(Suppl 2):99–107. doi: 10.1111/j.1463-1326.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavener DR, Gupta S, McGrath BC. PERK in beta cell biology and insulin biogenesis. Trends Endocrinol Metab. 2010;21(12):714–721. doi: 10.1016/j.tem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51(Suppl 3):S455–S461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, et al. PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Mol Cell Biol. 2012;32(24):5129–5139. doi: 10.1128/MCB.01009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, McGrath B, Cavener DR. PERK regulates the proliferation and development of insulin-secreting beta-cell tumors in the endocrine pancreas of mice. PLoS One. 2009;4(11):e8008. doi: 10.1371/journal.pone.0008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng D, Wei J, Gupta S, McGrath BC, Cavener DR. Acute ablation of PERK results in ER dysfunctions followed by reduced insulin secretion and cell proliferation. BMC Cell Biol. 2009;10:61. doi: 10.1186/1471-2121-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iida K, Li Y, McGrath BC, Frank A, Cavener DR. PERK eIF2 alpha kinase is required to regulate the viability of the exocrine pancreas in mice. BMC Cell Biol. 2007;8:38. doi: 10.1186/1471-2121-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, et al. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4(6):491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, et al. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya S, et al. Anti-tumorigenic effects of Type 1 interferon are subdued by integrated stress responses. Oncogene. 2013;32(36):4214–4221. doi: 10.1038/onc.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya S, et al. Role of p38 protein kinase in the ligand-independent ubiquitination and down-regulation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2011;286(25):22069–22076. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharya S, et al. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2010;285(4):2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, et al. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5(1):72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 44.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44(6):658–664. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- 46.Foulis AK, Farquharson MA, Meager A. Immunoreactive alpha-interferon in insulin-secreting beta cells in type 1 diabetes mellitus. Lancet. 1987;2(8573):1423–1427. doi: 10.1016/s0140-6736(87)91128-7. [DOI] [PubMed] [Google Scholar]
- 47.Tannir NM, Talpaz M, Ghazal H, Proothi S, Kantarjian HM. Acute pancreatitis associated with interferon alpha therapy for chronic myelogenous leukemia. Leuk Lymphoma. 2000;39(5-6):647–650. doi: 10.3109/10428190009113396. [DOI] [PubMed] [Google Scholar]
- 48.Eland IA, van Puijenbroek EP, Sturkenboom MJ, Wilson JH, Stricker BH. Drug-associated acute pancreatitis: twenty-one years of spontaneous reporting in The Netherlands. Am J Gastroenterol. 1999;94(9):2417–2422. doi: 10.1111/j.1572-0241.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 49.Morris DJ. Adverse effects and drug interactions of clinical importance with antiviral drugs. Drug Saf. 1994;10(4):281–291. doi: 10.2165/00002018-199410040-00002. [DOI] [PubMed] [Google Scholar]
- 50.Guerci AP, et al. Onset of insulin-dependent diabetes mellitus after interferon-alfa therapy for hairy cell leukaemia. Lancet. 1994;343(8906):1167–1168. doi: 10.1016/s0140-6736(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 51.Fabris P, et al. Type 1 diabetes mellitus in patients with chronic hepatitis C before and after interferon therapy. Aliment Pharmacol Ther. 2003;18(6):549–558. doi: 10.1046/j.1365-2036.2003.01681.x. [DOI] [PubMed] [Google Scholar]
- 52.Stewart TA, et al. Induction of type I diabetes by interferon-alpha in transgenic mice. Science. 1993;260(5116):1942–1946. doi: 10.1126/science.8100367. [DOI] [PubMed] [Google Scholar]
- 53.Li Q, McDevitt HO. The role of interferon alpha in initiation of type I diabetes in the NOD mouse. Clin Immunol. 2011;140(1):3–7. doi: 10.1016/j.clim.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Li Q, et al. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci USA. 2008;105(34):12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36(3):139–149. doi: 10.1016/j.it.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250(1):317–334. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya S, et al. Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol Med. 2014;6(3):384–397. doi: 10.1002/emmm.201303236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, et al. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun. 2008;367(2):388–393. doi: 10.1016/j.bbrc.2007.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, et al. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol Cell Biol. 2009;29(24):6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holland AM, Góñez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54(9):2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 61.Higgs BW, et al. A phase 1b clinical trial evaluating sifalimumab, an anti-IFN-α monoclonal antibody, shows target neutralisation of a type I IFN signature in blood of dermatomyositis and polymyositis patients. Ann Rheum Dis. 2014;73(1):256–262. doi: 10.1136/annrheumdis-2012-202794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McBride JM, et al. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis Rheum. 2012;64(11):3666–3676. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 63.Sheehan KC, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 64.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]