Significance
Microbial and animal rhodopsins share several striking structural features including a seven α-helix fold and a highly conserved active-site lysine in the seventh helix. However, these protein families lack significant similarity in their amino acid sequence. In this paper we address the question of why the sequence of animal rhodopsins, featuring an 11-cis chromophore, could have diverged from a microbial ancestor incorporating the more stable all-trans chromophore. We show that, by using light-responsive computer models of a eubacterial sensory rhodopsin and of a vertebrate visual rhodopsin, it is possible to identify a distinctive electronic character of the 11-cis chromophore that could have become an effective target for natural selection.
Keywords: rhodopsin, retinal chromophore, ultrafast isomerization, excited states, computational photobiology
Abstract
The functions of microbial and animal rhodopsins are triggered by the isomerization of their all-trans and 11-cis retinal chromophores, respectively. To lay the molecular basis driving the evolutionary transition from the all-trans to the 11-cis chromophore, multiconfigurational quantum chemistry is used to compare the isomerization mechanisms of the sensory rhodopsin from the cyanobacterium Anabaena PCC 7120 (ASR) and of the bovine rhodopsin (Rh). It is found that, despite their evolutionary distance, these eubacterial and vertebrate rhodopsins start to isomerize via distinct implementations of the same bicycle-pedal mechanism originally proposed by Warshel [Warshel A (1976) Nature 260:678–683]. However, by following the electronic structure changes of ASR (featuring the all-trans chromophore) during the isomerization, we find that ASR enters a region of degeneracy between the first and second excited states not found in Rh (featuring the 11-cis chromophore). We show that such degeneracy is modulated by the preorganized structure of the chromophore and by the position of the reactive double bond. It is argued that the optimization of the electronic properties of the chromophore, which affects the photoisomerization efficiency and the thermal isomerization barrier, provided a key factor for the emergence of the striking amino acid sequence divergence observed between the microbial and animal rhodopsins.
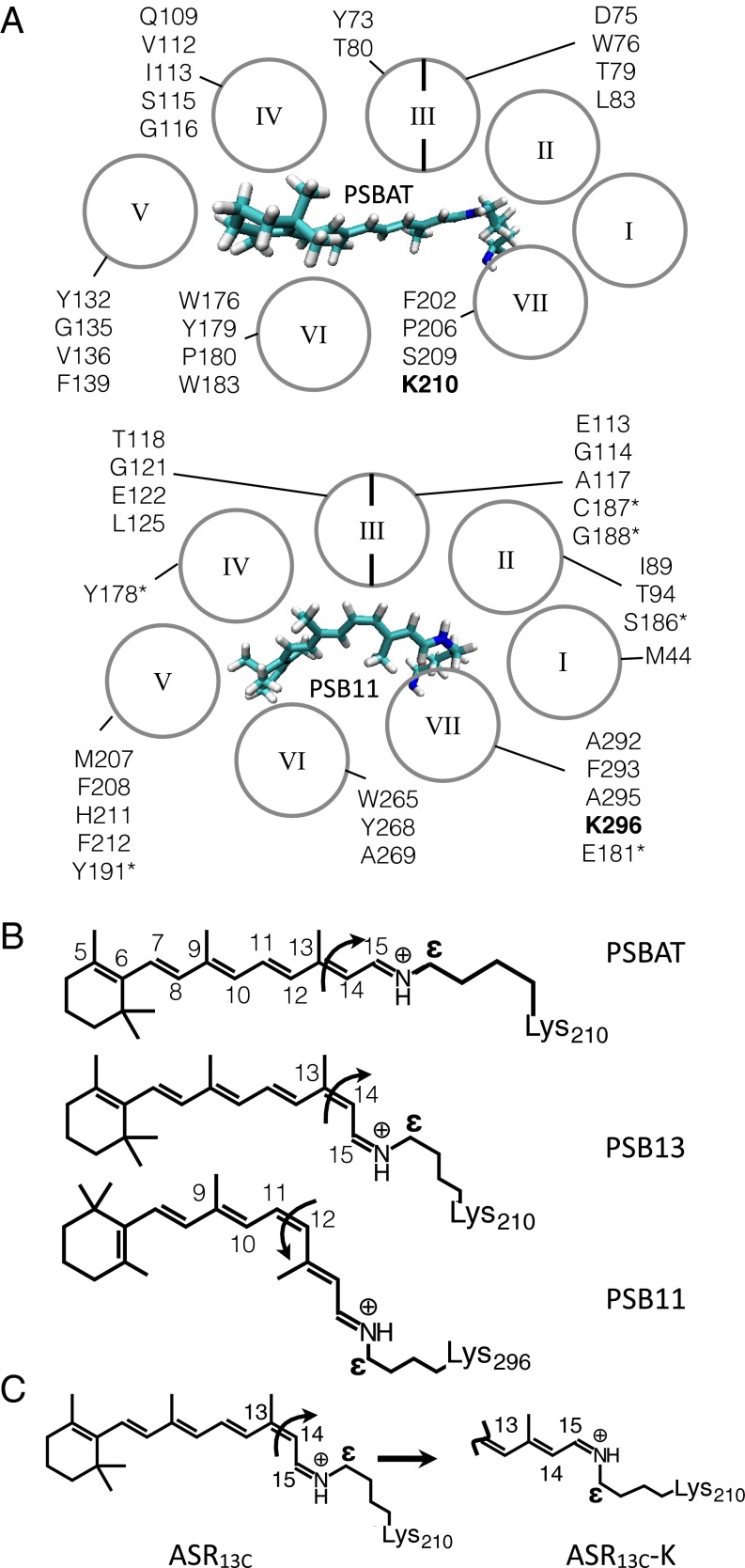
Rhodopsins (also called retinal or retinylidene proteins) are a family of membrane proteins found in all life domains (1). All members of the family bind a retinal chromophore and display either light-sensing, light-powered ion-pumping or light-gated ion-channeling activities. These functions are carried out by a common protein architecture featuring seven α-helices (I–VII in Fig. 1A) forming a cavity with a chromophore-binding lysine located in the middle of helix VII.
Fig. 1.
Rhodopsin structure. (A) Chromophore cavities of a type I (Upper, Anabaena PCC 7120) and a type II (Lower, Bos taurus) sensory rhodopsin. The cavity amino acids are given in one-letter code according to their α-helix location. Asterisk indicates the loop connecting helices IV and V. The chromophore-binding lysins are in bold. (B) Structures of the chromophores of ASRAT (Upper), ASR13C (Middle), and Rh (Lower). The curly arrows indicate the reactive double bond. (C) ASR13C→ASR13C-K chromophore isomerization reaction.
Despite their striking structural similarity, microbial (type I) and animal (type II) rhodopsins display a negligible amino acid sequence identity that is also reflected in dramatically different chromophore cavities. Although this would indicate that type I and type II rhodopsins have evolved in parallel from different ancestors, Theobald and coworkers (2, 3) have recently provided experimental evidence in favor of a single common ancestor. If this is the case, one is left with the problem of understanding the mechanisms that could have led to the drastic divergence in the amino acid sequences. Here, we investigate the role played by the transition to a different chromophore in such a process.
Type I and type II rhodopsins feature different retinal chromophores. Whereas type I rhodopsins have an all-trans chromophore (PSBAT in Fig. 1 A and B), type II rhodopsins incorporate an 11-cis chromophore (PSB11 in Fig. 1 A and B). PSBAT and PSB11 are responsible for the activation of the protein function that is triggered by the photoisomerization of their C13=C14 and C11=C12 double bond, respectively. However, the fact that PSB11 is thermodynamically unstable with respect to PSBAT indicates that PSB11 might have been selected to carry out an otherwise inaccessible function. In fact, given the basic role of the chromophore in the rhodopsin machinery, it is conceivable that the optimization of distinctive PSB11 functions would have led to an extreme divergence. To investigate whether such functions exist, we compare the isomerization mechanisms (see curly arrows in Fig. 1 B and C) of PSBAT, PSB13, and PSB11 at an atomic resolution. To do so we use computer models of microbial and vertebrate light-sensing rhodopsins (4, 5). The simulations unveil electronic effects not achievable by PSBAT and barely achievable in PSB13, directly related to the higher photoisomerization speed reported for type II rhodopsins and, in turn, to the control of the isomerization quantum efficiency (6–10).
The properties of the retinal chromophore are modulated by its interaction with the protein environment (11–16). For this reason, we simulate the chromophore isomerization in complete models of the sensory rhodopsin from the cyanobacterium Anabaena PCC 7120 (ASR) and of the visual pigment of the bovine retina (Rh), type I and a type II rhodopsins, respectively. ASR exists in two thermostable forms, ASRAT and ASR13C, featuring the 15-anti-PSBAT and 15-syn-PSB13 chromophores (Fig. 1B), respectively. Upon light irradiation, the chromophores isomerize about the C13=C14 double bond to generate the corresponding K intermediate (see Fig. 1C for ASR13C). Because ASRAT-K and ASR13C-K convert thermally to ASR13C and ASRAT, respectively, ASR displays a photochromic cycle (17). Such a photochromism is not observed in Rh, where irradiation causes the photoisomerization of PSB11 to PSBAT with formation of bathoRh. This is then thermally converted to states that interact with a transducer and hydrolyze the chromophore–lysine linkage (18).
The photoisomerization of Rh has been extensively investigated. Spectroscopic studies have shown that it occurs on a subpicosecond timescale with an S1 decay occurring in less than 100 fs (7, 19, 20). Furthermore, the observation of ground-state (S0) vibrational coherence (21) points to a direct transfer of the first excited state (S1) population to the photoproduct passing through a S1/S0 conical intersection (CI) without involving higher excited states (e.g., S2). Such a mechanism has been computationally documented (22–24) and experimentally tracked by infrared probing (24). ASRAT and ASR13C have been spectroscopically investigated by Ruhman and coworkers (25) and Haacke and coworkers (26, 27). The results indicate that in ASR13C the chromophore accesses the CI more quickly (0.10–0.15 ps) than in ASRAT (0.50–0.75 ps), showing a dynamics closer to that of Rh (0.08 ps). Such timescales have been found to be consistent with potential energy surface mapping and trajectory computations (23, 24, 28, 29).
The results revised above provide the basis for investigating the photoisomerization of type I and type II rhodopsins. Accordingly, we use multiconfigurational quantum chemistry (MCQC)-based quantum-mechanics/molecular-mechanics (QM/MM) models (30) of ASRAT, ASR13C and Rh to simulate their electronic and geometrical changes up to 200 fs after the electronic excitation. We show that the magnitude of the observed timescales correlate with the mixing of the S1 and S2 states and that such mixing does not occur in the PSB11 containing Rh. It is argued that this fact connects the optimization of the chromophore electronic properties to a higher light sensitivity of animal rhodopsins.
Results and Discussion
Absorption Maxima.
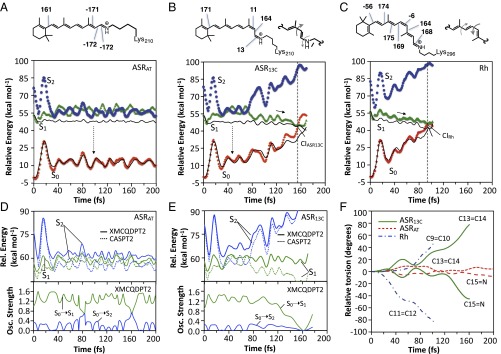
As documented in the SI Appendix [see SI Appendix, section 1 for the testing of alternative protocols ranked for their ability to yield ASR models of comparable quality to the Rh model (31)], the S0 models of ASRAT and ASR13C reproduce the vertical excitation energies (ΔES1-S0) associated with the observed absorption maximum value within a few kilocalories per mole (+1.5 and +1.0 kcal⋅mol−1 for ASRAT and ASR13C, respectively; see SI Appendix, section 2.1 for all computed and observed excitation energy values). The origin of the values is investigated by isolating the chromophores from their protein environment and recomputing their ΔES1-S0 without geometrical relaxation (this quantity is, usually, red-shifted compared with that of the protein due to the lack of effects stabilizing S0). We also looked at the vertical S1-S2 energy gap (ΔES2-S1) that will be discussed in the next sections. It is found that, for all models, the electrostatic effect of the protein is responsible for 7–8 kcal⋅mol−1 increase and 3–4 kcal⋅mol−1 decrease in ΔES1-S0 and ΔES2-S1, respectively. Since the protein has a systematic effect, the changes are due to the chromophore geometries that display excitation energy differences replicating those computed for the corresponding proteins. For instance, the isolated Rh chromophore has ΔES1-S0 and ΔES2-S1 values ∼5 and ∼3 kcal⋅mol−1 larger than the corresponding ASRAT chromophore values. These data parallel the ∼4 and ∼3 kcal⋅mol−1 differences obtained for the chromophore in Rh and ASR13C. These ΔES1-S0 and ΔES2-S1 changes are assigned to the out-of-plane twisting of the β-ionone ring (the C5–C6–C7–C8 dihedral angle is −56°) in Rh (32, 33), which decreases the conjugation with respect to the more planar ASR chromophores (see top structures in Fig. 2 A–C). Note that both the ΔES1-S0 and ΔES2-S1 values decrease when increasing the chromophore conjugation (SI Appendix, section 2.2).
Fig. 2.
QM/MM trajectories of ASRAT, ASR13C, and Rh computed at the two root state average scaled-CASSCF/Amber (black lines) level of theory and corrected at the CASPT2 (23) and XMCQDPT2 levels (35). (A) S0 (filled diamonds), S1 (filled triangles), and S2 (filled circles) CASPT2//CASSCF/Amber energy profiles along the ASRAT trajectory. The main out-of-plane (deviation larger than ±5°) dihedral angles of the chromophore S0 equilibrium structure are given at the top. The vertical dashed arrow represents weak fluorescence. (B) Same data for ASR13C with representation (at Top) of the bicycle pedal motion driving the isomerization on S1 in which the bond order is reversed. The vertical dashed arrow represents weak fluorescence. (C) Same data for the Rh trajectory. (D) View of the XMCQDPT2 (solid line) and CASPT2 (dotted line) S1 and S2 energy profiles (Upper) and oscillator strengths (Lower) along the ASRAT trajectory. (E) Same data for the ASR13C trajectory. (F) Geometrical progression of the reactive bond (C13=C14 for ASR and C11=C12 for Rh) and an adjacent coupled double bond (C15=N for ASR and C9=C10 for Rh) relative to the S0 equilibrium values.
Trajectory Calculations.
The photoisomerization dynamics of ASRAT and ASR13C are investigated by running single S1 trajectories starting from the corresponding S0 equilibrium structures with zero initial velocities (Franck–Condon trajectories) (34). The computations are carried on until the trajectories enter the region of a S1/S0 CI or reach 200 fs. We assume that during such an ultrashort time the trajectory describes the average evolution of the corresponding S1 population. This has been assessed for both Rh (23, 24, 31) and ASR models (SI Appendix, section 3). As shown in Fig. 2 A and B, ASRAT remains on S1 for the entire duration of the trajectory whereas ASR13C reaches the CI after ca. 150 fs. The estimated S1 lifetimes are consistent with the available transient spectroscopy measurements and computations (25, 29) showing that the ASRAT photoisomerizes at least five times more slowly than ASR13C. However, ASR13C seems to react in a timescale only slightly slower than that computed for Rh (31) (Fig. 2C), consistently with the observations (25, 26).
The slower photoisomerization of ASRAT is also consistent with its observed larger fluorescence quantum yield (26). Furthermore, the ASRAT model predicts a fluorescence maximum value consistent with the observations. In fact, starting 25 fs after excitation, we compute an average oscillator strength in the 0.8–1.6 range. Taking the average ΔES1-S0 values over the same 175-fs interval, we predict a fluorescence of 669 nm. A similar analysis can be carried out for ASR13C, yielding oscillator strength and fluorescence values of 1.1–1.5 and 665 nm, respectively. Both computed values are comparable with the observed values (i.e., a of 700–710 nm with a shoulder around 650 nm) (26). Also the ASRAT trajectory does not decay within the simulation time, consistent with the observed longer fluorescence lifetime with respect to ASR13C (26).
It is apparent from inspection of the energy profiles in Fig. 2 A–C and from the enlarged views in Fig. 2 D and E that both S1 and S2 are involved in the isomerizations of ASRAT and, partially, of ASR13C. Indeed, the corresponding energies become nearly degenerate ca. 25 fs after photoexcitation. It is observed that whereas ASRAT travels along the degeneracy for the full duration of the trajectory, ASR13C abandons the degeneracy region ca. 50 fs after entering it and then evolves toward the CI. In contrast, no degeneracy is found for Rh, which evolves toward the CI directly after photoexcitation. These data support a relationship between the magnitude of the estimated excited state lifetime (defined as the time it takes to reach the CI, τ) and the extent of the S2/S1 mixing. Inclusion of more excited states in the calculations does not change this conclusion (SI Appendix, section 8).
The geometrical progression of PSBAT, PSB13, and PSB11 chromophores in the protein are reported in Fig. 2F. The C12–C13–C14–C15 dihedral of ASR13C, which describes the isomerization of the C13=C14 bond, changes from 11° to 86°. This motion is coupled with a 13° to −32° change in the C14–C15–N–Cε dihedral describing the torsional deformation of the adjacent C15=N double bond. These results point to a ASR13C space-saving S1 bicycle-pedal isomerization mechanism (36, 37) shared by ASRAT in its initial attempt to isomerize (see first 50 fs in Fig. 2F) but distinct from the one reported for Rh (23) and other PSB11-hosting rhodopsins (31) involving the C11=C12 (7) and C9=C10 double bond pair (23, 24, 31). Notice that, in all cases, the bicycle-pedal motion is aborted upon S1 decay and only one double bond is accomplishing the isomerization (SI Appendix, Fig. S7C) (23, 28). The ASR and Rh isomerization mechanisms are also stereochemically distinct. In fact, whereas in ASR13C and ASRAT the reactive bond isomerizes in the counterclockwise direction (with respect to the Lys210 side chain), in Rh the isomerization occurs in the clockwise direction (with respect to the Lys296 side chain).
Origin of the S2/S1 Near Degeneracy.
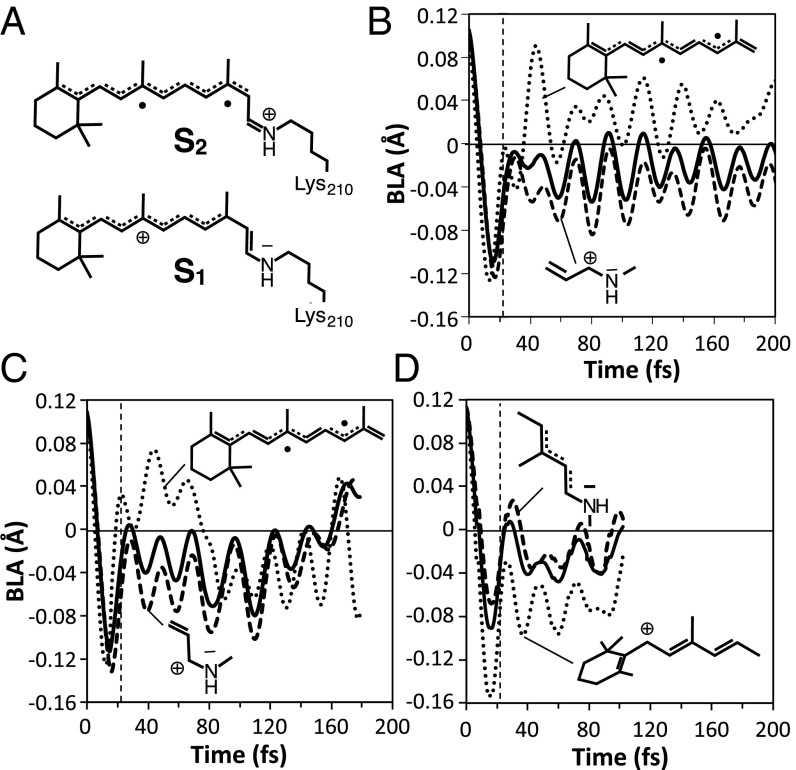
During the first 25 fs after photoexcitation, the rhodopsin chromophore undergoes an inversion in the bond length alternation (BLA) along the backbone (BLA is the difference between the average single bond and the average double bond lengths). For instance, in ASRAT, the C13=C14 bond expands from 1.37 to 1.51 Å whereas the C14–C15 bond contracts from 1.42 to 1.32 Å. Such a change is accompanied by changes in ΔES1-S0 and ΔES2-S1, which appear as oscillations in the corresponding S0, S1, and S2 energy profiles. We find that, upon BLA relaxation, both ASRAT and ASR13C enter the S2/S1 near degeneracy region (see Fig. 2 D and E, Upper). Because in the vertical excitation region S1 has a charge transfer character and S2 has a diradical character (Fig. 3A), the S1 electronic structure is perturbed along the trajectory and, for ASRAT, it mixes and even switches from a charge transfer to a diradical charge distribution. This is confirmed by the S0→S1 oscillator strength progression, which shows fast recursive variations (Fig. 2D, Lower) of a lower magnitude in ASR13C. The change in electronic character would modify the force field experienced by the S1 chromophore, resulting in a slower progression toward the CI (because the double bond character of C13=C14 is not completely lost in a diradical state and restrains the isomerization).
Fig. 3.
BLA changes in ASR and Rh. (A) Schematic representation of the electronic structure of the S1 and S2 states. (B) BLA changes for the ASRAT chromophore along the full conjugated chain (solid line), along the C5=C6–C7=C8-C9=C10–C11=C12–C13=C14 fragment (dotted line), and along the –C13=C14–C15=N fragment (dashed line). (C) Same data for the ASR13C chromophore. (D) The BLA along the full conjugated chain for the Rh chromophore (solid line), along the C5=C6–C7=C8–C9=C10–C11=C12 fragment (dotted line), and along the –C11=C12–C13=C14–C15=N fragment (dashed line).
Notice that, due to potential problems in using the complete active space second-order perturbation theory (CASPT2) method in near degeneracy regions, the S2/S1 mixings have been reexamined with a level of theory that does not have such issues (35, 38, 39). We have carried out, for each snapshot of the ASR trajectories, extended multiconfigurational quasi-degenerate second-order perturbation theory (XMCQDPT2) calculations (SI Appendix, section 4) on a model comprising the chromophore surrounded by the point charges of the entire apoprotein. As apparent from inspection of Fig. 2 A and B and Fig. 2 D and E, the XMCQDPT2 energy displays mostly recurrent avoided crossings rather than CASPT2 crossings.
To explain why a S2/S1 near degeneracy region is reached in ASR but not in Rh, we considered the effect of BLA on the different S2 and S1 electronic structures (see details in SI Appendix, section 5). Thus, we have investigated the BLA oscillations of specific fragments of the ASRAT, ASR13C chromophores. In Fig. 3 we compare the BLA values of the C5=C6–C7=C8–C9=C10–C11=C12–C13=C14 fragment of ASR (Fig. 3 B and C) with the corresponding C5=C6–C7=C8–C9=C10–C11=C12 fragment of Rh (Fig. 3D). These fragments show marked differences already starting 20 fs after photoexcitation. In particular, the ASR fragments have an average BLA value close to 0.0, indicating similar double and single bond lengths, whereas in Rh the corresponding fragment has negative BLA values consistent with a BLA inversion with respect to its positive S0 equilibrium value. Notice that ASR13C represents a “transition” between the ASRAT and Rh regimes and its BLA becomes similar to the one of Rh after exiting the S2/S1 near degeneracy.
The above observations support the hypothesis that the chromophore type, with its protein-imposed deformation, would control the S1 relaxations. In fact, the ASRAT relaxation leads to a fragment with four conjugated double bonds (C5=C6–C7=C8–C9=C10–C11=C12–) and an average BLA value consistent with effective delocalization (Fig. 3B). Such a pattern accommodates the diradical electronic structure better than the fragment generated during Rh relaxation (SI Appendix, section 2.2) which, due to the pretwisted β-ionone ring on one side and rapidly twisting C11=C12 (Fig. 2F) on the other, offers a shorter fragment with two conjugated double bonds (C7=C8–C9=C10–C11) with a negative (inverted) average BLA value (32). As a consequence, ΔES2-S1 will rapidly decrease along the photoisomerization coordinate in ASRAT and ASR13C but not in Rh.
Rhodopsin Isomerization in Different Organisms.
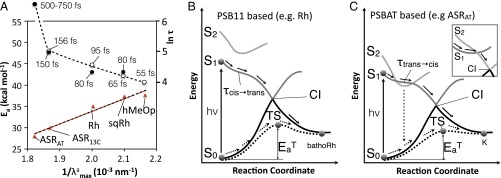
As discussed in a previous report (31), photoisomerization quantum yields and thermal isomerization rates are factors determining the level of light sensitivity of rhodopsins. To enhance sensitivity, quantum yields must be maximized whereas thermal rates, which create the “background noise” in the signal, must be minimized. According to a correlation proposed by Weiss and Warshel (6) and Mathies and coworkers (7, 8), for fast impulsive (rather than diffusive) dynamics, the larger the velocity of the S1 population moving toward the CI the higher the reaction quantum yield. Although the exact nature of such relationship is still under investigation (9, 27), it is apparent that a decrease in the time τ required to reach the CI enforces a coherent S1 dynamics/decay, which, in turn, allows controlling (40) the quantum yield. However, the trend in thermal isomerization rates may be estimated by computing the S0 isomerization barrier . Thus, the τ and values of ASR together with those previously reported for Rh, squid rhodopsin (sqRh), and human melanopsin (hMeOp) (31) can be used to model trends of light sensitivity. Indeed, the ASR13C, Rh, sqRh, and hMeOp τ values in Fig. 4A suggest a somehow exponential increase of τ when increases. However, the long S1 lifetime observed for ASRAT does not fit in. Thus, whereas an increase of τ may be associated with a decrease in slope of the S1 potential energy surface accelerating progression toward the CI (Fig. 4B), in ASRAT and other PSBAT-hosting rhodopsins the occurrence of the S2/S1 degeneracy changes the dynamics (Fig. 4C), which becomes slower and complex with an increased fluorescence lifetime. The S1 relaxation would thus be more diffusive with the chromophore exploring the rugged potential energy surface shaped by several S2/S1 avoided crossings (Fig. 3D, Lower) and, as a consequence, the velocity–quantum yield relationship loses its validity (which would indicate a different way of controlling the quantum yield). In other words, whereas a two-state isomerization model (Fig. 4B) applies to fast (<150 fs)-reacting systems incorporating the PSB11 or even PSB13 chromophores, a three-state model (Fig. 4C) would apply to slower rhodopsins hosting PSBAT.
Fig. 4.
PSBAT and PSB11 isomerization mechanism in distant rhodopsins. (A) CASPT2//CASSCF/Amber values of (triangles, left axis) and ln τ (circles, right axis) plotted as a function of the inverse of the value. Full circles indicate observed quantities [fluorescence lifetimes for ASR (26) and kinetic constants for Rh (24) and octopus rhodopsin (41) where octopus shares the same subclass (Coleoidea) with squid]. Open circles refer to computed quantities. (B) Schematic representation of the photochemical (solid arrows) and thermal (dashed arrows) isomerization paths for PSB11 in Rh. The CI, which is located energetically above the TS, features a different geometrical structure. τcis→trans and are quantities computed in the present work. (C) The same representation for PSBAT in ASRAT. The vertical dashed arrow indicates weak fluorescence. Inset shows the previously reported three-state model for the PSBAT-hosting Archaeal bacteriorhodopsins (bR) (41).
A three-state model has been previously reported (42) for the PSBAT-hosting Archaeal bacteriorhodopsins (bR), which displays a slow and complex excited state dynamics (42). However, in such model, the S2 diradical state ultimately drives the progression toward the CI (Fig. 4C, Inset). Although this seems to be the case in polyenes (43), it is inconsistent with the structure of protonated Schiff base CIs (44). Therefore, we propose that our ASR three-state model also applies to bR. This is not only consistent with the reported bR (37) and, recently, Channelrhodopsin (45) reaction paths, but also with the similar dynamics observed for ASRAT and light-adapted bR (hosting exclusively PSBAT) (25, 46) as well as for ASR13C and the corresponding PSB13-containing bR. The same three-state model may control the excited state dynamics of unsubstituted and certain substituted PSBAT in solution displaying S1 picosecond lifetimes (47). The presence of an S2/S1 degeneracy in solvated PSBAT models has been proposed (48) and held responsible for the observed solution dynamics of PSBAT (49). Similar data have been reported for PSB11 (50, 51), suggesting that the Rh cavity has important effects on the electronic structure of its chromophore.
The values in Fig. 4A (see SI Appendix for the computations) show a trend consistent with the Barlow correlation (52), an inverse proportionality between thermal activation rates and values proposed (52, 53) and computationally probed (54) for visual pigments. When taken together, the and τ trends lead to the hypothesis that PSB11 has been selected for achieving lower noise (higher values) and higher photoisomerization speed (lower τ values) with an impact on quantum efficiencies (6–10, 40).
During rhodopsin evolution, a PSBAT bearing ancestor may have acquired the PSB11 chromophore, possibly through a secondary photoisomerization. Our computations suggest that such putative ancestor of type I and type II rhodopsins may have then started to diverge rapidly to optimize features such as the fitting of a bent rather than straight molecular framework, the increasing of the degree of twisting of the β-ionone ring to avoid S2/S1 degeneracy and the pretwisting the C11=C12 bond to ensure a selective isomerization and, again, avoid the S2/S1 degeneracy by shortening the moiety available for radical/charge delocalization. The large twisting of the β-ionone ring and the selective C11=C12 isomerization would thus have consequences beyond contributing to the color modulation (33). The evolutionary process outlined above requires microbial rhodopsins capable of hosting both the all-trans and the 11-cis chromophores. A rhodopsin of this type has been recently found in the archaeal Haloquadratum walsbyi (55).
In conclusion, MCQC models have been used to compare the isomerization of ASR and Rh. It is found that the bicycle-pedal photoisomerization mechanism is a general feature of the S1 dynamics, although its regiochemistry and stereochemistry are different in microbial (ASR) and vertebrate (Rh) rhodopsins. The twisting of the β-ionone ring seems critical for differentiating the properties of the PSB11- and PSB13/PSBAT-hosting rhodopsins. It blue-shifts the by increasing the ΔES1-S0 value and increases the reaction speed (decreasing τ) by removing the S2/S1 mixing. The same effect also increases (via the Barlow correlation confirmed for the examined set). All these may create evolutionary advantages for improving light sensitivity.
Methods
The S0 equilibrium QM/MM models of ASRAT ad ASR13C were constructed starting from the crystallographic structure of ASR (56) available in the Protein Data Bank (PDB ID code 1XIO) by optimizing preprocessed structures (e.g., incorporating all hydrogens and counter ions) with the Molcas/Tinker code (57, 58). In all models the retinal chromophore was treated quantum mechanically using the complete-active-space self-consistent field (CASSCF) method (59) and embedded in a protein environment described by the AMBER force field. To account for the dynamic electron correlation, the optimized CASSCF geometries and wavefunctions were used for subsequent CASPT2 calculations (60). The XMCQDPT2 energy and oscillator strength calculations were performed using Firefly version 8.0.0 (61) based on the geometries of chromophore with full aproprotein residues treated as point charges. Further details of the QM/MM model building, related stability testing, and trajectory computations are documented in SI Appendix, section 1, and transition state computations are detailed in SI Appendix, section 6.
Supplementary Material
Acknowledgments
M.O. thanks the Center for Photochemical Sciences and School of Arts and Sciences of the Bowling Green State University. The authors thank NSF-XSEDE and Ohio Supercomputer Center for granted computer time. This work was supported in part by National Science Foundation Grants CHE-1152070 and CHE-1551416 and Human Frontier Science Program Organization Grant RGP0049/2012.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510262112/-/DCSupplemental.
References
- 1.Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev. 2014;114(1):126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devine EL, Oprian DD, Theobald DL. Relocating the active-site lysine in rhodopsin and implications for evolution of retinylidene proteins. Proc Natl Acad Sci USA. 2013;110(33):13351–13355. doi: 10.1073/pnas.1306826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackin KA, Roy RA, Theobald DL. An empirical test of convergent evolution in rhodopsins. Mol Biol Evol. 2014;31(1):85–95. doi: 10.1093/molbev/mst171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: Functional versatility and genetic mobility. Trends Microbiol. 2006;14(11):463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364(1531):2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss RM, Warshel A. A new view of the dynamics of singlet cis-trans photoisomerization. J Am Chem Soc. 1979;101(20):6131–6133. [Google Scholar]
- 7.Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. The first step in vision: Femtosecond isomerization of rhodopsin. Science. 1991;254(5030):412–415. doi: 10.1126/science.1925597. [DOI] [PubMed] [Google Scholar]
- 8.Kim JE, Tauber MJ, Mathies RA. Analysis of the mode-specific excited-state energy distribution and wavelength-dependent photoreaction quantum yield in rhodopsin. Biophys J. 2003;84(4):2492–2501. doi: 10.1016/S0006-3495(03)75054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schapiro I, et al. The ultrafast photoisomerizations of rhodopsin and bathorhodopsin are modulated by bond length alternation and HOOP driven electronic effects. J Am Chem Soc. 2011;133(10):3354–3364. doi: 10.1021/ja1056196. [DOI] [PubMed] [Google Scholar]
- 10.Weingart O, et al. Product formation in rhodopsin by fast hydrogen motions. Phys Chem Chem Phys. 2011;13(9):3645–3648. doi: 10.1039/c0cp02496a. [DOI] [PubMed] [Google Scholar]
- 11.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci USA. 1989;86(21):8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto K, Hasegawa J-Y, Hayashi S, Kato S, Nakatsuji H. Mechanism of color tuning in retinal protein: SAC-CI and QM/MM study. Chem Phys Lett. 2005;414:239–242. [Google Scholar]
- 13.Coto PB, Strambi A, Ferré N, Olivucci M. The color of rhodopsins at the ab initio multiconfigurational perturbation theory resolution. Proc Natl Acad Sci USA. 2006;103(46):17154–17159. doi: 10.1073/pnas.0604048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M, et al. Color tuning in rhodopsins: The mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J Am Chem Soc. 2006;128(33):10808–10818. doi: 10.1021/ja062082i. [DOI] [PubMed] [Google Scholar]
- 15.Ryazantsev MN, Altun A, Morokuma K. Color tuning in rhodopsins: The origin of the spectral shift between the chloride-bound and anion-free forms of halorhodopsin. J Am Chem Soc. 2012;134(12):5520–5523. doi: 10.1021/ja3009117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekharan S, Morokuma K. Why 11-cis-retinal? Why not 7-cis-, 9-cis-, or 13-cis-retinal in the eye? J Am Chem Soc. 2011;133(47):19052–19055. doi: 10.1021/ja208789h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL. Photochromicity of Anabaena sensory rhodopsin, an atypical microbial receptor with a cis-retinal light-adapted form. J Biol Chem. 2005;280(15):14663–14668. doi: 10.1074/jbc.M501416200. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann KP, et al. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem Sci. 2009;34(11):540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Kukura P, McCamant DW, Yoon S, Wandschneider DB, Mathies RA. Structural observation of the primary isomerization in vision with femtosecond-stimulated Raman. Science. 2005;310(5750):1006–1009. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 20.Kandori H, Shichida Y, Yoshizawa T. Photoisomerization in rhodopsin. Biochemistry (Mosc) 2001;66(11):1197–1209. doi: 10.1023/a:1013123016803. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. Vibrationally coherent photochemistry in the femtosecond primary event of vision. Science. 1994;266(5184):422–424. doi: 10.1126/science.7939680. [DOI] [PubMed] [Google Scholar]
- 22.Andruniów T, Ferré N, Olivucci M. Structure, initial excited-state relaxation, and energy storage of rhodopsin resolved at the multiconfigurational perturbation theory level. Proc Natl Acad Sci USA. 2004;101(52):17908–17913. doi: 10.1073/pnas.0407997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frutos LM, Andruniów T, Santoro F, Ferré N, Olivucci M. Tracking the excited-state time evolution of the visual pigment with multiconfigurational quantum chemistry. Proc Natl Acad Sci USA. 2007;104(19):7764–7769. doi: 10.1073/pnas.0701732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polli D, et al. Conical intersection dynamics of the primary photoisomerization event in vision. Nature. 2010;467(7314):440–443. doi: 10.1038/nature09346. [DOI] [PubMed] [Google Scholar]
- 25.Wand A, et al. Asymmetric toggling of a natural photoswitch: Ultrafast spectroscopy of Anabaena sensory rhodopsin. J Am Chem Soc. 2011;133(51):20922–20932. doi: 10.1021/ja208371g. [DOI] [PubMed] [Google Scholar]
- 26.Cheminal A, et al. Steady state emission of the fluorescent intermediate of Anabaena Sensory Rhodopsin as a function of light adaptation conditions. Chem Phys Lett. 2013;587:75–80. [Google Scholar]
- 27.Cheminal A, et al. 100 fs photo-isomerization with vibrational coherences but low quantum yield in Anabaena Sensory Rhodopsin. Phys Chem Chem Phys. 2015;17(38):25429–25439. doi: 10.1039/c5cp04353k. [DOI] [PubMed] [Google Scholar]
- 28.Strambi A, Durbeej B, Ferré N, Olivucci M. Anabaena sensory rhodopsin is a light-driven unidirectional rotor. Proc Natl Acad Sci USA. 2010;107(50):21322–21326. doi: 10.1073/pnas.1015085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schapiro I, Ruhman S. Ultrafast photochemistry of anabaena sensory rhodopsin: Experiment and theory. Biochim Biophys Acta. 2014;1837(5):589–597. doi: 10.1016/j.bbabio.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Warshel A, Levitt M. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103(2):227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 31.Rinaldi S, Melaccio F, Gozem S, Fanelli F, Olivucci M. Comparison of the isomerization mechanisms of human melanopsin and invertebrate and vertebrate rhodopsins. Proc Natl Acad Sci USA. 2014;111(5):1714–1719. doi: 10.1073/pnas.1309508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cembran A, Bernardi F, Olivucci M, Garavelli M. Counterion controlled photoisomerization of retinal chromophore models: A computational investigation. J Am Chem Soc. 2004;126(49):16018–16037. doi: 10.1021/ja048782+. [DOI] [PubMed] [Google Scholar]
- 33.Cembran A, et al. Structure, spectroscopy, and spectral tuning of the gas-phase retinal chromophore: The beta-ionone “handle” and alkyl group effect. J Phys Chem A. 2005;109(29):6597–6605. doi: 10.1021/jp052068c. [DOI] [PubMed] [Google Scholar]
- 34.Gozem S, et al. On the shape of multireference, EOM-CC, and DFT potential energy surfaces at a conical intersection. J Chem Theory Comput. 2014;10(8):3074–3084. doi: 10.1021/ct500154k. [DOI] [PubMed] [Google Scholar]
- 35.Granovsky AA. Extended multi-configuration quasi-degenerate perturbation theory: The new approach to multi-state multi-reference perturbation theory. J Chem Phys. 2011;134(21):214113. doi: 10.1063/1.3596699. [DOI] [PubMed] [Google Scholar]
- 36.Warshel A. Bicycle-pedal model for the first step in the vision process. Nature. 1976;260(5553):679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- 37.Altoè P, Cembran A, Olivucci M, Garavelli M. Aborted double bicycle-pedal isomerization with hydrogen bond breaking is the primary event of bacteriorhodopsin proton pumping. Proc Natl Acad Sci USA. 2010;107(47):20172–20177. doi: 10.1073/pnas.1007000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gozem S, et al. Dynamic electron correlation effects on the ground state potential energy surface of a retinal chromophore model. J Chem Theory Comput. 2012;8:4069–4080. doi: 10.1021/ct3003139. [DOI] [PubMed] [Google Scholar]
- 39.Malrieu J-P, Heully J-L, Zaitsevskii A. Multiconfigurational second-order perturbative methods: Overview and comparison of basic properties. Theor Chim Acta. 1995;90:167–187. [Google Scholar]
- 40.Brumer P, Shapiro M. Quantum interference in the control of molecular processes. Phil. Trans. R. Soc. A. 1997;355:2409–2412. [Google Scholar]
- 41.Yabushita A, Kobayashi T, Tsuda M. Time-resolved spectroscopy of ultrafast photoisomerization of octopus rhodopsin under photoexcitation. J Phys Chem B. 2012;116(6):1920–1926. doi: 10.1021/jp209356s. [DOI] [PubMed] [Google Scholar]
- 42.Gai F, Hasson KC, McDonald JC, Anfinrud PA. Chemical dynamics in proteins: The photoisomerization of retinal in bacteriorhodopsin. Science. 1998;279(5358):1886–1891. doi: 10.1126/science.279.5358.1886. [DOI] [PubMed] [Google Scholar]
- 43.Garavelli M, Celani P, Bernardi F, Robb MA, Olivucci M. Force fields for “ultrafast” photochemistry: The S2 (1Bu) → S1 (2Ag) → S0 (1Ag) reaction path for all-trans-Hexa-1,3,5-triene. J Am Chem Soc. 1997;119:11487–11494. [Google Scholar]
- 44.Cembran A, González-Luque R, Serrano-Andrés L, Merchán M, Garavelli M. About the intrinsic photochemical properties of the 11-cis retinal chromophore: Computational clues for a trap state and a lever effect in Rhodopsin catalysis. Theor Chem Acc. 2007;118(1):173–183. [Google Scholar]
- 45.Dokukina I, Weingart O. Spectral properties and isomerisation path of retinal in C1C2 channelrhodopsin. Phys Chem Chem Phys. 2015;17(38):25142–25150. doi: 10.1039/c5cp02650d. [DOI] [PubMed] [Google Scholar]
- 46.Briand J, Léonard J, Haacke S. Ultrafast photo-induced reaction dynamics in bacteriorhodopsin and its Trp mutants. J Opt. 2010;12(8):084004. [Google Scholar]
- 47.Bassolino G, et al. Synthetic control of retinal photochemistry and photophysics in solution. J Am Chem Soc. 2014;136(6):2650–2658. doi: 10.1021/ja4121814. [DOI] [PubMed] [Google Scholar]
- 48.Cembran A, Bernardi F, Olivucci M, Garavelli M. The retinal chromophore/chloride ion pair: Structure of the photoisomerization path and interplay of charge transfer and covalent states. Proc Natl Acad Sci USA. 2005;102(18):6255–6260. doi: 10.1073/pnas.0408723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zgrablić G, Voïtchovsky K, Kindermann M, Haacke S, Chergui M. Ultrafast excited state dynamics of the protonated Schiff base of all-trans retinal in solvents. Biophys J. 2005;88(4):2779–2788. doi: 10.1529/biophysj.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munoz-Losa A, Martín ME, Fernández Galván I, Sánchez ML, Aguilar M. Solvent effects on the radiative and non-radiative decay of a model of the rhodopsin chromophore. J Chem Theory Comput. 2011;7:4050–4059. doi: 10.1021/ct200295r. [DOI] [PubMed] [Google Scholar]
- 51.Muñoz-Losa A, Ignacio I, Aguilar MA, Martín ME. Simultaneous solvent and counterion effects on the absorption properties of a model of the rhodopsin chromophore. J Chem Theory Comput. 2013;9(3):1548–1556. doi: 10.1021/ct301090v. [DOI] [PubMed] [Google Scholar]
- 52.Barlow HB. Purkinje shift and retinal noise. Nature. 1957;179(4553):255–256. doi: 10.1038/179255b0. [DOI] [PubMed] [Google Scholar]
- 53.Ala-Laurila P, Donner K, Koskelainen A. Thermal activation and photoactivation of visual pigments. Biophys J. 2004;86(6):3653–3662. doi: 10.1529/biophysj.103.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gozem S, Schapiro I, Ferré N, Olivucci M. The molecular mechanism of thermal noise in rod photoreceptors. Science. 2012;337(6099):1225–1228. doi: 10.1126/science.1220461. [DOI] [PubMed] [Google Scholar]
- 55.Sudo Y, et al. A microbial rhodopsin with a unique retinal composition shows both sensory rhodopsin II and bacteriorhodopsin-like properties. J Biol Chem. 2011;286(8):5967–5976. doi: 10.1074/jbc.M110.190058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogeley L, et al. Anabaena sensory rhodopsin: A photochromic color sensor at 2.0 A. Science. 2004;306(5700):1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aquilante F, et al. MOLCAS 7: the next generation. J Comp Chem. 2010;31(1):224–247. doi: 10.1002/jcc.21318. [DOI] [PubMed] [Google Scholar]
- 58.Ponder JW, Richards FM. An efficient newton-like method for molecular mechanics energy minimization of large molecules. J Comp Chem. 1987;8(7):1016–1024. [Google Scholar]
- 59.Roos BO. 1987. The complete active space self-consistent method and its applications in electronic structure calculations. Ab Initio Methods in Quantum Chemistry, Advances in Chemical Physics, ed Lawley KP (Wiley, New York), Pt 2, Vol 69, pp 399–445.
- 60.Andersson K, Malmqvist PA, Roos BO, Sadlej AJ, Wolinski K. Second-order perturbation theory with a CASSCF reference function. J Phys Chem. 1990;94:5483–5488. [Google Scholar]
- 61.Granovsky AA. 2014 Firefly. Available at classic.chem.msu.su/gran/firefly/index.html. Version 8.0.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.