Significance
The gene interleukin-12 receptor β1 (IL12RB1) regulates susceptibility to several human diseases, including mycobacterial disease (e.g., tuberculosis). Here, we demonstrate that many of the mRNAs transcribed from IL12RB1 in primary immune cells contain RNA-DNA differences (RDDs). RDDs are nucleotide differences between RNA and its encoding DNA and are introduced posttranscriptionally; in the case of IL12RB1, RDDs are concentrated in cytokine-binding regions that are important for IL12RB1 function. This observation is significant, as it is the first demonstration to our knowledge that a mechanism of sequence diversification exists for a human cytokine receptor. Given IL12RB1’s importance to mycobacterial disease resistance, our data raise the intriguing possibility that individual differences in IL12RB1 RDD introduction contribute to differences in mycobacterial disease susceptibility.
Keywords: IL12RB1, IL-12, IL-23, RNA, RDD
Abstract
Human interleukin 12 and interleukin 23 (IL12/23) influence susceptibility or resistance to multiple diseases. However, the reasons underlying individual differences in IL12/23 sensitivity remain poorly understood. Here we report that in human peripheral blood mononuclear cells (PBMCs) and inflamed lungs, the majority of interleukin-12 receptor β1 (IL12RB1) mRNAs contain a number of RNA-DNA differences (RDDs) that concentrate in sequences essential to IL12Rβ1’s binding of IL12p40, the protein subunit common to both IL-12 and IL-23. IL12RB1 RDDs comprise multiple RDD types and are detectable by next-generation sequencing and classic Sanger sequencing. As a consequence of these RDDs, the resulting IL12Rβ1 proteins have an altered amino acid sequence that could not be predicted on the basis of genomic DNA sequencing alone. Importantly, the introduction of RDDs into IL12RB1 mRNAs negatively regulates IL12Rβ1’s binding of IL12p40 and is sensitive to activation. Collectively, these results suggest that the introduction of RDDs into an individual’s IL12RB1 mRNA repertoire is a novel determinant of IL12/23 sensitivity.
IL-12 and IL-23 (IL12/23) are proinflammatory cytokines that contribute to multiple aspects of human immunity, including the differentiation of human TH1, TH17, TFH, and TC subsets (1). Reflecting the multifaceted influence of IL12/23, individuals who are insensitive to IL12/23 are susceptible to a spectrum of intracellular pathogens, including Candida, Salmonella, and Mycobacteria species (2). Paradoxically, IL12/23 sensitivity also contributes to human autoimmunity, as demonstrated by the efficacy of an anti-IL12/23 monoclonal treatment for plaque psoriasis, psoriatic arthritis, and refractory Crohn’s disease (1). Maximal IL12 responsiveness can also lead to death (3). Given the detrimental outcomes associated with minimal and maximal IL12/23 responsiveness, it is important to understand the factors that balance IL12/23 sensitivity and underlie interindividual differences in IL12/23 responsiveness.
Interleukin-12 receptor β1 (IL12RB1) encodes IL12Rβ1, a type 1 transmembrane protein that positively regulates human IL12/23 sensitivity by binding the IL12p40-domain common to both cytokines (4). Consistent with this role, IL12RB1 polymorphisms produce IL12Rβ1 proteins of varying sensitivity to IL12/23 (5) and associate with susceptibility to numerous diseases regulated by IL12/23, including tuberculosis, nontuberculous mycobacterial infection, malaria, cancer, pediatric asthma, and atopic dermatitis (4). Here we report that in addition to containing genome-encoded polymorphisms, IL12RB1 mRNA transcripts expressed in peripheral blood mononuclear cells (PBMCs) and inflamed lungs also contain multiple RNA-DNA differences (RDDs) that concentrate in sequences encoding the IL12Rβ1 cytokine-binding region. RDDs are nucleotide differences between RNA and its encoding DNA and are detectable by both deep sequencing and classical Sanger sequencing (6). Importantly, the extent to which the IL12Rβ1 cytokine-binding region is altered by RDDs is suppressed by activation and is inversely related to the amount of IFNγ secreted. Functional evidence suggests this is a result of amino acid substitutions that interfere with IL12p40 binding. Collectively, our results demonstrate that the introduction of RDDs into human IL12RB1 mRNA (i.e., IL12RB1 RDD introduction) negatively regulates IL12p40 binding and points to RDD introduction as a novel mechanism underlying individual differences in IL12/23 sensitivity.
Results
IL12RB1 mRNAs Contain Multiple RDDs, the Appearance of Which Decrease with Activation.
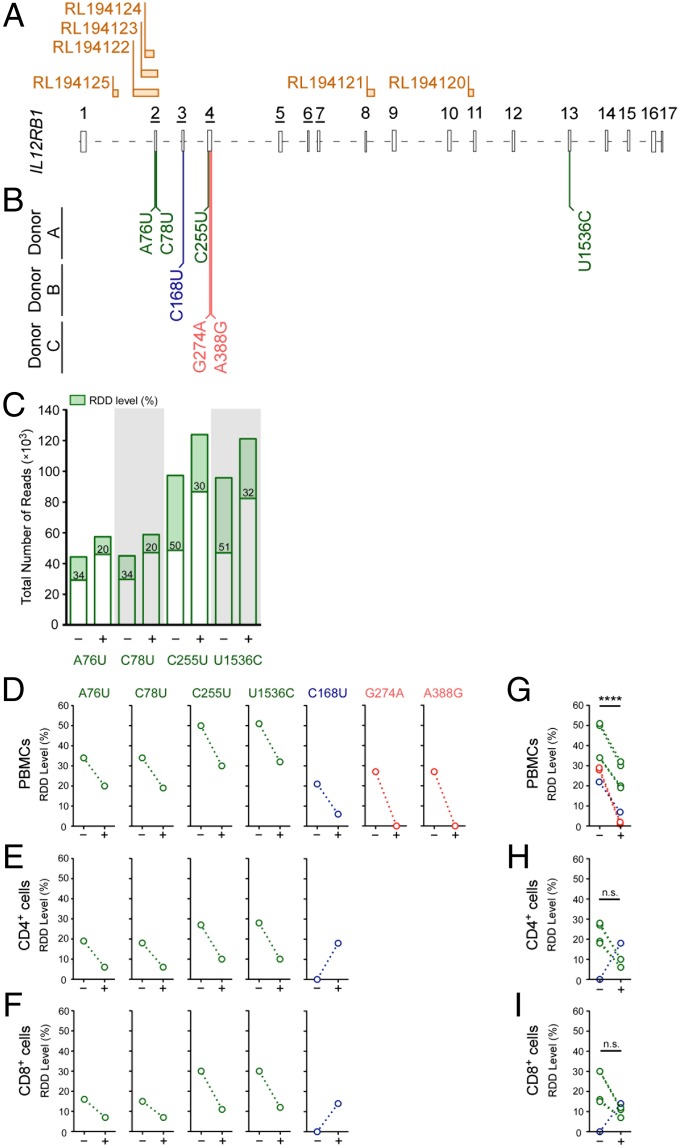
RDDs are present throughout the human transcriptome (6), and their introduction is coupled to R-loop formation (7). IL12RB1, a gene that regulates the development of both pathogen- and autoimmune-driven inflammatory responses in humans, contains several predicted R-loop-forming sequences (8) (Fig. 1A). To determine whether RDDs are introduced into human IL12RB1 transcripts, IL12RB1 mRNAs from PBMCs of immunocompetent donors were sequenced to high depth, using Ion Torrent semiconductor sequencing. IL12RB1 mRNAs were prepared from PBMCs both before and after activation with phytohemagglutinin (PHA); RDDs were identified by comparing the mRNA sequences with the genomic DNA (gDNA) IL12RB1 sequence of the same PBMC preparation (Datasets S1 and S2). The results of this analysis demonstrated that across three healthy PBMC donors, we observed seven distinct IL12RB1 RDDs that varied in abundance, depending on the PBMCs’ activation state (Fig. 1 and Dataset S3). Specifically, RDDs were found in exon 2 (A76U, C78U), exon 3 (C168U), exon 4 (C255U, G274A, A388G), and exon 13 (U1536C) (Fig. 1B). For donor A, the levels of each RDD declined with activation, as evidenced by the coverage data for A76U, C78U, C255U, and U1536C in PHA− and PHA+ conditions (Fig. 1C). The pattern of activation-driven IL12RB1 RDD-decline in donor A was also true of each IL12RB1 RDD expressed in PBMCs of donors B and C (Fig. 1D). To determine whether IL12RB1 RDD introduction can be observed in more than one PBMC subpopulation, CD4+ and CD8+ cells were purified from donor A and donor B PBMC preparations before and after PBMC activation; the IL12RB1 mRNAs expressed by each subset were then sequenced similar to PBMC IL12RB1 mRNA (Dataset S3). We observed that both donors’ CD4+ and CD8+ cells expressed the same RDDs as those present in total PBMCs, and that their presence in both lineages was also affected by activation (Fig. 1 E and F). Collectively, these data demonstrate that in human PBMCs, the mRNAs encoding IL12Rβ1 contain multiple RDDs, the levels of which decline with activation.
Fig. 1.
IL12RB1 mRNAs contain multiple RDDs, the appearance of which decrease with activation. (A) IL12RB1 mRNA comprises 17 exons, the relative sizes and orientation of which are depicted 5′-3′ alongside the location of six putative R-loop-forming regions: RL194, RL194122, RL194123, RL194124, RL194121, and RL194120. The exon numbers that are underlined (exons 2–7) encode the IL12Rβ1 CBR. (B) The position and nature of the IL12RB1 RDDs in PBMCs of three healthy, immunocompetent donors (donors A, B, and C), as identified by next-generation sequencing. In the top row are the IL12RB1 RDDs present in donor A (A76U, C78U, C255U, and U1536C); the center and bottom rows, respectively, list the IL12RB1 RDDs present in donor B (C168U) and donor C (G274A, A388G). (C) Sequencing read coverage data and relative appearance of each donor A RDD in the absence (−) or presence (+) of an activation signal (PHA); each donor A RDD is listed along the x-axis. Each bar represents the total number of reads at that specific site in the IL12RB1 transcript; the RDD level is indicated in the green portion of each bar. (D) The levels of each individual IL12RB1 RDD found in donors A–C PBMCs in the presence or absence of activation. Each open circle represents one of the RDDs listed in B, using the same color scheme for each donor. An identical analysis is shown for (E) CD4+ and (F) CD8+ cells from the same donor PBMC preparations. (G–I) IL12RB1 RDD-level data were combined from donors A–C to show (G) cumulative RDD levels in donor A–C PBMCs, (H) cumulative RDD levels in donors A and B CD4+ cells, and (I) cumulative RDD levels in donor A and B CD8+ cells, both in the presence and absence of PHA. For A and B, exon boundaries and ribonucleotide number assignments, we used the IL12RB1 ribonucleotide numbering system described in Dataset S6. R-loop sites were identified by Wongsurawat et al. (8); these sites and their designations are taken from their publically available R-loop database found at rloop.bii.a-star.edu.sg/. For statistical comparisons of PHA− and PHA+ data in G and H, donor data were pooled and significance (P) determined using a paired t-test. ****P < 0.0001.
The Extent of IL12RB1 RDD Introduction Varies Significantly Between Individuals.
The high-throughput nature of deep sequencing has raised concerns that a majority of RDDs identified by this approach are either artifacts of complex alignment algorithms or errors introduced by recombinant polymerases (9–11). To determine whether IL12RB1 RDDs were artifacts of alignment or polymerase error, PBMC mRNA was used to generate and Sanger sequence IL12RB1 cDNA libraries from eight separate donors (donors A, B, D, and G–K). Each clone within a library contained the entire length of the IL12RB1 isoform 1 cDNA (∼2,000 bp). Individual clones were randomly selected and Sanger sequenced with two to three times coverage (average, 15 clones per donor). RDDs were initially identified by software alignment of donors’ cDNA sequences to their gDNA sequence; to control for any error in alignment, all RDDs identified were confirmed by visual examination of Sanger sequencing traces. To empirically determine the number of errors introduced by recombinant polymerases, a synthetic IL12RB1 mRNA of known sequence was generated, reverse transcribed, amplified in both the presence and absence of nonspecific carrier DNA (herring sperm DNA), and Sanger sequenced using protocols identical to those used for PBMC mRNA.
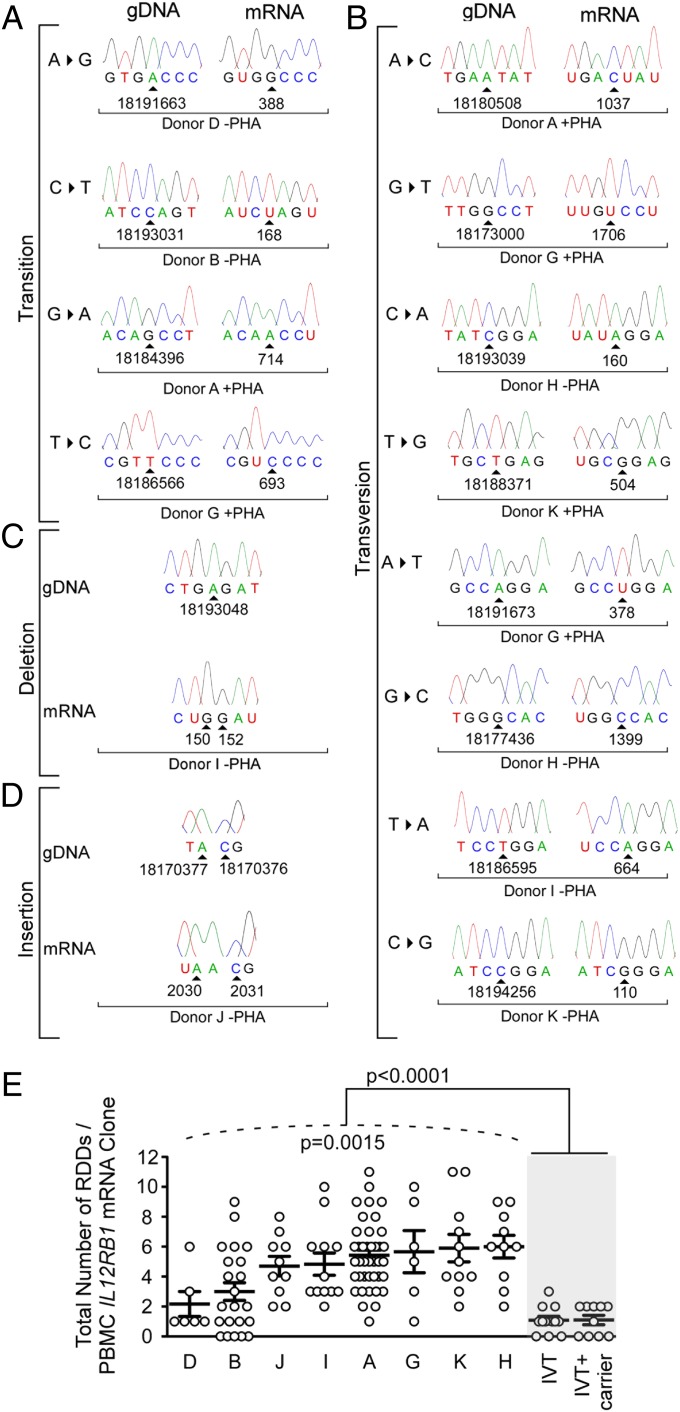
The results of this analysis demonstrate that across eight donors, all possible RDD types were detectable in IL12RB1 mRNAs (Fig. 2 and Dataset S4). These RDD types included transitions (Fig. 2A), transversions (Fig. 2B), insertions (Fig. 2C), and deletions (Fig. 2D). As depicted in Fig. 2E, there was significant interindividual variation in the extent to which IL12RB1 RDDs were present (P = 0.0015, as tested using a mixed-effects Poisson regression model), with some donors exhibiting low numbers of IL12RB1 RDDs (e.g., donor D) and some having high numbers of IL12RB1 RDDs (e.g., donor K). Importantly, when the data from all eight donors were pooled, the number of RDDs present in PBMC IL12RB1 cDNAs is significantly higher than the number of differences introduced as a result of recombinant polymerase error (P < 0.0001; Fig. 2E). Activation did not affect how frequent each RDD type occurred donor-wide (Fig. S1). Collectively, these data demonstrate that IL12RB1 RDDs are detectable by Sanger sequencing, vary in transcript-wide frequency depending on the donor, and are not artifacts of complex alignment algorithms or errors introduced by recombinant polymerases.
Fig. 2.
The extent of IL12RB1 RDD introduction varies significantly between individuals. After their purification from the PBMCs of eight different donors (A, B, D, and G–K), IL12RB1 mRNAs were amplified and cloned into the sequencing vector pUC19. Individual clones from each PBMC IL12RB1 library were randomly chosen for Sanger sequencing (two to three times coverage; average of 15 mRNA clones per donor). (A–D) The IL12RB1 RDDs detected by Sanger sequencing comprised multiple RDD types, including (A) transitions, (B) transversions, (C) deletions, and (D) insertions. Shown for each RDD type in (A and B) are sequencing chromatograms of the IL12RB1 gDNA sequence (Left) next to the corresponding IL12RB1 mRNA clone sequence from the same donor (Right). (C and D) The sequence of a donor gDNA (Top) and the corresponding mRNA clone (Bottom) sequence. (A–D) The genomic location of the deoxyribonucleotide encoding the affected ribonucleotide is indicated at the bottom of each sequencing trace, as is the ribonucleotide number and associated mRNA clone information. (E) The extent to which RDDs occurred in IL12RB1 mRNA clones expressed by her or his PBMCs (PHA− and PHA+ data are combined; Dataset S4). Each circle represents a single IL12RB1 mRNA clone and its IL12RB1 RDD frequency (i.e., the number of RDDs present divided by the mRNA clone length). The extent to which donors’ individual RDD frequency data significantly differed from one another is indicated below the hatched line (P = 0.0015, as measured using a mixed-effects Poisson regression model); the extent to which the RDD frequency data from all donors differed from recombinant polymerase error rate (in vitro transcription), which was determined both in the presence and absence of nonspecific carrier DNA, is indicated at the top of the graph (P < 0.0001, using Student’s t test).
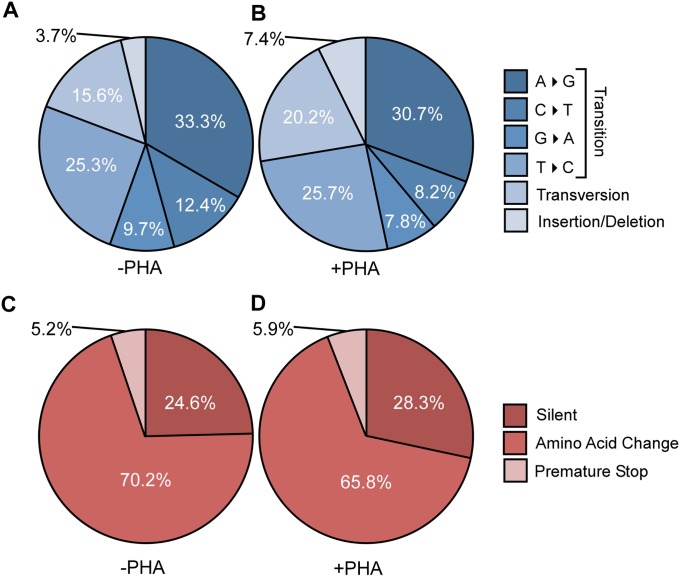
Fig. S1.
The representation of each IL12RB1 RDD type is unaffected by activation. (A and B) Pie charts depicting the relative representation of transitions, transversions, insertions, and deletions in IL12RB1 mRNAs from (A) PHA− and (B) PHA+ PBMC preparations. (C and D) The relative representation of RDDs that are silent, result in an amino acid change, or introduce a premature stop codon in IL12RB1 mRNAs from (C) PHA− and (D) PHA+ PBMC preparations. Percentages are taken from the pooled Sanger sequencing data of all donors.
RDD-Containing IL12RB1 Transcripts Are Expressed in Inflamed Human Lungs.
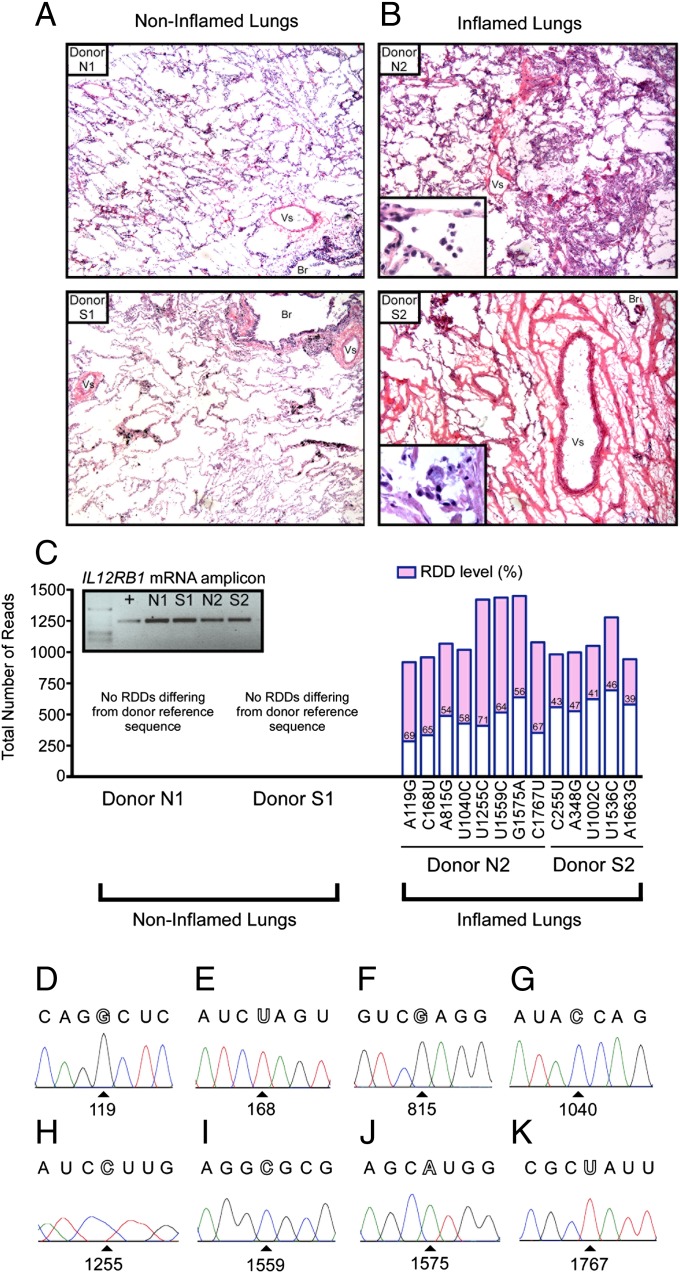
Human lungs are protected by IL12RB1-dependent inflammatory responses (12). To determine whether inflammatory responses in the lung were associated with IL12RB1 RDD introduction, we measured the extent of RDD introduction present in the lungs of individuals with differing degrees of pulmonary inflammation at time of death. Specifically, postmortem lung tissue from four different individuals (donors N1, N2, S1, and S2; Dataset S5) were screened for the presence or absence of inflammation by H&E staining (Fig. 3 A and B); IL12RB1 mRNA and gDNA from adjacent tissue was then used to assay for the presence of IL12RB1 RDDs introduction by Ion Torrent sequencing (Fig. 3C) and Sanger sequencing (Fig. 3 D–K). The results of this analysis demonstrate that although IL12RB1 mRNA was expressed in the lungs of each donor (Fig. 3C, Inset, agarose gel image of IL12RB1 cDNA amplicons), only the mononuclear- and polymorphonuclear-leukocyte-infiltrated lungs of donors N2 and S2 contained IL12RB1 RDDs differing from reference sequence (Fig. 3 A–C); the lungs of donors N1 and S1 were unremarkable in their histological appearance, had no obvious inflammatory infiltration, and did not contain any detectable IL12RB1 RDDs. Notably, one of the IL12RB1 RDDs observed in donor N2’s lung was identical to one found in PBMC donor B (C168U; Fig. 1B), and two of the IL12RB1 RDDs observed in donor S2’s lung were identical to those found in PBMC donor A (C255U, U1536C; Fig. 1B). Sanger sequencing of individual IL12RB1 mRNA clones was used to confirm the presence of each of the eight RDDs observed in donor N2 IL12RB1 mRNAs (Fig. 3 D–K). Collectively, these data demonstrate that IL12RB1 RDDs differing from reference sequence are found in the inflamed human lung.
Fig. 3.
IL12RB1 mRNAs expressed in inflamed human lungs contain RDDs. Lung tissue from four different individuals (donors N1, N2, S1, and S2) was scored for the presence or absence of pulmonary inflammation by H&E analysis; tissue from areas adjacent to the sectioned areas were then used for IL12RB1 RDD identification by Ion Torrent and Sanger sequencing. (A and B) Representative 10× magnification images of (A) noninflamed lung tissue from donors N1 and S1 and (B) inflamed lung tissue from donors N2 and S2 (Br, bronchiole; Vs, blood vessel). Inset within the images B are 40× magnifications of the same sections, which demonstrate that inflammatory infiltrates make up both polymorphonuclear and mononuclear lineages. (C) Listed for each lung along the horizontal axis are the RDDs identified in IL12RB1 transcripts expressed in each lung. Each bar represents the total number of reads at that specific site in the IL12RB1 transcript; the RDD level at each specific site is indicated in the pink portion of each bar. IL12RB1 transcripts in donors N1 and S1 did not contain any RDDs differing from the reference sequence. Inset within C is a gel image of the IL12RB1 mRNA amplicons from each tissue sample alongside a positive control, as visualized by agarose gel electrophoresis. (D–K) Sanger sequencing traces of IL12RB1 mRNA clones from donor N2 containing the RDDs (D) A119G, (E) C168U, (F) A815G, (G) U1040C, (H) U1255C, (I) U1559C, (J) G1575A, and (K) C1767U.
IL12RB1 RDDs Are Concentrated in the IL12Rβ1 Cytokine-Binding Region and Function to Attenuate IL12p40 Binding.
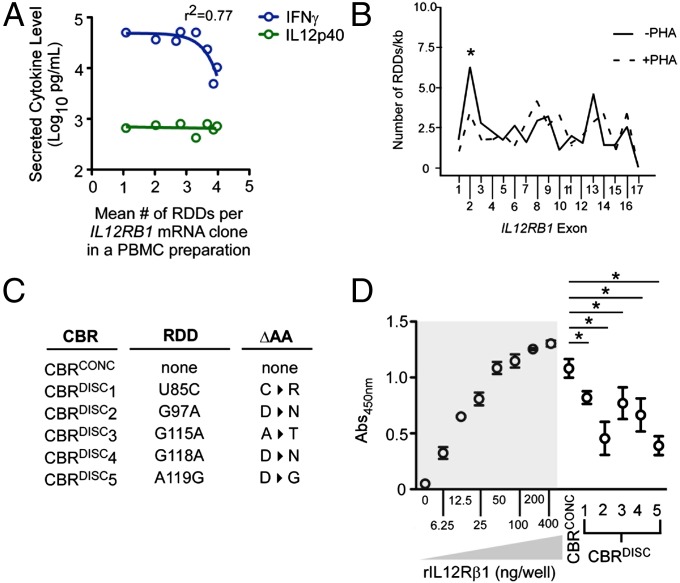
IL12Rβ1 and IL12p40 are principal drivers of activated human PBMC IFNγ secretion (13, 14). Over the course of sequencing PBMC IL12RB1 transcripts, we noted an inverse relationship between the mean RDD frequency across IL12RB1 transcripts in a PBMC preparation and the amount of IFNγ secreted by the same PBMC preparation (Fig. 4A). Because no such correlation was observed with the amount of IL12p40 secreted (Fig. 4A), these data raised the possibility that RDD introduction negatively regulates IL12p40 bioactivity. To gain insight into the mechanism by which this may occur, we determined whether IL12RB1 RDDs were spread evenly over the IL12RB1 mRNA sequence or whether RDDs concentrated in sequences known to influence IL12Rβ1 function (15). The results of this analysis demonstrate that RDDs are not evenly distributed over the length of IL12RB1 transcripts (Fig. 4B and Fig. S2). Rather, RDDs are significantly enriched in exon 2, which contributes to the IL12Rβ1 cytokine-binding region (CBR) (16). After PBMC activation, there were fewer RDDs located in exon 2-encoded sequences (Fig. 4B and Fig. S2). Notably, the errors we observed in in vitro transcription mRNA clones begin ∼1,100 bases into the IL12RB1 mRNA (corresponding to sequences encoded by exon 10); therefore, the RDD enrichment we observe in exon 2-encoded sequences is not likely to be a result of recombinant polymerase error.
Fig. 4.
IL12RB1 RDDs concentrate in the IL12Rβ1 CBR and negatively affect IL12p40-binding. (A) The relationship between the mean IL12RB1 RDD frequency in a given PBMC preparation (i.e., the average number of RDDs per IL12RB1 mRNA clone in PBMCs of a given donor) and the amounts of IL12p40 (green) and IFNγ (blue) secreted by the same PBMC preparation. A nonlinear regression analysis is shown for IFNγ. (B) The RDD rate for each exon was calculated by dividing the number of RDDs that occurred in that exon by the exon length. The mean RDD rate for each exon (RDD/kb); data are combined from nine donors. The significance of an exon enrichment was determined by comparing each individual exon RDD rate with the mean rate of all exons combined (fixed effects analysis); a significant departure from the mean rate is indicated by an asterisk (P ≤ 0.05). (C) The RDDs and associated amino acid changes present in each IL12RB1 mRNA clone selected. Five clones contained RDDs in exon 2 that changed the protein sequence by one amino acid (U85C, C→R; G97A, D→N; G115A, A→T; G118A, D→N; A119G, D→G), and thus produced CBRs that were discordant with the gDNA sequence (CBRDISC). One clone contained no RDDs, and was thus used to produce a CBR concordant with that of the IL12RB1 genome sequence (CBRCONC). (D) Recombinant CBRs were used to assess IL12p40 binding via ELISA; full-length recombinant IL12Rβ1 (rIL12Rβ1) was used as a positive control with which to compare the binding ability of CBRCONC and CBRDISC. Shown are the binding capacities (Abs450nm) of each preparation when bound to wells coated with a saturating amount of IL12p40. rIL12Rβ1 was added to IL12p40-coated wells at varying amounts (0, 6.25, 12.5, 25, 50, 100, 200, and 400 ng/well) to determine what amount confers maximal IL12p40 binding; given the results of this determination (highlighted gray), all recombinant IL12RB1 CBRs (CBRCONC and CBRDISC1–5) were added to IL12p40-coated wells at 200 ng/well. Significant differences (P ≤ 0.05) between CBR preparations’ abilities to bind IL12p40 were determined by ANOVA analysis.
Fig. S2.
RDD exon enrichment in IL12RB1 mRNA clones before and after activation with PHA. Shown for each PBMC donor is the extent to which RDDs are exon-enriched in their IL12RB1 mRNA clones, both before and after activation with PHA. The length of each IL12RB1 mRNA was divided into 17 portions based on exon boundaries (horizontal axis), and the “RDD rate” for each portion is plotted for each individual (vertical axis). Each PBMC donor is represented by a different line color; the left half of the figure shows the RDD rates in each mRNA portion before PBMC activation (PHA−), whereas the right half of the figure shows the RDD rates in each mRNA portion after PBMC activation (PHA+). Listed adjacent to the figure legend is the IFNγ level in the supernatant of each PBMC preparation after its activation with PHA. The average RDD rate for each IL12RB1 mRNA portion across all donors was used to generate the graph shown in Fig. 4B.
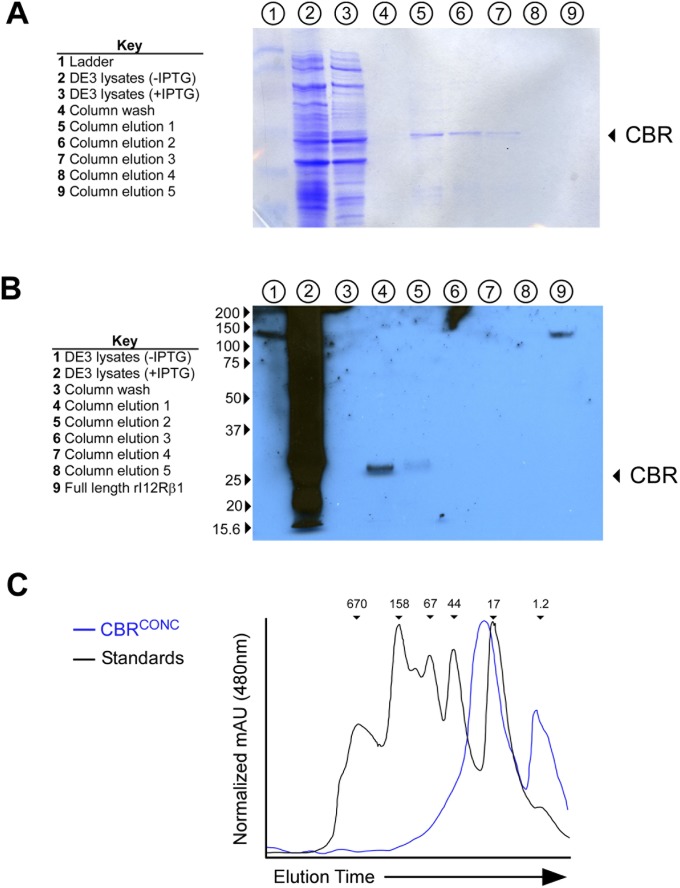
To directly measure the effect of IL12RB1 RDD introduction on IL12Rβ1’s ability to bind IL12p40, recombinant CBRs from RDD-containing mRNA clones were produced and tested in an IL12p40-binding assay. Specifically, CBRs from five RDD-containing transcripts were cloned into bacterial expression vector pET28 (Fig. 4C), and a C-terminal 6×His-tag was introduced onto each of these five discordant CBRs (CBRDISC1–5) for purification purposes. A control CBR whose amino acid sequence was unaffected by RDD introduction, which was thus concordant with the originating donor’s gDNA (CBRCONC), was generated in an identical manner. Each CBR produced a protein near that of its predicted size of 30.2 kDa (Fig. S3 A and C), and CBRs were also recognized by polyclonal anti-IL12Rβ1 (Fig. S3B). Each recombinant CBR was then used in an ELISA to assess binding to plate-bound IL12p40. The results of these binding experiments are shown in Fig. 4D. Normalizing to recombinant full-length IL12Rβ1 (rIL12Rβ1), we observed that CBRCONC binds to IL12p40 at ∼80% of rIL12Rβ1’s binding capacity, indicating that the amino acids in this region of IL12Rβ1 are responsible for the majority of IL12p40 binding (Fig. 4D). The capacities of CBRDISC1–5 to bind IL12p40 were significantly below those of CBRCONC, indicating the amino acid differences present in CBRDISC1–5 disrupt IL12p40 binding (Fig. 4D). Collectively, when combined with RDD enrichment data (Fig. 4B), these data demonstrate that RDDs are preferentially introduced into PBMC IL12Rβ1 CBR-encoding sequences in a manner that is environmentally adaptive and that attenuates IL12p40-binding.
Fig. S3.
Production of recombinant IL12Rβ1 CBRs. (A) Purification fractions from recombinant CBR from DE3 cells were collected and used for SDS/PAGE/Coomassie stain analysis: total lysate from uninduced (IPTG−) DE3 cells, total lysate from induced (IPTG+) DE3 cells, nickel column wash (after passing over bound CBR), and serial elutions with imidazole-containing buffer (five total). The lanes containing each of these samples are indicated above the gel image (key is provided to the left of the gel image); the location of the CBR is indicated by an arrow. (B) The same fractions were used for Western blot analysis of reactivity with polyclonal anti-IL12Rβ1. Shown is a representative blot containing each fraction, as well as rIL12Rβ1 as a positive control. (C) The analytical FPLC spectra of CBRCONC alongside the spectra of six recombinant standards (1.2, 17, 44, 67, 158, and 670 Kda). The relative locations of each standard are indicated on the black spectra; the spectra of CBRCONC is blue. The results in A–C) are representative of the purification data from all CBRs used for binding studies.
Discussion
Here we have demonstrated that IL12RB1 mRNAs expressed by PBMCs and inflamed lungs of immunocompetent individuals contain a number of RDDs that affect sensitivity to IL12p40. These RDDs are not derived from individuals’ genomic IL12RB1 sequence and are concentrated in CBR-encoding sequences. Because the extent to which RDDs are introduced into IL12RB1 mRNAs is variable between individuals, we refer to the IL12RB1 mRNA transcripts expressed in an individual as her or his “IL12RB1 mRNA repertoire.” The introduction of RDDs into an individual’s IL12RB1 mRNA repertoire occurs in proximity to putative R-loop-forming sequences, is sensitive to activation, can result in amino acid changes, and attenuates IL12p40 binding to the IL12Rβ1 CBR. These data are consistent with previous studies of RDDs (6, 7) and support a model in which RDD introduction is a mechanism underlying variable IL12/23 sensitivity in humans.
The extent to which RDD introduction occurs in the human mRNA transcriptome is a contentious subject, and is likely to remain contentious, as RDD introduction challenges the basic assumption that DNA is transcribed into mRNA with near-perfect fidelity (i.e., limited only by the error rate of human RNA polymerase II, which is estimated to range between 2 × 10−6 and 3 × 10−4) (17). The first RDDs were discovered ∼30 y ago (18–20) and are known now to be the result of adenosine deaminase acting on RNA (ADAR, which catalyzes A-to-I editing, the latter nucleotide being recognized as a G) and apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC, which catalyzes C-to-U editing) (21, 22). Given the substrate specificity of ADAR and APOBEC’s catalytic domain, it was widely believed that few human mRNAs contain RDDs. This belief was challenged by the results of Li et al. (6), who, over the course of deep-sequencing mRNA and gDNA of immortalized human B cells from 27 individuals, observed 10,210 sites of mismatch between a given individual’s mRNA and gDNA sequence. These mismatches made up all 12 possible transitions and transversions, as well as insertions and deletions. A follow-up study demonstrated that these RDDs were enriched in R-loop-forming regions and were diminished in an individual with senataxin-deficiency (SETX, which encodes a DNA/RNA helicase that aids in R-loop resolution) (7). Given the significance of claiming that the gDNA and mRNA sequences from the same cells are not always identical, it should perhaps come as no surprise that several groups raised concerns with the methods of Li et al. (9–11). These concerns are summarized as follows, with each one being a source of “false-positive” RDDs: (i) the use of random hexamers to produce cDNA can result in mispriming; (ii) low gDNA sequence coverage, which would result in not knowing all of the gDNA SNPs present in an individual; (iii) erroneous alignment of mRNA sequences to paralogous regions of the gDNA; and (iv) that the majority of RDDs were enriched at the termini of RNA sequencing reads (i.e., the first or last base of a read), suggesting they are technical artifacts.
For our own studies, we were keen to avoid the four concerns described here, and thus addressed them in the following ways: (i) all cDNAs were generated using oligodT primers, which ensured we were examining mature mRNAs; (ii) IL12RB1 gDNA coverage was >500× per individual and corroborated by Sanger sequencing to ensure we identified all SNPs; (iii) mRNA sequences were solely aligned to IL12RB1, ruling out the effect of any homologous pseudogenes; and (iv) any RDDs occurring at read termini were excluded from our analysis. RDDs were also confirmed by Sanger sequencing. Thus, in addition to uncovering a novel aspect of IL12RB1 biology, we believe our data inform the discourse concerning the genome-wide RDD frequency by demonstrating that for one gene, there is significant interindividual variation in the extent of RDD introduction in primary human tissues, and not just immortalized B cells. That distinct T-cell lineages (i.e., CD4+ and CD8+ T cells, which diverge during thymic selection) express the same RDDs within the same individual also suggests that RDD introduction begins early in lineage development. Finally, that common IL12RB1 RDDs can be identified across different individuals increases the likelihood that future RDD studies will identify specific RDDs that associate with a given human trait.
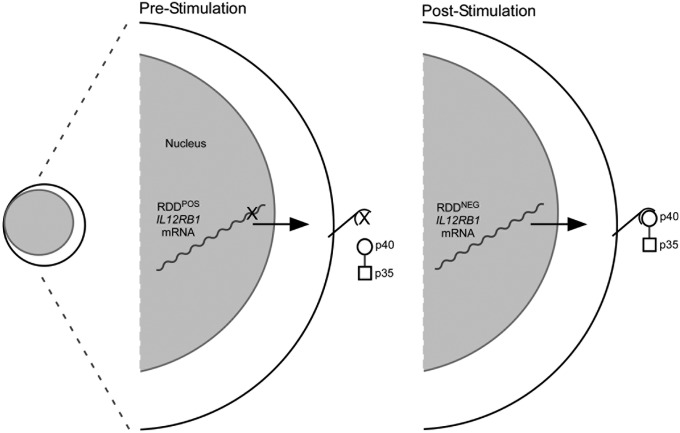
Given that IL12Rβ1 is essential for human resistance to intramacrophagic pathogens (2), and IL12RB1 sequence variation associates with susceptibility to infectious disease, cancer, pediatric asthma, and dermatological disease (4), our data raise the possibility that RDD introduction into an individual’s IL12RB1 mRNA repertoire contributes to her/his susceptibility to disease. By decreasing the number of RDD-containing IL12RB1 transcripts after activation, either through decreased RDD introduction or increased degradation of RDD-containing transcripts, lymphocytes may poise themselves to better receive the developmental and survival signals provided by IL12/IL23 (Fig. 5). In addition to altering the IL12Rβ1 amino acid sequence, it is also possible that RDDs introduce mRNA sequences or form secondary structures that promote IL12RB1 mRNA degradation. On the basis of this model, we predict that lower IL12RB1 RDD levels and/or frequencies are beneficial in the context of infectious disease (where IL12/IL23 bioactivity promotes disease resistance), and higher IL12RB1 RDD levels and/or frequencies are beneficial in the context of autoimmune diseases (where IL12/IL23 bioactivity promotes disease progression). Our future efforts will focus on determining whether certain IL12RB1 mRNA repertoires associate with conditions regulated by IL12/IL23 bioactivity (e.g., tuberculosis) and whether transgenic overexpression of different IL12RB1 mRNA repertoires confers varying susceptibility to experimental disease. The importance of mRNA sequence diversity is well-recognized for two other genes that regulate disease resistance: TCRA and TCRB. Although the underlying mechanism of sequence diversification is different for TCRA/TCRB and IL12RB1, the consequence of this diversification is the same: the expression of proteins of variable amino acid sequence and variable immunological function.
Fig. 5.
Proposed model for how IL12RB1 RDDs regulate IL12/IL23 responsiveness. Before PBMC stimulation, the IL12RB1 gene is transcribed into RDD-containing (RDDPOS) mRNAs that are translated into IL12Rβ1 proteins with diminished IL12p40-binding capacity; as a consequence, unstimulated PBMCs are less responsive to IL12/IL23 (only IL12 is depicted). After PBMC stimulation, the introduction of RDDs into IL12RB1 mRNAs declines, producing IL12RB1 mRNAs that lack RDDs (RDDNEG) and allowing for translation of IL12Rβ1 proteins that confer IL12/IL23-responsiveness.
Materials and Methods
Human Tissues.
All studies using human tissues were approved by the Medical College of Wisconsin Institutional Review Board. Informed consent was given by each donor before her or his participation in our study. Units of blood from healthy adult donors were collected into sodium citrate-treated bags at the Blood Center of Wisconsin; donors were excluded if they were taking (or had taken within 2 wk before collection) any of the following categories of immunosuppressant medications: antineoplastic agent, antiviral agent, corticosteroid (either dermatological or nondermatological), a disease-modifying antirheumatic drug, or immunosuppressive mAb drugs. Human lung tissue specimens were supplied by the National Resource Center of the National Disease Research Interchange; donor information is presented in Dataset S5. Between 1 and 5 h postmortem, lung tissues were recovered and snap frozen for the National Disease Research Interchange repository. On transfer to our laboratory, these tissue specimens were divided into smaller pieces for scoring the degree of inflammation in each specimen (H&E analysis), and purification of RNA and genomic DNA from adjacent sections.
Identification of IL12RB1 RDDs.
IL12RB1 mRNA (isoform 1) was purified and amplified from PBMC and lung lysates, using our previously established protocols (23). gDNA from the same cell preparations was isolated using the DNeasy method (Qiagen); the primers and conditions used to amplify each IL12RB1 exon (17 total) are provided in Dataset S1. Purified IL12RB1 mRNA and gDNA amplicons were used to generate libraries for next-generation sequencing, using the Ion Torrent Personal Genome Machine (Life Technologies). The preparation of each amplicon library for Ion Torrent sequencing was carried out per our previously reported protocols (24). IL12RB1 RDDs were identified by comparing the IL12RB1 mRNA sequence data of a given tissue preparation with the IL12RB1 gDNA sequence of the same preparation, using STAR software for splicing alignment (25) and Samtools for variant calling (26). IL12RB1 ribonucleotides were categorized as either truly representing the donor gDNA or being the result of RDD introduction, based on the presence or absence, respectively, of the corresponding deoxyribonucleotide in the donor gDNA. Depending on the sample sequenced, the total number of reads at the each site varied between 1,628 and 124,669 for PBMCs (Dataset S3) or 916 and 17,347 for lungs (Dataset S3). In the samples determined to have RDDs, at least 20% of the mapped reads correspond to the RDD or the nonreference allele. Genome sequence alignment of RDD-containing reads was used to ensure that sequences mapped solely to IL12RB1, and not homologous genes (e.g., GP130) or pseudogenes. Any RDDs occurring at read termini were excluded from our analysis. Whether a donor’s genomic sequence at each site was monomorphic or polymorphic was confirmed by Sanger sequencing of all 17 IL12RB1 exon amplicons; depicted in Dataset S2 is the degree of monomorphism/polymorphism in each PBMC donor for all known IL12RB1 exonic polymorphisms (27). For Sanger sequencing of IL12RB1 mRNA clones, amplicons were cloned into the sequencing vector pUC19 and Sanger sequenced (two to three times coverage) on an ABI 3730xl DNA Sequencer, using our previously reported protocols (23); RDDs were identified using Sequencher DNA Sequence Analysis software (Gene Codes Corporation) and confirmed by visual inspection of individual chromatograms. To determine the number of RDDs introduced solely as a result of recombinant polymerase error, a synthetic IL12RB1 mRNA of known sequence was generated by in vitro transcription with the Riboprobe system (Promega); mRNA was then reverse transcribed, amplified both in the presence and absence of nonspecific carrier DNA (herring sperm DNA), and Sanger sequenced in a manner identical to that done for PBMC IL12RB1 mRNAs. To indicate the site of each IL12RB1 RDD identified by Ion Torrent or Sanger sequencing, we used the IL12RB1 ribonucleotide numbering system of van de Vosse et al. (15) (Dataset S6). The term “RDD level” is used to describe the number of RDD-bearing transcripts divided by the total number of transcripts at one specific site, whereas the term “RDD frequency” is used to describe the number of RDDs over a defined region.
Additional experimental details can be found in SI Materials and Methods.
SI Materials and Methods
PBMC Isolation and Activation.
After de-identification, blood units were delivered to our laboratory via courier the same day for PBMC isolation. For this, blood was diluted 1:2 in sterile PBS (Ca2+/Mg2+ free) and immediately spun over a room-temperature Ficoll-Histopaque 1107 gradient (GE Healthcare). Buffy coats were washed twice in PBS, counted, and then used either immediately for total RNA/DNA purification (PHA−) or in vitro activation (PHA+). For in vitro activation, PBMCs were cultured in complete RPMI 1640 containing CD2-activator PHA-P (1 μg/mL final concentration; Sigma-Aldrich). After 3 d, activated PBMCs were washed and lysed for RNA/DNA purification. For indicated experiments, CD4+ and CD8+ subsets were magnetically purified from PHA− and PHA+ PBMCs. Briefly, PBMCs were suspended in a slurry of magnetic bead–Ab conjugates recognizing either human CD4 (clone L200) or CD8 (clone SK1), according to the manufacturer’s protocols (BD Biosciences). After positive selection and two rounds of washing in PBS, subsets were counted and immediately lysed for total RNA extraction.
Production of Recombinant IL12Rβ1 CBRs.
Recombinant IL12Rβ1 CBRs were produced by amplification and cloning of the corresponding mRNA sequence (nucleotides 70–729 of the IL12RB1 isoform 1 mRNA) into the bacterial expression vector pET28 (Novagen). The primer sequences used to amplify and clone the CBR are listed in Dataset S1. CBR-containing pET28 clones were transformed into DE3 Escherichia coli and expression induced with isopropyl β-D-1-thiogalactopyranoside (IPTG). The soluble fraction of DE3 lysates was placed over a nickel column (Qiagen) to purify 6×His-tagged CBRs (a tag introduced by pET27); CBRs were eluted with imidazole and dialyzed into PBS/Tween. The concentrations of each CBR were normalized to one another; FPLC and SDS/PAGE analysis were used to verify purity and confirm that CBRs were near their predicted size (30.2 kDa; Fig. S3).
IL12p40 Binding ELISA.
High-protein-binding 96-well plates (Corning) were coated overnight at 4 °C with a saturating amount of recombinant human IL12p40 (R&D Systems; 200 ng/well). After washing with PBS/Tween, recombinant CBRs were added at 200 ng/well and allowed to bind overnight at 4 °C. rIL12Rβ1 (R&D Systems) was used as a positive control for IL12p40 binding. After again washing with PBS/Tween, CBR binding was detected using HRP-conjugated anti-6×His antibody and associated substrate/detection solutions; the Abs450nm of each well was measured on a Viktor Microplate Reader (Perkin-Elmer).
Statistical Analysis.
Statistical analyses were designed and performed in consultation with the Medical College of Wisconsin Biostatistics Consulting Service. For measuring the significance of interindividual variation in RDD rates (Fig. 2E), we used a mixed-effects Poisson regression with the number of IL12RB1 RDDs as the response variable, and the log-transformed length of each clone as an offset to compare the RDD rate between groups. This approach treats the observed number of RDDs as a count variable, but allows modeling of the number of RDDs per kilobase through the use of the offset term. Donor pool and IL12RB1 RDD number (nested within donor) were the random effects in the model, whereas the presence of PHA was a fixed effect. For RDD enrichment analysis (Fig. 4B), the “RDD rate” for each exon was calculated by dividing the number of RDDs within a given exon by the exon length [using the IL12RB1 exon boundaries defined by National Center for Biotechnology Information (NCBI) and reproduced in Dataset S6]. The RDD rate for each IL12RB1 exon was determined in this manner, for each individual in the PHA− and PHA+ cohorts. Because our main comparison of interest was between exons (rather than between individuals), we performed a fixed-effects analysis of RDD rate per exon, with rate data pooled from each individual in a group. The distribution of these pooled exon RDD rates was then compared with the mean rate of all exons. Because the IL12Rβ1 CBR is encoded by exons 2–8, a significant (P < 0.05) departure in exons’ 2–9 RDD rates from the mean rate of all exons was considered an enrichment in the CBR. All other data comparisons were performed using either t test or ANOVA analysis, as indicated in each figure legend.
Online Supplemental Material.
Supplemental material includes data regarding the representation of each IL12RB1 RDD type in the presence or absence of PHA stimulation (Fig. S1); the extent of RDD exon enrichment in IL12RB1 mRNA clones before and after activation with PHA, stratified by donor (Fig. S2); biochemical data pertinent to the production of recombinant IL12Rβ1 CBRs (Fig. S3); sequence information for the primers used in our studies (Dataset S1); the IL12RB1 genotype of PBMC and lung tissue donors (Dataset S2); the IL12RB1 RDDs identified in human tissues by Ion Torrent sequencing (Dataset S3); the IL12RB1 RDDs identified in human tissues by Sanger sequencing (Dataset S4); lung tissue donor information (Dataset S5); and the IL12RB1 gDNA/mRNA nucleotide numbering system used for our studies (Dataset S6).
Supplementary Material
Acknowledgments
We thank Dr. Nicole Ford and Allison Reeme for their contributions to our Sanger sequencing efforts, as well as Dr. Kathleen Boyle and Matthew Bluma for their assistance with recombinant CBR purification, Dr. Aniko Szabo for her assistance with our statistical analysis, and Gail Hecox for proofreading our manuscript. This work was supported by funds from the Medical College of Wisconsin, the Children’s Research Institute, Advancing a Healthier Wisconsin, and the National Institutes of Health (R21AI099661 to R.T.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515978112/-/DCSupplemental.
References
- 1.Teng MW, et al. IL-12 and IL-23 cytokines: From discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21(7):719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: Genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 2014;26(6):454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard JP, et al. Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-gamma production. Blood. 1997;90(7):2541–2548. [PubMed] [Google Scholar]
- 4.Robinson RT. IL12Rβ1: The cytokine receptor that we used to know. Cytokine. 2015;71(2):348–359. doi: 10.1016/j.cyto.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Vosse E, de Paus RA, van Dissel JT, Ottenhoff TH. Molecular complementation of IL-12Rbeta1 deficiency reveals functional differences between IL-12Rbeta1 alleles including partial IL-12Rbeta1 deficiency. Hum Mol Genet. 2005;14(24):3847–3855. doi: 10.1093/hmg/ddi409. [DOI] [PubMed] [Google Scholar]
- 6.Li M, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333(6038):53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang IX, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Reports. 2014;6(5):906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wongsurawat T, Jenjaroenpun P, Kwoh CK, Kuznetsov V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012;40(2):e16. doi: 10.1093/nar/gkr1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335(6074):1302. doi: 10.1126/science.1209658. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, Piskol R, Tan MH, Li JB. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335(6074):1302. doi: 10.1126/science.1209658. [DOI] [PubMed] [Google Scholar]
- 11.Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335(6074):1302. doi: 10.1126/science.1210484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Garra A, et al. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 13.de Beaucoudrey L, et al. Revisiting human IL-12Rβ1 deficiency: A survey of 141 patients from 30 countries. Medicine (Baltimore) 2010;89(6):381–402. doi: 10.1097/MD.0b013e3181fdd832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prando C, et al. Inherited IL-12p40 deficiency: Genetic, immunologic, and clinical features of 49 patients from 30 kindreds. Medicine (Baltimore) 2013;92(2):109–122. doi: 10.1097/MD.0b013e31828a01f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Vosse E, et al. IL-12Rβ1 deficiency: Mutation update and description of the IL12RB1 variation database. Hum Mutat. 2013;34(10):1329–1339. doi: 10.1002/humu.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Vosse E, Lichtenauer-Kaligis EG, van Dissel JT, Ottenhoff TH. Genetic variations in the interleukin-12/interleukin-23 receptor (beta1) chain, and implications for IL-12 and IL-23 receptor structure and function. Immunogenetics. 2003;54(12):817–829. doi: 10.1007/s00251-002-0534-9. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26(8):345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SH, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 19.Powell LM, et al. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 20.Benne R, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 21.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2(5):594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford NR, et al. Inflammatory signals direct expression of human IL12RB1 into multiple distinct isoforms. J Immunol. 2012;189(9):4684–4694. doi: 10.4049/jimmunol.1200606. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt EG, et al. IL-10 produced by induced regulatory T cells (iTregs) controls colitis and pathogenic ex-iTregs during immunotherapy. J Immunol. 2012;189(12):5638–5648. doi: 10.4049/jimmunol.1200936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.