Abstract
An organism with a single recessive loss-of-function allele will typically have a wild-type phenotype while individuals homozygous for two copies of the allele will display a mutant phenotype. Here, we develop a method that we refer to as the mutagenic chain reaction (MCR), which is based on the CRISPR/Cas9 genome editing system for generating autocatalytic mutations to generate homozygous loss-of-function mutations. We demonstrate in Drosophila that MCR mutations efficiently spread from their chromosome of origin to the homologous chromosome thereby converting heterozygous mutations to homozygosity in the vast majority of somatic and germline cells. MCR technology should have broad applications in diverse organisms.
Keywords: CRISPR/Cas9, Drosophila, mutagenic chain reaction, MCR, gene-drive
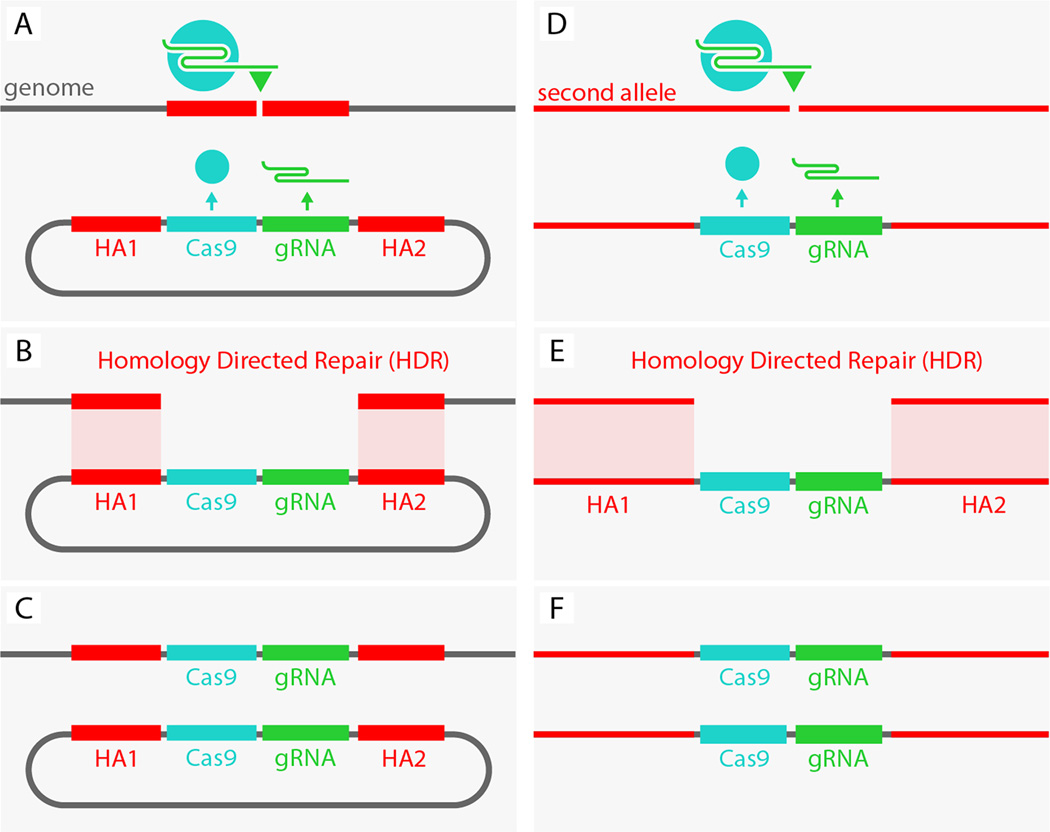
It is often desirable to generate recessive loss-of-function mutations in emergent model organisms, however, scoring for such mutations in the heterozygous condition is challenging. Taking advantage of the CRISPR/Cas9 genome editing method (1,2), we have developed a strategy to convert a Drosophila heterozygous recessive mutation into a homozygous condition manifesting a mutant phenotype. We reasoned that autocatalytic insertional mutants could be generated with a construct having three components: 1) a Cas9 gene (expressed in both somatic and germline cells), 2) a guide-RNA (gRNA) targeted to a genomic sequence of interest, and 3) homology arms flanking the Cas9/gRNA cassettes that match the two genomic sequences immediately adjacent to either side of the target cut site (Fig. 1A). In such a tripartite construct, Cas9 should cleave the genomic target at the site determined by the gRNA (Fig. 1A) and then insert the Cas9/gRNA cassette into that locus via homology-directed repair (HDR) (Fig. 1B,C). Cas9 and the gRNA produced from the insertion allele should then cleave the opposing allele (Fig. 1D), followed by HDR-driven propagation of the Cas9/gRNA cassette to the companion chromosome (Fig. 1E,F). We refer to this trans-acting mutagenesis scheme as a mutagenic chain reaction (MCR).
Figure 1.
Scheme outlining the Mutagenic Chain Reaction (MCR). A plasmid consisting of a core cassette carrying a Cas9 transgene, a gRNA targeting a genomic sequence of interest, and flanking homology arms corresponding to genomic sequences abutting the target cleavage site (A) inserts the core Cas9/gRNA cassette into the targeted locus via HDR (B,C). In turn, the inserted cassette expresses both Cas9 and the gRNA leading to cleavage (D) and HDR-mediated insertion of the cassette into the second allele, thereby rendering the mutation homozygous (E,F). HA1 and HA 2 denote the two homology arms that directly flank the gRNA-directed cut site.
We expected that autocatalytic allelic conversion by MCR should be very efficient in both somatic and germline precursor cells given the high frequency and specificity of mutagenesis (3) and efficacy of homology based integration (4) mediated by separate genome encoded Cas9 and gRNA genes observed in previous studies. We tested this prediction in D. melanogaster using a characterized efficient target sequence (y1) (5) in the X-linked yellow (y) locus as the gRNA target and a vasa-Cas9 transgene as a source of Cas9 (Fig. 2C) since it is expressed in both germline and somatic cells (4). As the defining element of our MCR scheme, we also included two ~1 kb homology arms flanking either side of the central elements (Fig. 2C) that precisely abut the gRNA-directed cut site. Wild-type (y+) embryos were injected with the y-MCR element (see Materials and Methods) and emerging F0 flies were crossed to a y+ stock. According to Mendelian inheritance, all F1 female progeny of such a cross should have a y+ phenotype (i.e., F1♀s inherit a y+ allele from their wild-type parent).
Figure 2.
Experimental demonstration of MCR in Drosophila. A) Mendelian male inheritance of an X-linked trait. B) Theoretical MCR-based inheritance results in the initially heterozygous allele converting the second allele generating homozygous female progeny. C) Diagram of y-MCR construct. Two y locus homology arms flanking the vasa-Cas9 and y-gRNA transgenes are indicated and the locations of the PCR primers used for analysis of the genomic insertion site (listed in the Methods section). D) PCR analysis of a y+ MCR-derived F2♂ (lanes 1–3, see Fig. S1 for sequence), yMCR F1♀ (lanes 4–6) and ♂ (lanes 7–9) showing junctional bands corresponding to y-MCR insertion into the chromosomal y locus (lanes 2–3,5–6,8–9) and an approximately wild-type size band from the y locus (lanes 1,4,7). Although the yMCR F1♂ (carrying a single X-chromosome) displays only MCR derived PCR products (lanes 8–9), yMCR F1♀s generate both MCR and non-insertional amplification products. E) Summary of F2 progeny obtained from crosses described in detail in Table S1. F) Low magnification view of F2 progeny flies from an yMCR X y+. Nearly all female progeny display a y- phenotype. G) High magnification view of a full body yMCR F1♀. H) A rare 50% left-right mosaic female. I) A y+ control fly.
From 2 independent F0 male (♂) X y+ female (♀) crosses and 7 F0♀ X y+♂ crosses we recovered y- F1♀ progeny, which should not happen according to Mendelian inheritance of a recessive allele. Six such yMCR F1♀ were crossed individually to y+♂ resulting in 95–100% (average = 97%) of their F2 progeny exhibiting a full-bodied y- phenotype (Table S1, Fig. 2E,G) in contrast to the expected rate of 50% (i.e., only in males). We similarly tested MCR transmission via the germline in two y- F1♂ recovered from an F0♀ cross that also yielded y- female siblings. These y- F1♂ were considered candidates for carrying the y-MCR construct and were crossed to y+ females. All but one of their F2 female progeny had a full body y- phenotype (Fig. 2E,F). Occasionally among yMCR F2♀ we also recovered mosaics (~ 4%) with few small y+ patches as well as a lone example of a 50% chimeric female (Fig. 2H), and in two instances, we recovered y+ male progeny from a yMCR F1♀ mother (Fig. 2E, Table S1). These infrequent examples of imperfect y-MCR transmission indicate that while HDR is a highly efficient at this locus in both somatic and germline lineages, the target occasionally evades conversion.
PCR analysis of the y locus in individual y- F1 progeny confirmed the precise gRNA/HDR-directed genomic insertion of the y-MCR construct in all flies giving rise to y- female F2 progeny (Fig. 2D). Males carried only this single allele, as expected, whereas females in addition possessed a band corresponding to the size of the wild-type y locus (Fig. 2D, lane 4), which varied in intensity between individuals, indicating that females were mosaic for MCR conversion. The left and right y-MCR PCR junction fragments were sequenced from y- F1 progeny from 5 independent F0 parents. All had the precise expected HDR-driven insertion of the y-MCR element into the chromosomal y locus. In addition, sequence analysis of a rare non-converted y+ allele recovered in a male offspring from a yMCR F1♀ (Fig. 2E) revealed a single nucleotide change at the gRNA cut site (resulting in a T->I substitution), which most likely resulted from non-homologous end-joining repair, as well as an in-frame insertion/deletion in a y+♀ sibling of this male (Fig. S1, Table S1). The high recovery rate of full bodied y- F1 and F2 female progeny from single parents containing a yMCR allele detectable by PCR indicates that the conversion process is remarkably efficient in both somatic and germline lineages. Phenotypic evidence of mosaicism in a small percent of MCR carrying females and the presence of wild-type size y locus-derived PCR products in all tested y- F1 females suggests that females may all be mosaic to varying degrees. In summary, both genetic and molecular data demonstrate that the y-MCR element efficiently drives allelic conversion in somatic and germline lineages.
MCR technology should be applicable to different model systems and a broad array of situations including: enabling mutant F1-screens in pioneer organisms, accelerating genetic manipulations and genome engineering, providing a potent gene drive system for delivery of transgenes in disease vector or pest specie populations, and potentially serving as a disease-specific delivery system for gene therapy strategies. Although we provide an example in this study of an MCR element causing a viable insertional mutation within the coding region of a gene, by including two gRNAs in the MCR construct targeting separated sequences and appropriate flanking homology arms, one should also be able to efficiently generate viable deletions of coding or non-coding DNA. MCR using the simple core elements tested in this study is applicable to generating homozygous viable mutations, creating regulatory mutations of essential genes, or targeting other non-essential sequences. The method may also be adaptable to targeting essential genes if an in-frame recoded gRNA-resistant copy of the gene providing sufficient activity to support survival is included. In addition to these positive applications of MCR technology, we are also keenly aware of the substantial risks associated with this highly invasive method since the failure to take stringent precautions could lead to the unintentional release of MCR organisms into the environment. We therefore concur with others (6, 7) that a dialogue on this topic should become an immediate high priority issue. Perhaps, in analogy to the famous Asilomar meeting that assessed the risks of recombinant DNA technology, a similar conference could be convened to consider Biosafety measures and institutional policies appropriate for limiting the risk of engaging in MCR research while affording workable opportunities for positive applications of this concept.
Supplementary Material
Acknowledgements
We thank M. Yanofsky, W. McGinnis, S. Wasserman, R. Kolodner, H. Bellen and members of the Bier lab for helpful discussions and comments on the manuscript. We thank M. Harrison, K. O'Connor-Giles, J. Wildonger, and S. Bullock for providing CRISPR/Cas9 reagents and information. We also thank J. Vinetz and A. Lubar for granting us access to their BSL2 Insectary. These studies were supported by NIH Grants: # R01 GM067247, R56 NS029870 and by a generous gift from Drs. Sarah Sandell and Michael Marshall.
References
- 1.Zhang F, Wen Y, Guo X. Hum Mol Genet. 2014 Sep 15;23:R40. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Cell. 2014 Jun 5;157:1262. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Port F, Chen HM, Lee T, Bullock SL. Proc Natl Acad Sci U S A. 2014 Jul 22;111:E2967. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratz SJ, et al. Genetics. 2014 Apr;196:961. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett AR, Tibbit C, Ponting CP, Liu JL. Cell Rep. 2013 Jul 11;4:220. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oye KA, et al. Science. 2014 Aug 8;345:626. [Google Scholar]
- 7.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Elife. 2014 Jul 17;:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.