Significance
The patterns of lateral branching, including tillers and inflorescence branches, determine grain yields of many cereals. In this study, we characterized a regulatory network composed of microRNAs and transcription factor that coordinately regulate vegetative (tiller) and reproductive (panicle) branching in rice. The findings hold tremendous promise for application in rice genetic improvement and may also have general implications for understanding branching regulation of grasses.
Keywords: Oryza sativa, lateral branch, panicle, spikelet, microRNA
Abstract
Grasses produce tiller and panicle branching at vegetative and reproductive stages; the branching patterns largely define the diversity of grasses and constitute a major determinant for grain yield of many cereals. Here we show that a spatiotemporally coordinated gene network consisting of the MicroRNA 156 (miR156/)miR529/SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) and miR172/APETALA2 (AP2) pathways regulates tiller and panicle branching in rice. SPL genes negatively control tillering, but positively regulate inflorescence meristem and spikelet transition. Underproduction or overproduction of SPLs reduces panicle branching, but by distinct mechanisms: miR156 and miR529 fine-tune the SPL levels for optimal panicle size. miR172 regulates spikelet transition by targeting AP2-like genes, which does not affect tillering, and the AP2-like proteins play the roles by interacting with TOPLESS-related proteins (TPRs). SPLs modulate panicle branching by directly regulating the miR172/AP2 and PANICLE PHYTOMER2 (PAP2)/Rice TFL1/CEN homolog 1 (RCN1) pathways and also by integrating other regulators, most of which are not involved in tillering regulation. These findings may also have significant implications for understanding branching regulation of other grasses and for application in rice genetic improvement.
The architecture of grasses is largely determined by the branching patterns. Tillers and inflorescence branches are produced at vegetative and reproductive stages, respectively, and their patterns greatly contribute to the diversity of grasses and constitute a major determinant of grain yield of major cereals.
Rice branching has attracted much attention because of its importance in food production. Axillary buds produce tillers during the vegetative stage. However, only the early ones formed from the unelongated internodes outgrow as tillers, whereas later ones formed from the upper internodes remain dormant. After reproductive transition, the shoot apical meristem is converted to inflorescence meristem to produce panicle. Rice panicle morphology is largely determined by the timing of identity transition among the different types of meristems (SI Appendix, Fig. S1). Therefore, fine-tuning of meristem phase change at reproductive stage defines the size and architecture of the rice panicle (1).
Many genes have been identified as regulators of rice branching. Generally, genes involved in axillary bud initiation control both vegetative and reproductive branching, whereas genes under axillary bud outgrowth have specific roles only at certain stages (2, 3). LAX PANICLE 1 (LAX1) and MONOCULM1 control axillary bud initiation; mutation in either of them results in reduction of both tiller and panicle branches (4, 5). Other genes such as Grain number, plant height, and heading date7 exclusively control panicle branching (6). As a third class, many genes, including Ideal Plant Architecture 1 (IPA1)/Wealthy Farmer’s Panicle (WFP) and genes related to strigolactone, play opposite roles in tiller and panicle branches (7–9). Therefore, there are both commonalities and distinctions in the mechanisms regulating vegetative and reproductive branching. An interesting and fundamental question is how the tillers and panicle branches are coordinately regulated. Elucidating the shifting gene regulatory networks underlying branch outgrowth following the developmental stages should provide understanding of the coordinated regulation and offer guidance for plant breeding practice.
MicroRNA 156 (miR156) targets the plant-specific transcription factor SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) gene families. In Arabidopsis, miR156/SPL plays vital roles in both vegetative and reproductive phase changes (10, 11), whereas miR172 shows an opposite role in phase change by targeting APETALA2 (AP2)-like transcription factors (10). The sequential actions of miR156 and miR172 in regulating vegetative phase change has been reported in many plant species (12). Compared with Arabidopsis, grass inflorescence development and phase changes are more complicated, involving different types of meristems. Whether these transitions are also related to the miR156/miR172 pathway is still unknown. Both miR156 and miR172 play as regulators of inflorescence and tiller development in rice and maize (13–16). Unlike Arabidopsis, SPL genes are also regulated by miR529 in grasses (17). However, further studies are required to understand the regulatory network and coordination of these three miRNAs in lateral branching.
In this study, we elucidated the roles of miR156, miR172, miR529 and their target genes in regulating rice tiller and panicle branching. Our findings suggest that the miRNAs and transcription factors in coordination regulate the vegetative and reproductive branching by shifting gene regulation networks.
Results
Effects of miR156 and miR529a on Tiller and Panicle Branching.
Two groups of genes exhibited complementary expression profiles from early to late stages of panicle development (18). Among them, SPL7, SPL14, and SPL17 showed decreased expression from early to late stages. They are targets of miR156 and miR529, which together with miR172 were reported to control developmental timing in plants (12). Thus, we analyzed these three miRNAs and their target genes in branching.
Compared with wild-type (WT) plants, the miR156 overexpressors (designated as miR156OE) had shorter plastochron length (SI Appendix, Fig. S2 A and B). Accompanied with higher leaf initiation rate, the tiller bud was produced as early as 7 d after germination in miR156OE plants, whereas it was 15 d in WT (SI Appendix, Fig. S3A). A tiller bud was produced from the axil of each leaf except the flag leaf, and the ones from the elongated internode were usually dormant (SI Appendix, Fig. S3 B and D), whereas ectopic tiller bud from the axil of flag leaf and higher-order tillers were produced in miR156OE plants, resulting in many more tillers (Fig. 1A and SI Appendix, Fig. S3 C–G). Thus, miR156 regulates both initiation and outgrowth of vegetative branching. The panicles of miR156OE were very small (Fig. 1B), with the number of spikelets only 6.95% of the WT on average (SI Appendix, Table S1). This phenotype was consistent with previous results (15). Smaller inflorescence meristem and fewer primary branch primordia were produced in miR156OE plants than WT, as revealed by scanning electron microscopy (Fig. 1 G and H). The meristem marker gene OSH1 (3) was expressed in much fewer primordia in miR156OE plants than in WT, as detected by using in situ hybridization (Fig. 1 I–K). Therefore, miR156 negatively regulates inflorescence meristem activity and the initiation of reproductive branching. Manually removing the newborn tillers every other day from seedling to maturation stages led to bigger panicles in WT, but not miR156OE, plants (Fig. 1 C and D), implying that the smaller panicles in miR156OE plants were not the trade-off of the higher number of tillers. Thus, miR156 regulates tiller and panicle branching through distinct pathways.
Fig. 1.
Tillering and panicle branching regulated by miR156 and miR529. (A and B) The plants (A) and panicles (B) at adult stage of WT, MIM156, miR156OE, and the hybrid between miR156OE and MIM156 plants. (C and D) Main panicles of WT (C) and miR156OE (D) plants with or without tiller removal every other day. Values are means ± SEM (n = 15). (E and F) Plants (E) and panicles (F) of WT and miR529aOE plants at adult stage. (G and H) Scanning electron microscopic images of the panicles at the primary branch initiation stage in WT (G) and miR156OE (H) plants. The red and blue asterisks indicate inflorescence and primary branch meristems, respectively. (Scale bars: 50 μm.) (I–K) In situ hybridization of OSH1 in WT (I and K) and miR156OE (J) plants by using antisense (I and J) and sense (K) probes. [Scale bars: 200 μm (I) and 100 μm (J and K).]
The target mimicry approach (19) was used to interfere the activity of miR156, which reduced the level of miR156 significantly in the transgenic plants (MIM156) (SI Appendix, Fig. S4A), accompanied by elevated levels of the target SPL genes (SI Appendix, Fig. S4B). The rates of leaf and tiller production in MIM156 plants were slower than WT (SI Appendix, Figs. S2 A and B and S3 A and F), resulting in fewer tillers with bigger panicles (Fig. 1 A and B and SI Appendix, Table S1), exactly opposite from the phenotype of miR156OE. Moreover, the defects of miR156OE could largely be corrected by MIM156 (Fig. 1 A and B and SI Appendix, Fig. S4 C–H), suggesting that overexpression and target mimicry of miR156 could counteract each other in planta.
miR529 sharing 14-nt homology with miR156 also targets SPL genes, mostly at later panicle stage (SI Appendix, Fig. S5A) (17). The reporter gene firefly luciferase (LUC) fused with the miR529 binding site of SPL17 was significantly repressed by miR529a, but not miR529b or AtmiR172b, in a transient expression assay demonstrating that miR529a regulated SPL in planta (SI Appendix, Fig. S5 B and C). Accordingly, miR529a, but not miR529b, overexpressor (miR529aOE) produced similar phenotypes to miR156OE in Zhonghua 11 (ZH11), although to a lesser extent (Fig. 1 E and F). Transcript levels of SPL14 and SPL17 were also reduced in miR529aOE (SI Appendix, Fig. S5 D and E), suggesting that SPLs were regulated by miR529a as well.
Effects of SPL7, SPL14, and SPL17 on Tillering and Panicle Branching.
Among the SPL gene family (SI Appendix, Fig. S6), SPL7, SPL14, and SPL17 showed the highest expression in panicles, as revealed in microarray data (18), and the patterns could be confirmed by quantitative RT-PCR (qRT-PCR) in 16 other tissues (SI Appendix, Fig. S7). A T-DNA insertion mutant 4A-00131 for SPL7 in Dongjin genetic background (spl7-1) and RNAi lines of SPL14 and SPL17 in ZH11 (SPLxRi) were obtained (SI Appendix, Fig. S8 A–G). Not much change in branching and leaf emergence rate was observed in spl7-1 (SI Appendix, Figs. S2C and S8 H and I and Table S1), whereas the lateral organ initiation rates and overall architectures of SPL14Ri and SPL17Ri plants were reminiscent of miR156OE plants (SI Appendix, Figs. S2B, S3A, and S8 J–O). Panicle branching and spikelets were heavily reduced in both SPL14Ri and SPL17Ri plants (SI Appendix, Table S1), indicating that SPLs positively regulated the activities of inflorescence and branch meristems. Each of the RNAi transformants showed gene-specific repression as expected, whereas SPL7 showed slight, but insignificantly, increased expression in both RNAi lines (SI Appendix, Fig. S8 E–G). Double RNAi lines of SPL14 and SPL17 enhanced phenotypic effects compared with individual RNAi plants, but still were weaker than the miR156OE plants (SI Appendix, Fig. S8 P and Q), implying that SPL genes play a redundant function in rice development.
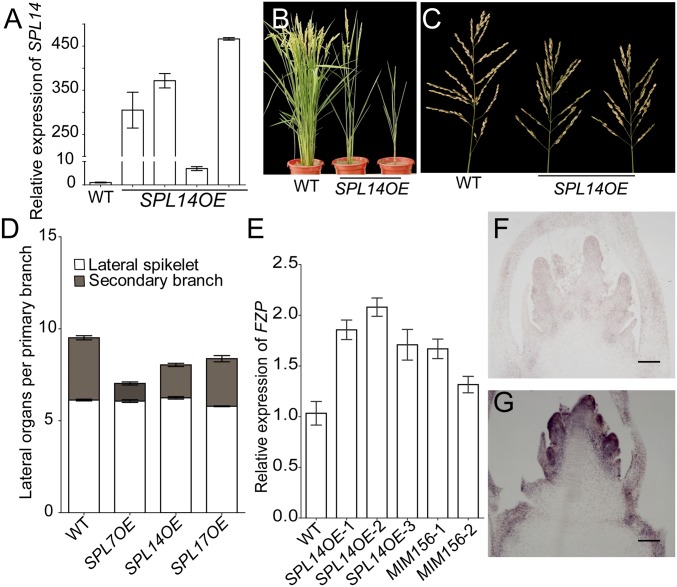
To further dissect the roles of SPL genes, we overexpressed SPL7, SPL14, and SPL17 in ZH11 (SPLxOE). The overexpressors of all three genes greatly reduced tiller numbers (Fig. 2 A–C and SI Appendix, Fig. S9 A–F), of which SPL7 had the strongest effect, such that many of the positive transgenic plants were of monoculm and died before maturation. Panicle branching and spikelets also decreased significantly in the overexpressors (SI Appendix, Table S1), such that secondary branches, rather than the lateral spikelets produced on the primary branches, were reduced (Fig. 2D), implying that the early arising lateral meristems on the primary branches were precociously converted to spikelets. Correspondingly, the expression level of Frizzy Panicle (FZP), the marker of spikelet meristem in grasses (20, 21), was elevated in SPL14OE and MIM156 lines (Fig. 2E). FZP was transiently expressed in the spikelet meristem in WT (Figs. 2F and 3O) (20). However, in situ hybridization showed that FZP was ectopically expressed in the branch meristem of SPL14OE plants (Fig. 2G), suggesting that SPL genes promoted the transition from branch to spikelet meristem. The resistant SPL7 (rSPL7) produced by changing the recognition site of miR156 and miR529, but not the protein sequence driven by its native promoter (designated rSPL7HA), also showed similar phenotypes to the SPL7OE plants (SI Appendix, Fig. S9 G, H, and L–O). GFP fused to the C-terminal SPL14 (termed as SPL14GFP) showed a similar phenotype to SPL14OE plants (SI Appendix, Fig. S9 I–K and P–S). Together, SPL genes promoted the conversion of branch to spikelet meristem and had a general function in reducing branching.
Fig. 2.
The transition from branch to spikelet meristem promoted by SPL genes. (A) Relative expression level of SPL14 in flag leaf of its overexpressing plants. Values are means ± SEM (n = 3). (B and C) The plants (B) and panicles (C) of SPL14OE compared with WT. (D) Numbers of secondary branches and lateral spikelets produced by primary branches in SPL overexpressors. Values are means ± SEM (n = 15). (E) Relative expression level of FZP in the young panicle (<1 mm) of SPL14OE and MIM156 plants compared with WT. Values are means ± SEM (n = 3). (F and G) In situ hybridization of FZP in the panicles at the primary branch initiation stage in WT (F) and SPL14OE (G) plants. (Scale bars: 100 μm.)
Fig. 3.
Regulation of panicle branching, but not tillering, by the miR172/AP2 pathway. (A–L) The plants (A, D, G, and J), panicles (B, E, H, and K), and primary branches (C, F, I, and L) of miR172OE (A–C), MIM172 (D–F), rSNBOE (G–I), and OsTOE1 (J–L) transformants. (M and N) Scanning electron microscopic images of developing panicles ∼1 mm in length of WT (M) and MIM172 (N) plants. (Scale bars: 100 μm.) (O and P) In situ hybridization of FZP in panicles ∼1 mm in length of WT (O) and MIM172 (P) plants. (Scale bars: 200 μm.)
Effects of miR172 and its Target Genes on the Transition of Spikelet Meristem Identity.
In rice, five AP2-like transcription factors (SNB, OsIDS1, SHAT1, OsTOE1, and OsGL15) are targeted by miR172 (SI Appendix, Fig. S10A) (16). miR172 overexpressor (miR172OE) did not show marked difference in plant height and tillering (Fig. 3A and SI Appendix, Fig. S10B), but the panicle branching was greatly reduced (Fig. 3B and SI Appendix, Table S2). Like the overexpressors of SPL genes, secondary branches, rather than lateral spikelets on primary branches, decreased significantly in miR172OE plants (Fig. 3C and SI Appendix, Fig. S10 F and G).
The target mimicry approach was also used to interfere the miR172 in ZH11 (MIM172) (SI Appendix, Fig. S10 C–E). The MIM172 plants were markedly shorter than the WT, whereas the numbers of tillers were comparable (Fig. 3D). Strikingly, the panicle architecture changed drastically in MIM172 (Fig. 3 E and F and SI Appendix, Table S2). Larger numbers of secondary branches, rather than lateral spikelets on primary branches, were produced in MIM172 (SI Appendix, Fig. S10 F and G), and even tertiary branches occasionally occurred, suggesting that the transition from branch to spikelet meristem was delayed. Consistently, the scanning electron microscopic image revealed that the flower primordia were differentiated in the panicles ∼1-mm in length in WT, but not in MIM172, plants (Fig. 3 M and N). In addition, the lack of FZP expression in MIM172 revealed by in situ hybridization also showed that the spikelet transition was delayed (Fig. 3 O and P). Together, miR172, like SPL genes, also positively promoted the transition from branch to spikelet meristem. Given that miR172 inhibited the transition from spikelet to floret meristem (16), it apparently had dual roles in establishing and maintaining the spikelet meristem identity.
To investigate the roles of miR172 targeted genes, we analyzed the effects of SNB and OsTOE1 as the representative of AP2-like genes. Consistent with previous results (16), RNAi lines of SNB and OsTOE1 in ZH11 produced smaller panicles but normal tiller branching (SI Appendix, Fig. S10 H–K and Table S2). Constructs of OsTOE1 and miR172-resistant SNB (rSNB by changing the miR172 binding site, but not protein sequence, and fused with GFP) both driven by the 35S promoter were also introduced into ZH11 (designated as OsTOE1OE and rSNBOE) (SI Appendix, Fig. S10A). Overexpressors of both genes showed similar, but weaker, phenotypic changes compared with MIM172 (Fig. 3 G–L)—i.e., dense panicle with significant increase in numbers of branches and spikelets (SI Appendix, Table S2) and the secondary branches, but not the lateral spikelets, increased on the primary branches (SI Appendix, Fig. S10 F and G). Our results, together with the fact that FZP was ectopically expressed in the double mutant of snb osids1 (16), suggested that OsTOE1 and SNB dampened the transition from branch to spikelet meristem. Together, miR172 and its target genes had crucial roles in regulating reproductive, but not vegetative, branching in rice.
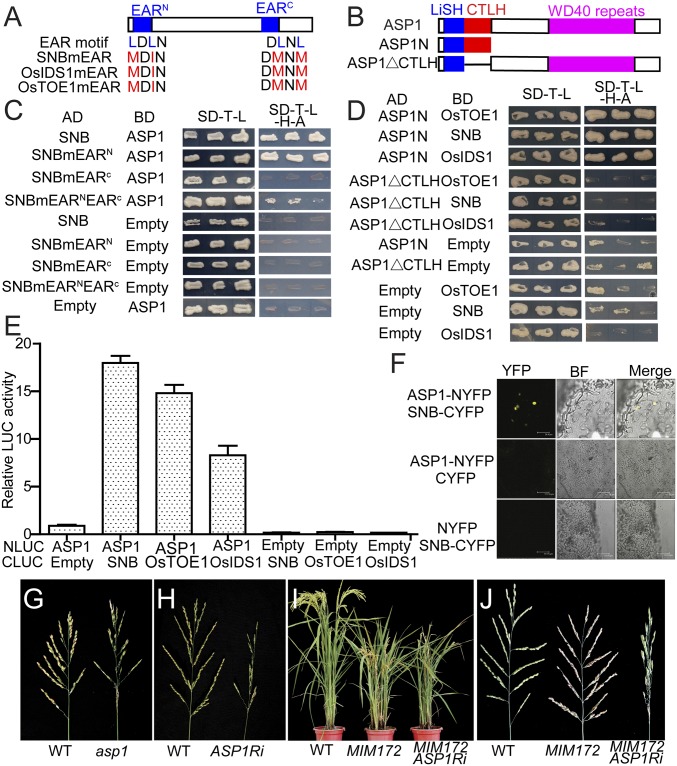
SNB and OsIDS1 fused to the GAL4 DNA binding domain attenuated the activity of the reporter 4×UAS:LUC in the transient expression assay, suggesting that SNB and OsIDS1 might act as transcriptional repressors (SI Appendix, Fig. S11 A and B). Accordingly, two putative ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs, which usually interacted with the transcriptional corepressor TOPLESS-related proteins (TPRs), were found in the proteins of SNB, OsIDS1, and OsTOE1 at the N and C termini (termed as EARN and EARC respectively) (SI Appendix, Fig. S11C). One of the three rice TPRs, TPR2—also termed ABERRANT SPIKELET AND PANICLE1 (ASP1) or Lissencephalytype-1-like1 (OsLIS-L1)—was involved in regulating panicle development (22, 23). Yeast two-hybrid assay showed that SNB, OsIDS1, and OsTOE1 interacted with TPR1 and ASP1 in yeast, and a series of protein mutation and truncation assays showed that the EARC motif of SNB, OsIDS1, and OsTOE1, and the LisH (lissencephaly type I-like homolog) domain of ASP1 were responsible for the interactions (Fig. 4 A–D and SI Appendix, Fig. S12). Both firefly LUC complementation imaging assay (LCI) (24) and bimolecular fluorescence complementation (BiFC) further substantiated the physical interactions of SNB, OsTOE1, and OsIDS1 with ASP1 in planta (Fig. 4 E and F).
Fig. 4.
Interactions of SNB, OsIDS1, and OsTOE1 with TPRs. (A) Schematic representation of the EAR motif in SNB, OsIDS1, and OsTOE1 proteins. Leu (L) to Met (M) or Ile (I) substitution mutations in the EAR motif were used to investigate its role in the interaction with TPRs. EARN and EARC indicate the N and C termini of the EAR motif, respectively. (B) Schematic representation of the protein domains of ASP1. ASP1N and ASP1ΔCTLH are the truncated versions of ASP1 used for investigating the roles of each domain in the protein– protein interaction. (C) Yeast two-hybrid assay showing the EARC of SNB to be essential for the interaction with ASP1 in the yeast strain AH109. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. (D) Yeast two-hybrid assay showing the CTLH domain of APS1 to be essential for the interaction with SNB, OsIDS1, and OsTOE1 in yeast. (E) LCI assay showing interactions of ASP1 with SNB, OsIDS1, and OsTOE1 in Arabidopsis protoplast. Data are normalized to the internal control 35S:GUS cotransformed in the assay. Values are means ± SEM (n = 3). (F) BiFC assay confirming the interaction between ASP1 and SNB in tobacco leaf cells. BF, bright field. (G and H) Panicles of asp1 (G) and ASP1Ri (H) plants compared with WT. (I and J) Plants (I) and panicles (J) of WT, MIM172, and the ASP1Ri in the MIM172 genetic background.
The panicles of the ASP1 RNAi lines in ZH11 were reminiscent of its null-allele mutant asp1 (oslis-l1-1), in which the panicle branching and spikelet were heavily perturbed (Fig. 4 G and H). The same panicle defects were also observed in the ASP1Ri plants in MIM172 background (Fig. 4 I and J), suggesting that ASP1 was essential for the function of miR172 and its target genes in regulating rice panicle development.
Interactions Between SPL Genes and miR172 in Regulating Panicle Branching.
The results that both SPL genes and miR172 promoted spikelet transition led us to investigate their possible interactions in regulating panicle branching. miR156 and miR172 had spatially–temporally complementary expression patterns in a range of tissues (SI Appendix, Fig. S13 A and B), implying that they might play complementary roles in many developmental processes beyond the vegetative phase change (12). The collective levels of the precursors of miR156, miR172, and miR529 were similar to their mature miRNAs (SI Appendix, Fig. S13 C–G), suggesting that the complementary expression patterns of mature miRNAs were modulated at the transcriptional level.
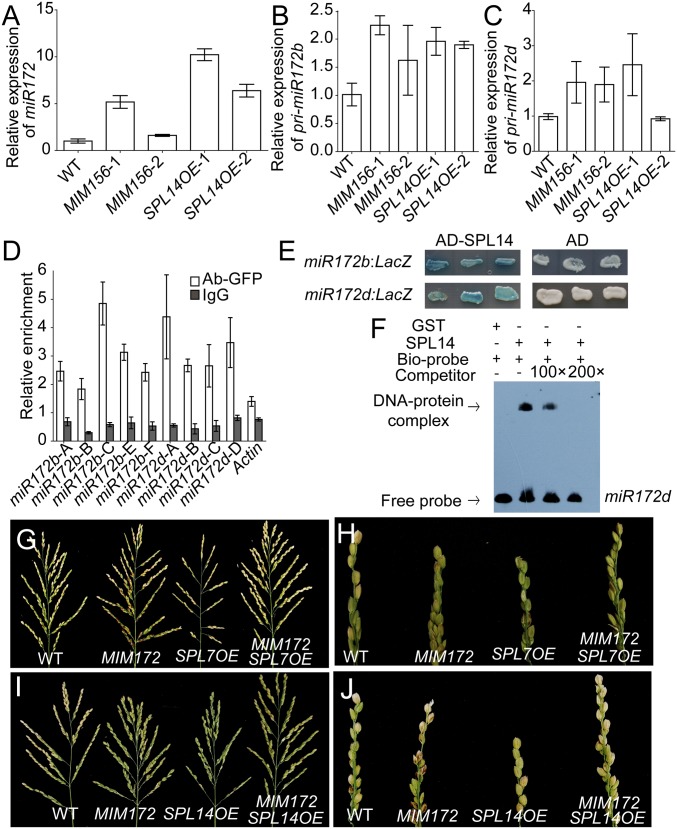
miR172 and its precursors were elevated in MIM156 and SPL14OE plants (Fig. 5 A–C), suggesting that SPLs regulated miR172 expression. Several GTAC motifs, the binding site of SPL proteins, were found in the promoters of pri-miR172b and -miR172d (SI Appendix, Fig. S13H). Chromatin immunoprecipitation (ChIP)-PCR showed that SPL14 bound to the promoter fragments of both pri-miR172b and -miR172d, but not the control actin gene, in vivo (Fig. 5D). Moreover, yeast one-hybrid (Y1H) assay showed that SPL14 bound the promoter fragments of pri-miR172b and -miR172d, thus activating the expression of reporter gene LacZ in yeast (Fig. 5E). Also, electrophoresis mobility shift assay (EMSA) showed that the recombinant protein GST-SPL14N but not GST alone physically bound the promoter fragment of pri-miR172d in vitro (Fig. 5F). Thus, SPL14 might regulate miR172 expression directly.
Fig. 5.
Regulation of spikelet transition by SPL14 via miR172. (A–C) Relative expression levels of mature miR172 (A), pri-miR172b (B), and pri-miR172d (C) in the young panicles (<1 mm) of SPL14OE and MIM156 plants compared with WT. Values are means ± SEM (n = 3). (D) ChIP assays of pri-miR172b and -miR172d in the young panicles (<10 mm) collected from Ubi:GFP and SPL14GFP plants. Samples were precipitated with anti-GFP antibody and IgG protein. The values were first normalized to the input values, then divided by the Ubi:GFP value to get the enrichment fold. Values are means ± SEM (n = 3). The amplification fragments are shown in SI Appendix, Fig. S13H. (E) Y1H assay of SPL14 with the promoters of pri-miR172b and -miR172d in yeast. (F) EMSA of GST and GST-SPL14N recombinant proteins incubated with biotin-labeled probes of pri-miR172d. (G–J) Panicles (G and I) and primary branches (H and J) of WT, SPL7OE, SPL14OE, MIM172, and the corresponding hybrids.
To investigate the possibly genetic interaction between miR156 and miR172 in rice panicle development, we knocked down both miRNAs simultaneously (referred as MIM156–172). The transgenic plants showed similar tiller phenotypes to MIM156 at vegetative stage, whereas their panicle morphology was similar to MIM172 plants (SI Appendix, Fig. S13 I–J), suggesting that miR156 and miR172 regulated rice vegetative and reproductive branches in coordination. The panicle branching defects and precocious transition of spikelet in SPL7OE and SPL14OE plants could largely be corrected to normal by MIM172 (Fig. 5 G–J and SI Appendix, Fig. S13 K and L). Similar results were also obtained by crossing rSNBOE with SPL7OE and SPL14OE plants (SI Appendix, Fig. S13 M–P). Together, the results suggested that the spikelet transition promoted by SPL genes was largely through miR172.
Other Elements in the SPL Pathway in Branching Regulation.
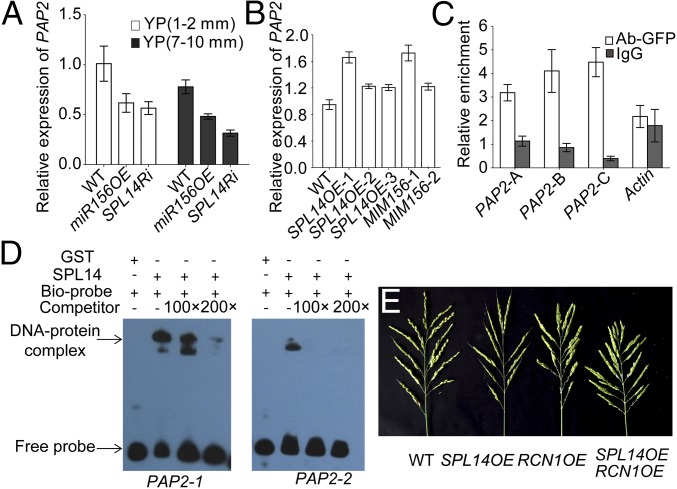
PANICLE PHYTOMER2 (PAP2) encoding MADS34 was another positive regulator of spikelet meristem identity in rice by suppressing Rice TFL1/CEN homolog (RCNs) (25–27), and this genetic module was conserved in Arabidopsis in determining its inflorescence architecture (27). In Arabidopsis, SPL genes controlled flowering time by regulating MADS genes (11, 28); thus, we speculated that SPL genes might also regulate spikelet transition by PAP2/MADS34-RCN besides the miR172-AP2 pathway in rice. Indeed, PAP2 was down-regulated in both miR156OE and SPL14Ri plants and up-regulated in SPL14OE and MIM156 plants (Fig. 6 A and B), whereas RCN1 was up-regulated in the panicles of miR156OE plants (SI Appendix, Fig. S15B). Several GTAC motifs were found in the promoters of PAP2 (SI Appendix, Fig. S14A), ChIP assay showed that SPL14 directly bound to the PAP2 promoter, but not the control, in vivo (Fig. 6C). Although Y1H assay could not reveal the interaction of SPL14 with the promoter fragments of PAP2 due to their high autoactivation activity in yeast, physical interactions in vitro were indicated by EMSA (Fig. 6D). These results demonstrated that PAP2 was also directly regulated by SPL14 in rice.
Fig. 6.
SPL14 also regulated spikelet transition by PAP2/MADS34-RCN pathway. (A and B) Relative expression levels of PAP2 in the panicles (<1 mm) of miR156OE, SPL14Ri (A), SPL14OE, and MIM156 (B) plants. Values are means ± SEM (n = 3). (C) ChIP assay of PAP2 in the young panicles (<10 mm) collected from Ubi:GFP and SPL14GFP plants. The details are as in Fig. 5D. Values are means ± SEM (n = 3). (D) EMSA of GST and GST-SPL14N recombinant proteins incubated with biotin-labeled probes of PAP2. (E) The panicles of WT, SPL14OE, RCN1OE, and the corresponding hybrid.
Because of unavailability of overexpressor or mutant of PAP2 in ZH11 genetic background, we used RCN1 to test the genetic interaction with miR156-SPL. Overexpressing RCN1 in ZH11 greatly delayed spikelet meristem transition, thus producing much more secondary branches and spikelets (SI Appendix, Fig. S14 B and C). Strikingly, tillering was also reduced in RCN1OE plants (SI Appendix, Fig. S14 D–K), suggesting that RCN1 regulated both tiller and panicle branching in rice. The high number of tillers of SPL14Ri and SPL17Ri plants could be corrected to normal by RCN1OE or vice versa (SI Appendix, Fig. S14 D–K). The defects of secondary, but not primary, branches of SPL14Ri, SPL17Ri, and SPL14OE plants could be rescued by RCN1OE (Fig. 6E and SI Appendix, Fig. S14 D–K), suggesting that SPL genes regulated spikelet transition, but not inflorescence meristem activity by RCN1.
To further investigate the downstream genes regulated by the miR156-SPL pathway in rice panicle development, we compared the transcriptomes of very young panicles (<1 mm) between miR156OE and WT plants and found that expression of a large number of genes, including the ones related to transcriptional regulation, were altered in miR156OE plants (P < 0.05, cutoff >1.5-fold; SI Appendix, Fig. S15A and Dataset S1). Besides PAP2/MADS34 and the SPL genes, which were down-regulated in miR156OE plants as expected, many other known genes related to rice panicle development were also down-regulated, including LAX1, LONELY GUY (LOG), and Rice Leafy Homolog (RFL), all of which are important regulators of rice panicle development (4, 29, 30). Some differentially expressed panicle regulators were further confirmed by qRT-PCR (SI Appendix, Fig. S15B).
Besides spikelet transition, SPL genes also regulated rice panicle primary branching. LAX1 and RFL, as the important genes for rice panicle branch development (4, 30), were highly coexpressed with SPL7, SPL14, and SPL17 (18) and down-regulated in the panicles of the miR156OE plant. Thus, we also investigated the functions of these two genes in the miR156-SPL pathway. The larger panicles of MIM156 could be recovered by RNAi of LAX1 and RFL (SI Appendix, Fig. S15 C and D). EMSA and Y1H assay revealed that SPL14 bound to the LAX1 promoter (SI Appendix, Fig. S15 E–G), implying that LAX1 might also be directly regulated by SPL14. Moreover, Leafy, as the ortholog of RFL, was directly regulated by SPL genes in Arabidopsis (28). These results suggested that miR156-SPL might also regulate panicle branching by LAX1 and RFL.
Discussion
Our study revealed the gene networks regulated by miR156, miR172, miR529, and their target genes that coordinately control rice vegetative and reproductive branching, which we attempted to summarize as a model in SI Appendix, Fig. S16. At early vegetative stage, miR156 promotes tillering by inhibiting SPLs. Our results revealed that RCN1 and SPL genes might be involved in the same genetic pathway in regulating tillering. It should be noted in this connection that TEOSINTE BRANCHED 1 (TB1), another negative regulator of tillering, is also directly regulated by SPL14 (31). Therefore, the miR156/SPL pathway regulates tillering at least involving RCN1 and TB1 as the downstream elements, although details of the actions and their relationships with other known genes are still lacking for constructing the pathway.
After reproductive transition, SPL genes reach the highest levels at the early panicle stage. It is remarkable that panicle branches were reduced in both RNAi and overexpression lines of SPL genes, indicating that the expression of SPL genes must be fine-tuned to optimal levels for reproductive branching: Either above or below the optimal levels would reduce branching. Therefore, the miR156 and miR529 at reproductive stage would maintain the SPL expression to the more optimal levels, thus promoting panicle branching. At the downstream, the miR172/AP2 pathway is used in regulating panicle, but not tiller, branching. Therefore, the activities of the miR156/miR529/SPL and miR172/AP2 pathways harmoniously coordinate vegetative and reproductive branching by shifting gene networks in different developmental stages.
Our results showed that miR156 negatively regulated inflorescence meristem activity; therefore, SPLs had positive roles in maintaining the activity of inflorescence meristem. LOG encoding an enzyme for activating the cytokinin is directly regulated by SPL14 (29, 31), suggesting that SPL genes may play these roles by regulating cytokinin signaling in rice. It was reported that more branches and spikelets were produced in rice genotype containing the allele IPA1/WFP of SPL14 (7, 8). In light of our results, SPL14 has negative roles on panicle branches by promoting the transition of spikelet meristem, thus negatively regulating secondary branches. Therefore, the total number of spikelets represents a balance among the multiple roles of SPL14. The SPL genes could increase spikelets only when their level is optimal. Plants with moderately high activity of SPL14, such as the ones containing the IPA1/WFP allele or MIM156, produce more spikelets, because the higher activity of inflorescence meristem results in more primary branches, which compensates the negative effect of precocious transition of spikelet meristem identity. Conversely, very high activity of SPL genes reduces total spikelets by precocious transition of spikelet meristem. Therefore, SPLs have multiple roles in regulating panicle branches, depending on the expression levels. Fine-tuning of the expression of SPL genes to the most favorable levels may provide a strategy for increasing rice productivity in breeding application.
Both molecular and genetic evidence demonstrated that SPL14 positively regulates the transition of spikelet from branch meristem via both miR172/AP2 and PAP2/RCN1 pathways. Natural variations in miR172 and its targets AP2-like genes have crucial roles in the domestication and evolutionary processes of grasses (14, 32, 33). However, the relationship of natural variation of these genes with agronomic traits had not been demonstrated in rice. We showed that more spikelets were produced in MIM172 and rSNBOE plants, which may be used for rice breeding by genome manipulating technologies. SNB and OsIDS1 have intrinsic transcriptional repressor activity, and they interacted with the transcriptional corepressor TPRs, which usually recruit the histone deacetylase to modify the epigenetic states of their downstream genes (34); thus, AP2-like genes might repress the expression of their target genes. ASP1 might serve as another link between the miR156/SPL and miR172/AP2 pathways, because it was also directly targeted by SPL14 (31).
Some of the SPL and AP2 genes had been studied in maize, barley, and wheat (14, 32, 33, 35), and our results uncovered that functions of these genes are mostly conserved in rice. Moreover, miR156-regulated vegetative phase change via miR172 is well known to be conserved in many plant species, including maize and rice (12), and the gene regulatory networks revealed in this study may shed light for understanding the ancient and conserved mechanisms. However, there are also distinctions. For example, the branch meristems were converted to spikelet meristems in the mutant asp1 in rice (22), whereas exactly the reverse was the case in the mutant of its maize ortholog RAMOSA ENHANCER LOCUS 2, in which spikelet meristems were converted to branch meristems (36). Thus, functional divergence of the pathways had taken place during evolution. Furthermore, there are questions remaining to be addressed in future studies, regarding whether the panicle branching regulation by physically interaction between AP2 and TPRs is conserved and how miR156/SPLs and PAP2/RCN1 pathways are related in other grasses.
Materials and Methods
The rice variety Zhonghua 11 (ZH11) was used for transformation in most of this study. The T-DNA insertion mutant spl7-1 was obtained from the Postech rice mutant library (37). Details of experimental methods are given in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Lizhong Xiong, Changyin Wu, and Gynheung An for providing rice seeds; Jianmin Zhou, Jian Xu, Shiping Wang, and Rongcheng Lin for providing the constructs; and Weijiang Tang for the great assistance in the luciferase-related experiments. This work was supported by National 863 Project Grant (2012AA10A303) and a grant from the Bill & Melinda Gates Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521949112/-/DCSupplemental.
References
- 1.Kyozuka J, Tokunaga H, Yoshida A. Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr Opin Plant Biol. 2014;17:110–115. doi: 10.1016/j.pbi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 2.McSteen P, Leyser O. Shoot branching. Annu Rev Plant Biol. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Yuan Z. Molecular control of grass inflorescence development. Annu Rev Plant Biol. 2014;65:553–578. doi: 10.1146/annurev-arplant-050213-040104. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu K, et al. LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci USA. 2003;100(20):11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, et al. Control of tillering in rice. Nature. 2003;422(6932):618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- 6.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 7.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 8.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42(6):545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 9.Jiang L, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504(7480):401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Poethig RS. Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39(4):544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 14.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39(12):1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 15.Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142(1):280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee DY, An G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012;69(3):445–461. doi: 10.1111/j.1365-313X.2011.04804.x. [DOI] [PubMed] [Google Scholar]
- 17.Jeong DH, et al. Massive analysis of rice small RNAs: Mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell. 2011;23(12):4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61(5):752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 19.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130(16):3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- 21.Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 2002;298(5596):1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY. Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012;70(2):327–339. doi: 10.1111/j.1365-313X.2011.04872.x. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, et al. OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta. 2012;235(4):713–727. doi: 10.1007/s00425-011-1532-7. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146(2):368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010;51(1):47–57. doi: 10.1093/pcp/pcp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X, et al. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 2010;153(2):728–740. doi: 10.1104/pp.110.156711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013;24(6):612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi A, et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17(2):268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012;69(1):168–180. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- 31.Lu Z, et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell. 2013;25(10):3743–3759. doi: 10.1105/tpc.113.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair SK, et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA. 2010;107(1):490–495. doi: 10.1073/pnas.0909097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172(1):547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312(5779):1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 35.Chuck GS, Brown PJ, Meeley R, Hake S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci USA. 2014;111(52):18775–18780. doi: 10.1073/pnas.1407401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallavotti A, et al. The control of axillary meristem fate in the maize ramosa pathway. Development. 2010;137(17):2849–2856. doi: 10.1242/dev.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong DH, et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45(1):123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.