Abstract
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system and common cause of non-traumatic neurological disability in young adults. The likelihood for an individual to develop MS is strongly influenced by her or his ethnic background and family history of disease, suggesting that genetic susceptibility is a key determinant of risk. Over 100 loci have been firmly associated with susceptibility, whereas the main signal genome-wide maps to the class II region of the human leukocyte antigen (HLA) gene cluster and explains up to 10.5% of the genetic variance underlying risk. HLA-DRB1*15:01 has the strongest effect with an average odds ratio of 3.08. However, complex allelic hierarchical lineages, cis/trans haplotypic effects, and independent protective signals in the class I region of the locus have been described as well. Despite the remarkable molecular dissection of the HLA region in MS, further studies are needed to generate unifying models to account for the role of the MHC in disease pathogenesis. Driven by the discovery of combinatorial associations of Killer-cell Immunoglobulin-like Receptor (KIR) and HLA alleles with infectious, autoimmune diseases, transplantation outcome and pregnancy, multi-locus immunogenomic research is now thriving. Central to immunity and critically important for human health, KIR molecules and their HLA ligands are encoded by complex genetic systems with extraordinarily high levels of sequence and structural variation and complex expression patterns. However, studies to-date of KIR in MS have been few and limited to very low resolution genotyping. Application of modern sequencing methodologies coupled with state of the art bioinformatics and analytical approaches will permit us to fully appreciate the impact of HLA and KIR variation in MS.
Keywords: multiple sclerosis, genetics, human leukocyte antigen, killer-cell immunoglobulin-like receptor
1. Introduction
Multiple sclerosis (MS) is a chronic neurological disease associated with central nervous system (CNS) inflammation and neurodegeneration mediated by the adaptive and innate arms of an unregulated immune response [1, 2]. MS pathology is characterized by well-demarked inflammatory infiltrates, breakdown of myelin sheaths, microglia activation, proliferation of astrocytes and gliosis, and variable grades of axonal degeneration linked to oxidative stress and mitochondrial injury [3, 4]. Demyelinated lesions are disseminated through the CNS, involving both the white and gray matter. MS is a common cause of progressive neurological deficits in young adults, but disease expression is heterogeneous, varying from a mild illness to a rapidly evolving, incapacitating disease requiring profound lifestyle adjustments.
The individual and socioeconomic consequences of this debilitating and unpredictable disease are stunning. Fifteen years after diagnosis more than 80% of patients have functional and/or cognitive limitations, and approximately half require assistance to walk [5]. Twelve FDA-approved treatments for MS are now available, and several others are in late phases of development. However, the effects of these therapies on the long-term prognosis of the disease are largely unknown [6]. Furthermore, these therapies have diverse safety and toxicity profiles, and in effect no comparative data exist to guide when to initiate, change, or even how to select amongst the available options. Furthermore, no therapy exists for the progressive forms of MS, the subtypes most responsible for disability and debilitation [7].
2. MS epidemiology
In Europeans and their descendants, MS is the most common cause of non-traumatic neurological disability in young adults, affecting approximately 2.5 million people worldwide and more than 400,000 individuals in the US [8]. The prevalence of MS varies with geography and ethnicity. Indeed, with some notable exceptions, MS is more frequent in high latitude regions and northern European populations [9]. Notwithstanding difficulties in surveillance, MS is almost nonexistent in black Africans and native populations of the Americas and Oceania. Remarkably, the incidence of MS seems to have increased considerably over the last century, and this increase may have occurred primarily in women [10, 11] and in populations traditionally considered to be at low-risk, such as Hispanics, Asians, and African Americans. For example, early estimates suggested that the disease is significantly less prevalent in African Americans than in European Americans (relative risk of 0.64 [12]). In contrast, contemporary incidence studies are challenging the long-held belief that African Americans are at a reduced risk for developing MS [13, 14]. Remarkably, compared to individuals with predominately European ancestry, African Americans are more likely to have a more severe disease course, which at least in part appears to be genetically determined [15, 16].
3. MS genetics
Evidence for a genetic component in MS pathogenesis is found in the clustering of affected individuals in families, high disease concordance rate in monozygotic twins, and differences in disease prevalence among different ancestral groups [17–21]. However, a simple model of inheritance for all MS is unlikely since neither the recurrence rate nor the twin concordance supports the presence of a Mendelian trait. On the other hand, there is a broad consensus that the disease is multifactorial and the MS-prone genotype results from multiple independent or interacting polymorphic genes, with risk alleles common in the population, and each exerting a small or at most a moderate effect to the overall risk. Several epidemiologic risk factors have been consistently reported; these include vitamin D deficiency, exposure to the Epstein Barr virus (EBV) during childhood with manifestations of infectious mononucleosis, and cigarette smoking, among others [22, 23].
The polygenic model of MS genetics provided the rationale and drive for assembling large DNA datasets to pursue genome-wide association studies (GWAS), which have been highly successful in uncovering variants influencing susceptibility. Currently, a total of 110 polymorphisms in 103 discrete loci outside the major histocompatibility complex (MHC) have been firmly associated with susceptibility (Summarized in [24]). In aggregate, the proportion of the genetic variance accounting for disease risk explained by these polymorphisms is roughly 30%, but the mapping of additional risk variants is likely to proceed rapidly through ongoing multi-center initiatives utilizing dense, specialized arrays and very large sample collections. It is not inconceivable, however, that the potential for the discovery of additive risk variance extractable from large DNA screens will be quickly exhausted. Similar to other complex genetic diseases, multiple explanations for the missing heritability in MS have been proposed, including gene by gene and gene by environment interactions, cis/trans regulators of allelic expression, unidentified rare and penetrant semi-private variants, population and/or disease heterogeneity, neglecting the analysis of sex chromosomes, and hidden epigenetic effects. Regardless, genes encoding antigen-presenting molecules within the MHC region account for the largest component of the genetic risk for MS.
4. The human leukocyte antigen [HLA] region and MS
A search for “multiple sclerosis” and “HLA” in Pubmed reveals over two thousand entries. The HLA (Box 1) association with MS, which was first described several decades ago [25, 26], is consistent with the idea that MS is, at its core, an antigen-specific autoimmune disease. The association of the HLA locus with MS risk has been observed across all populations studied, and in both primary progressive and relapsing-remitting patients. The primary signal within the MHC maps to the HLA-DRB1 gene, or more specifically to the DRB1*15:01 allele, in the class II segment of this locus. Complex allelic hierarchical lineages, cis/trans haplotypic effects, and independent protective signals in the class I region of the locus have been described as well and are summarized below.
Box 1. The human Major Histocompatibility Complex.
Located on the short arm of chromosome 6p21, the human Major Histocompatibility Complex (MHC) is a remarkably gene-dense region with numerous immune response loci. These include the HLA genes, which were first recognized with respect to their critical role in histocompatibility, and are the major determinants of transplant outcome. Additionally, more than 100 infectious, autoimmune, inflammatory and pharmacological disease phenotypes and cancers are associated with HLA variation. The classical HLA class I and class II loci are the most polymorphic loci in the human genome and serve as a model for the study of genetic variation in human health and disease [27]. The extensive allelic diversity seen at these loci has been well documented [28, 29], with more than 10,000 alleles identified to date, and their critical role in disease predisposition and transplant outcome has long been recognized.
There are two major classes of HLA-encoding genes. The telomeric region contains the class I genes, whereas the centromere proximal region encodes HLA-class II genes. HLA class I and class II encoded molecules are cell surface glycoproteins whose primary role in an immune response is to display and present short antigenic peptide fragments to peptide/MHC-specific T cells. The classical HLA-class I molecules, HLA-A, -B, and -C, are found on most nucleated cells as heterodimers, and bind and present peptides primarily derived from endogenous synthesized proteins (e.g. viral and tumor peptides) to CD8+ T cells; HLA class I also serve as ligands for the killer immunoglobulin-like receptors (KIR) on the surface of natural killer cells (Box 3). These heterodimers consist of an HLA-encoded alpha chain associated with the chromosome 17-encoded monomorphic polypeptide, β2 microglobulin. The classical class II molecules, HLA-DR, -DQ, and DP consist of an alpha and beta chain, both HLA encoded, associated as heterodimers on the cell surface of antigen presenting cells such as B cells, dendritic cells, and macrophages. Class II molecules also serve as receptors for processed peptides; however these peptides are derived predominantly from membrane and extracellular proteins (e.g. bacterial peptides), and they are presented primarily to CD4+ T-lymphocytes. A third group of genes collectively known as class III, cluster between the class I and II regions and include genes coding for complement proteins, 21 -hydroxylase, tumor necrosis factor and heat shock proteins.
4.1. Association with DRB1*15:01∼DQA1*01:02∼DQB1*06:02. An historical perspective
The influence of HLA in MS susceptibility was first recognized prior to the molecular description of the class II molecules, and based on association with the serological specificity LD-7a determinant [25, 30–32], which was initially linked to HLA-A3 and -B7; with the latter giving the strongest association signal. Subsequently, it was demonstrated that these class I alleles were part of an extended class I and class II haplotype, and that the serological determinant was likely recognizing Dw2, whose serological specificity is now known as DR2 [33–35]. The continuous investigation of HLA polymorphism over the course of the last four decades, and each incremental technological advance that led to improved genotyping resolution, has provided further insights into the role of HLA in health and disease [35–38]. With the advent of molecular genotyping methods, the DR2 specificity was understood to encompass two distinct molecular allotypes, DR*15 and DR*16, and the association with MS was refined to DRB1*15 [39] and more specifically DRB1*15:01 [40]. As investigation into the genetic underpinnings of MS has continued, and genotyping methodologies have continued to evolve with ever-increasing power of resolution, the vast majority of results have remained strikingly consistent in implicating DRB1*15:01. A HuGE (Human Genome Epidemiology) review of 72 papers published from 1993–2004 found that in all but a very few studies, the frequency of DRB1*15:01 was greater in cases than controls, and that this was by a wide margin the most consistent finding [41]. The studies where DRB1*15:01 was not associated with MS were, in nearly every instance, conducted in non-European populations. More recently, GWAS have demonstrated that the main susceptibility signal maps to the HLA-DRB1 gene in the class II region of the MHC, and explains up to 10.5% of the genetic variance underlying risk [42]. HLA-DRB1*15:01 has the strongest effect with an average odds ratios (OR) of 3.08, and all additional DRB1 associations appear to account for less than 2% of the remaining variance.
Association of DRB1*15:01 in MS adheres to an additive model, with clear dose response to 0,1, or 2 copies of the risk allele [43, 44]. This follows a pattern similar to that observed in other autoimmune diseases. For example, in celiac disease, homozygosity for the DQB1*02 risk allele is associated with increased risk and severity of disease [45]. Similar evidence for dose effect of HLA has been reported for narcolepsy [46] and rheumatoid arthritis [47]. A comparable dose effect is observed in type I diabetes (T1D), where the primary risk genotypes are those with two copies of the predisposing variants DR3 or DR4 [48]. In addition to the increased risk for disease observed for DRB1*15:01 homozygous genotypes, there appears to be an epistatic effect for the DRB1*15:01/*08:01 heterozygous genotype, where increased risk relative to other heterozygous *15:01 genotypes has been observed [49]. DRB1*08:01 does not appear to be predisposing on its own, but rather enhances the effect of DRB1*15:01 when the two alleles are present in the same genotype. However, an independent association for DRB1*08:01 has been observed in an Ashkenazi cohort when patients were subdivided into clinical subgroups, with a weak but significant association reported for primary progressive patients only [50]. Finally, DRB1*15:01 has frequently been associated with markers of disease severity. MS patients carrying the DRB1*15:01 haplotype are more likely to be female, and have earlier age of onset [51, 52]. Further, this haplotype is associated with presence of oligoclonal bands and IgG levels in the cerebrospinal fluid of MS patients. In contrast, there have been no consistent findings for the other MS associated DRB1 alleles with respect to disease severity or progression [43, 53].
4.2. DRB1*15:01 haplotypic associations
DRB1*15:01 is most often found in European populations as part of an extended haplotype including DQA1*01:02 and DQB1*06:02, and thus it is difficult to distinguish the primary predisposing locus or allele. Extensive work in fine-mapping the HLA contribution to MS susceptibility localized the primary risk to DRB1 in Europeans [54] when classical HLA alleles were imputed from SNP data. In that study, all effect attributed to DQB1*06:02 could be explained by association with DRB1*15:01. However, an earlier, smaller study aimed at fine-mapping the HLA association in Sardinian MS patients assigned risk to both DRB1 and DQB1 [55]. Aside mapping via SNPs, various strategies can be employed to isolate the primary risk locus, including admixture mapping and examination of risk in non-DRB1*15:01 families and individuals. The finding that heterozygosity for DRB1*15:01 with *08:01 increases risk has also been interpreted as evidence of a role for the DQ heterodimer [49]; the rationale being that because the DRA molecule [in the DRα∼DRβ antigen presenting heterodimer] is essentially invariant, any trans effect of the heterozygous haplotype would most likely be mediated by DQA∼DQB interactions, where both the alpha and beta chains are relatively polymorphic. This explanation is in keeping with that proposed for the observed increased risk for DR3/4 heterozygotes in T1D [56, 57]. In celiac disease, where the primacy of the DQ molecule in conferring risk is more firmly established, a similar role for DQA/DQB associations with risk in trans has also been suggested [58].
Because different populations exhibit varying patterns of linkage disequilibrium (LD) in the HLA region, cross-population analysis can be especially informative in teasing out the causative locus from a multilocus association. This approach has been particularly successful in narcolepsy where the initial observation of HLA association with disease was identical to that for MS: the extended DRB1*15:01∼DQA1*01:02∼DQB1*06:02 haplotype. Subsequently, investigation in an African American narcolepsy dataset revealed that the association was entirely attributable to DQB1*06:02, because lower levels of LD between DRB1 and DQB1 in this population enabled examination of DQB1*06:02 haplotypes with DRB1 alleles other than *15:01 [59]. Conversely, examination of MS in African American individuals showed risk to be firmly attributed to DRB1 [60], either *15:01 or *15:03. In these individuals, examination of alternate DQB1*06:02 haplotypes (without DRB1*15:01) revealed no difference between cases and controls, ruling out DQB1*06:02 as the causative allele of the association signal. This conclusion echoed a finding in a cohort from Martinique, where there is a corresponding influence of African ancestry [61]. Confounding interpretation of these results studies in Sardinians [55], Afro Brazilians [35], and Canadian families [62] suggest a possible secondary role DQ variation in MS susceptibility, underscoring the difficulty in definitively assigning risk to one locus of MHC multilocus haplotypes in extremely strong LD.
4.3. Additional DRB1 risk alleles
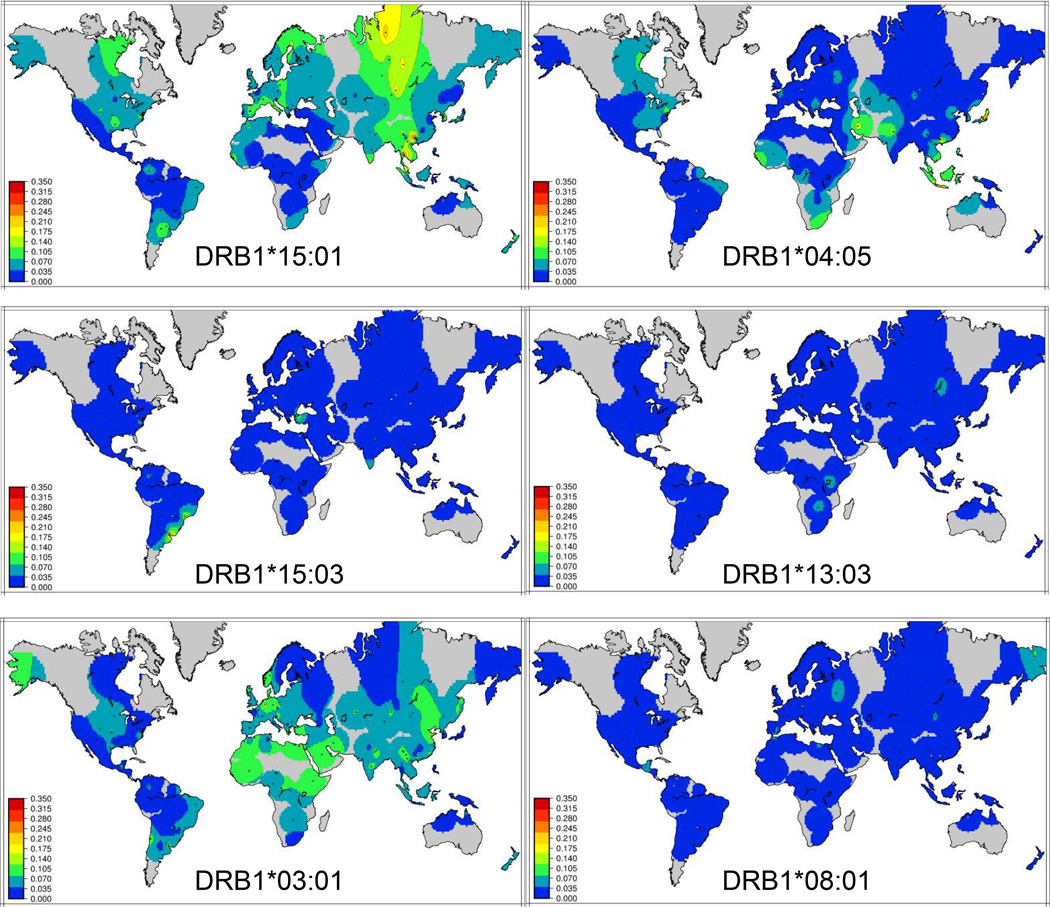
The vast majority of studies investigating the relationship between HLA polymorphism and MS have focused on individuals of European descent; this can be explained by the fact that most MS is observed in these individuals. The incidence of MS in other worldwide populations is significantly lower, and thus well-powered cohorts are few and far between. DRB1*15:01 is found at relatively high frequencies (>10%) in European and Asian populations, but is relatively rare elsewhere (Figure 1). Most studies without a significant association in MS for DRB1*15:01 have been conducted with non-European populations. Investigations in these populations have yielded important clues to the details underlying HLA association in MS. For example, in Japanese individuals, an association of DRB1*15 was originally recognized as predisposing to “Western type” or “conventional” MS, while the clinically distinct opticospinal or “Asian type” showed no association with that allele. Later, association of DRB1*04:05 with “Asian “ MS was recognized as a risk variant for MS, but with a clinically distinct disease course characterized by earlier age of onset, reduced severity [63] and a lack of brain lesions [64]. DRB1*04:05 has also been found to be associated with MS in Sardinia, where the incidence of MS is quite high [65, 66], refining an early observation of an association with DR4. Finally, DRB1*04:05 appears to confer risk for MS in African Americans [67] as well as Sicilians [66]. This allele is relatively common in both the Japanese and Sardinian populations, but much less so in other Europeans. Thus, limited statistical power may explain a lack of clear association of DRB1*04:05 in other populations.
Figure 1.
Global frequency distributions of MS predisposing alleles of HLA-DRB1
Additional DRB1 risk alleles that have been reported consistently include *03:01 and *13:03. DRB1*03:01, the primary risk allele in T1D [48], appears to have a recessive effect in MS [68]. The association of DRB1*03:01 was first observed in a cohort from Sardinia [69], but this allele is very common across Europe, Africa and Asia (Figure 1). Additional associations with DRB1*03:01 may be reflected in some reports of association with DRB1*17. These reports include an apparent nomenclature error, as it implies that there is a first field HLA-DRB1 numbered 17 (there is not); rather, it likely refers to alleles with serological specificity DR17, which include DRB1*03:01. GWAS studies combining several European populations confirmed this association through imputation of classical HLA alleles [54, 70], although the effect size was relatively small (OR=1.26). Owing to the likely recessive effect of this allele, it is inconsistently associated with disease across studies, despite its generally high frequency in many populations.
In contrast to DRB1*03:01, DRB1*13:03 is seldom observed at population frequencies greater than 3% worldwide (Figure 1), but the association with MS appears to be more robust with a stronger effect size. The association of DRB1*13:03 with MS was first identified through analysis of an Ashkenazi and non-Ashkenazi Jewish cohort from Israel using traditional HLA genotyping methods [50]. A study of Israeli families with MS and with low frequencies of DRB1*15:01 also provided evidence for association of DRB1*13:03 with disease, with a trend toward transmission disequilibrium [71]. Lincoln et al. [62] observed over-transmission of the DRB1*13∼DQA1*05:01∼DQB1*03:01 haplotype in Canadian multiplex MS families. While this study did not examine the DRB1 locus at high resolution, this haplotype is nearly always associated with the DRB1*13:03 allele in Europeans, while the other common DRB1*13 allele, *13:01 and *13:02 are generally found on other DQA1∼DQB1 haplotypes; these alternate DR*13 haplotypes were not found to be over-transmitted in the Canadian study. DRB1*13:03 is also associated with MS in the Sardinian population [72]. Examination of larger cohorts using statistical imputation of classical HLA alleles from SNPs has confirmed these associations [54, 70] in Europeans, despite the fact that this allele is generally seen at very low frequency in European populations. The most likely explanation for the inconsistent identification of DRB1*13:03 in MS risk is low power due to the relative rarity of this allele in most populations. Where it is seen at higher frequencies, such as in the Israeli population, or in studies with thousands of individuals, DRB1*13:03 is revealed clearly as a risk allele.
4.4. Risk vs. non-risk alleles: amino acid substitution patterns and functional considerations
While DRB1*15:01 is associated [although much more weakly] with MS in African Americans, the predominant DR2 allele in this population is *15:03. Importantly, this DR*15 subtype is also associated with MS [60]. Association of DRB1*15:03 with MS has also been observed in cohorts from Iran [73], Brazil [74] and Martinique [61]. The clear association of both DRB1*15:01 and *15:03 suggests that risk can be attributed to variation common within the DR*15 allele group, which is distinguished from other HLA alleles by Alanine at position 71. This residue imparts a distinctive characteristic on the P4 pocket of the HLA-DRβ1 molecules, as its smaller side renders the pocket larger and more hydrophobic, resulting in a unique peptide binding profile [75]. DRB1*15:01 and *15:03 differ only at amino acid position 30 and have an identical peptide binding motif, and both have the capacity to present the 85–89 immunodominant peptide of myelin basic protein (MBP) [61]. In contrast, DRB1*15:02 is found at high frequencies primarily in Southeast Asia and Oceania, but is also frequently observed in other populations at low frequency and has not been found to be associated with MS. This allele differs from the predisposing *15:01 allele by a single amino acid substitution, Val -> Gly at position 86. This mirrors findings in several other autoimmune diseases, where risk vs. neutral or protective alleles have been found to differ only with respect to this valine-glycine dimorphism at position 86. For example, in Juvenile Idiopathic Arthritis (JIA), the common European allele DRB1*11:01 is present on the same DQA1∼DQB1 haplotype as the highly predisposing DRB1*11:03 allele, yet it is neutral with respect to disease [76]; likewise DRB1*13:02 is not associated with disease in JIA, while *13:01 is a clear risk allele. In each of these cases, the predisposing DRB1 allele differs from the related neutral allele only with respect to the valine at position 86. Valine at position 86 has been shown to impact peptide binding and dimer stability [77], and thus its presence may be determinative of T cell response to bound antigen. However, while another MS risk allele, DRB1*03:01 also has valine at position 86, DRB1*13:03 does not, suggesting that this variant in the P1 pocket of the peptide binding groove is neither necessary nor sufficient for development of MS. Further, differential susceptibility between DRB1*15:02 and the risk alleles *15:01 and *15:03 may not be related to ability to present MBP; Finn et al. [78] demonstrated that several myelin peptides previously shown to bind to *15:01 were capable of presentation by *15:02 and provoking strong T-cell responses. Notably, aside from the closely related DR*15 alleles, each predisposing DRB1 allele has been identified as part of a different DR supertype [79, 80], which have divergent profiles for bound peptide, and distinctly different sets of residues at key peptide contact sites (Table 1), challenging the notion of a single autoantigen in HLA-mediated susceptibility to MS.
Table 1. Amino acid residues at peptide binding contact sites of DRB1.
Residues for the primary predisposing allele, DRB1*15:01, are shown at top. Difference at sites between DRB1*15:01 and other predisposing DRB1 alleles are shown below. At bottom, the corresponding pocket of the peptide-binding groove is given
| Position | 9 | 11 | 13 | 26 | 28 | 30 | 37 | 47 | 57 | 61 | 67 | 70 | 71 | 74 | 78 | 85 | 86 | 89 | 90 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRB1*15:01 | W | P | R | F | D | Y | S | F | D | W | I | Q | A | A | Y | V | V | F | T |
| DRB1*15:03 | - | - | - | - | - | H | - | - | - | - | - | - | - | - | - | - | - | - | - |
| DRB1*04:05 | E | V | H | - | - | Y | Y | Y | S | - | L | - | R | - | - | - | - | - | - |
| DRB1*13:03 | E | S | S | - | - | Y | Y | Y | S | - | I | D | K | - | - | - | - | - | - |
| DRB1*08:01 | E | S | G | - | - | Y | Y | Y | S | - | F | D | R | L | - | - | - | - | - |
| DRB1*03:01 | E | S | S | Y | - | Y | N | F | D | - | L | - | K | R | - | - | - | - | - |
| 9 | 6 | 4 | 4 | 4/7 | 6 | 9 | 7 | 9 | 7 | 7 | 4/7 | 4 | 4 | 4 | 1 | 1 | 1 | 1 |
4.5. Summary of DRB1 risk associations
Recognized for over forty years, the strong and consistent association of HLA-DRB1*15:01∼DQA1*01:02∼DQB1*06:02 remains the quintessential predisposing association in MS. The observation that this HLA haplotype also likely influences disease severity, whether assessed indirectly through age of onset of through clinical or radiological findings, suggests that it plays a specific, singular role in disease pathogenesis, distinct from other associated HLA alleles and haplotypes. It is possible that this role is shared by DRB1*13:03, where the association with disease, when observed, has a nearly equivalent effect size to the DRB1*15:01 haplotype. However, given the rarity of this allele in European populations, efforts to examine its role in disease severity are hampered by lack of power. In contrast, the remaining associated HLA alleles may contribute to disease risk through alternative mechanisms. The observation in Asian populations that DRB1*04:05 is associated with a distinct clinical phenotype supports this contention. Importantly, each of these alleles (DRB1*15:01, *15:03, *13:03, and *04:05) appears to act in a dominant fashion and have relatively strong and equivalent effect sizes (Table 2). That alleles other than DRB1*15:01 have been inconsistently reported as predisposing in MS may be attributed to their relative rarity in European populations (Figure 1), in whom the majority of MS cases are reported and studies conducted. In contrast, the other DRB1 alleles that have consistent findings of risk in MS, DRB1*03:01 and *08:01, are relatively common in European populations (Figure 1), contribute to risk through recessive modes or interaction effects, and do not appear to impact disease severity, again suggesting a dissimilar role in disease.
Table 2.
Summary of HLA Class II risk alleles in MS
| Risk alleles | Mode | Frequency in European populations |
|---|---|---|
| DRB1*15:01 | Dominant/Dose effect | Common |
| DRB1*15:03 | Dominant | Rare |
| DRB1*04:05 | Dominant | Rare |
| DRB1*13:03 | Dominant | Rare |
| DRB1*08:01 | Interaction only with *15:01 | Rare |
| DRB1*03:01 | Recessive | Common |
4.6. Other DRB genes in MS
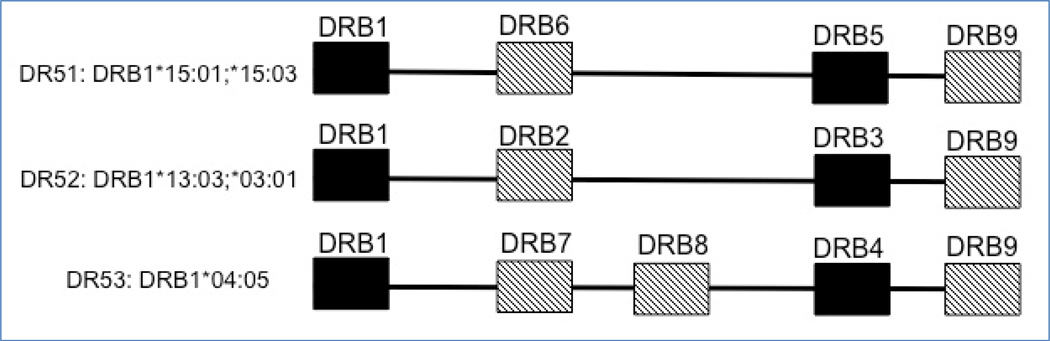
Beside DRB1, an individual may bear other functional DRB genes, which are present in a haplotypic-specific manner, with several present on MS associated DR haplotypes (Figure 2). Additional genetic interactions attributable to these other DRB genes have been identified, with perhaps the most interesting being related to HLA-DRB5 [81], a class II gene immediately telomeric of HLA-DRB1 and found exclusively in DRB1*15 and DR*16 haplotypes, which form the DR51 haplotype lineage [82]. HLA-DRB5/DRA heterodimers appear to be effective myelin antigen-presenting molecules, and elegant experiments using triple DRB1-DRB5-hTCR transgenic mice support functional epistasis between HLA-DRB1 and HLA-DRB5 genes whereby DRB5 modifies the T cell response driven by DRB1 through activation-induced cell death, resulting in a milder and relapsing form of autoimmune demyelinating experimental disease [83]. Similarly, in African Americans with MS, carriers of the HLA-DRB1*15:01 with HLA-DRB5*null haplotypes have a more severe disease course than HLA-DRB5 wild types [81]. Although based on a small number of individuals with the rare DRB5*null mutation, the convergence of findings obtained from HLA humanized EAE mice with human MS genetic data supports a modulatory role of HLA-DRB5 gene products on the progression of autoimmune demyelinating disease. Finally, the functional DRB4 gene is present on the DR53 haplotype with DRB1*04:05, while both DRB1*03:01 and *13:03 reside on the DR52 haplotype, which also bears the functional and polymorphic DRB3 locus, and thus consideration of the impact of these additional DRB genes might be considered as plausible contributors to disease risk.
Figure 2.
Extended DR haplotype structures for MS predisposing DRB1 alleles, showing additional functional DRB gene [black]. DRB pseudogenes may also be present [hatched]
4.7. HLA-DPB1
While risk for MS in the HLA class II region is generally attributed to alleles of DR∼DQ, an independent association with DPB1 has been observed in the course of GWAS in several cohorts [54, 70, 84]. Patsopoulos et al. [54] mapped the risk SNP to DPB1*03:01, although this allele alone was insufficient to explain association with the SNP; it is possible that other DPB1 alleles in LD with the risk SNP are associated with disease, but because only one or a few DPB1 alleles are generally present at any appreciable frequency in a population, there may be insufficient power to detect associations with additional alleles. An earlier study examining DPB1 variation using traditional HLA genotyping methods in Australian DQB1*06:02-negative MS patients and controls also implicated DPB1*03:01 [85]. Likewise in Japanese [86], and Sardinians [87]. Another allele, DPB1*05:01, has been found to be associated with MS in some Asian populations [86, 88]; however, this allele appears to be specifically associated with anti-AQP4 positive NMO in Japanese patients [89].
4.8. Protection from MS mediated by HLA class II
The class II allele most consistently associated with protection from MS in European populations is DRB1*14:01 [44, 49, 90]. Protection mediated by DRB1*14 appears to be dominant, abrogating susceptibility attributed to DRB1*15:01 [44]. Additional protective class II alleles have been identified in smaller studies. In a Brazilian [91] and Canadian [92] cohorts, DRB1*11 was also found to be protective. Protection mediated by DRB1*13∼DQB1*06:03 has also been observed in a Finnish cohort [93] and Canadian families with MS [62]. Although these studies did not include high-resolution genotyping for DRB1, the DRB1*13 allele most often found on this haplotype in Europeans is *13:01. As noted above, these alleles have a valine occupying position 86, further undermining the notion that this residue is predisposing to MS. While DRB1*13:01 differs from *13:02 only at position 86, *13:03, which confers risk in MS, is significantly more divergent, with several additional variants relative to *13:01. Whether similar high-resolution variation is present with respect to protection mediated by DRB1*11 is not known, as studies to-date have been conducted at low resolution. While the mechanism for class II-mediated protection in MS is not known, engagement of MHC-promiscuous, auto-reactive thymocytes, and resultant Treg formation, has been suggested as an explanation for these associations [94].
4.9. HLA class I associations
Several HLA class I alleles have been implicated in MS. Increased risk due to HLA-A3 was first observed in 1972 [30, 32] although many subsequent studies determined that this association was due to LD with DRB1*15:01∼DQB1*06:02. However, a possible epistatic interaction between DRB1*15:01 and HLA-A*03:01 was identified in a Norwegian study [95], and it is also possible that the presence of A*03:01 may be marking a more refined DRB1*15:01 susceptibility haplotype. Fogdell Hahn et al. [96] reported that HLA-A*03:01 was predisposing in MS, and that this effect was independent of DRB1*15:01∼DQB1*06:02. However, analysis of haplotype transmission in Canadian families suggested that any effect attributable to A*03:01 is secondary to LD with the class II. Likewise, HLA-A3 is in strong LD with HLA-B7, and thus it is likely that associations observed for HLA-B*07 are due to LD with the extended DRB1*15:01∼DQB1*06:02 haplotype [97].
In contrast, the protective effect of HLA-A*02:01 has been observed in multiple independent studies and does not appear to be secondary to class II associations. Analysis of SNPs in the class I region identified protection mediated by HLA-A*02:01 as the only class I effect after controlling for all class II associations [54, 70]. Examination of cohorts from Sweden [96, 98], Norway [95] and Italy [99] using traditional HLA genotyping methods demonstrated a similar protective role for A*02:01.
Additional roles for class I molecules in MS susceptibility have been observed for HLA-B and HLA-C. These two loci are physically close within the class I region and in extremely tight LD. HLA*B44 appears to afford protection in MS [100]. A higher resolution genotype, B*44:02, was inferred through imputation from SNPs and was also identified as protective [101]. However, in the latter finding, HLA-B*44:02 was observed to be in LD with HLA-C*05, which has independently been shown to confer protection in MS in the absence of DRB1 risk alleles [102], and thus it is difficult to distinguish whether these results mirror a single effect through strong LD. Bergamaschi et al. [99] also observed association with HLA-C*05, in the absence of the DRB1*03:01 risk allele. Further associations are observed when class I alleles are analyzed with respect to their role as ligand for killer immunoglobulin-like [KIR] receptors, discussed in detail below.
It is important to note that many of the HLA associations identified in MS, aside from the canonical DRB1*15:01 haplotype, have been detected through imputation of HLA alleles from SNP data (Box 2), rather than direct assay of HLA sequence. While statistical imputation of HLA alleles is dependent on patterns of LD across the MHC, these patterns and the strength of the association can vary by locus or allele, particularly on specific haplotypes. In particular, due to the extensive structural variation around DRB1, where the additional DRB loci (DRB3, DRB4 and DRB5) are present to varying degrees depending on the specific haplotype, imputation accuracy may suffer. An examination of HLA imputation concordance with sequence-based genotyping results in Parkinson’s disease found accuracy to be poorest for DRB1 [103]. In a Finnish study making a similar comparison, DRB1 imputation accuracy was reported to be less than 30% [104]. In non-European populations accuracy for DRB1 has also been found to be limited, and likewise HLA-B [105, 106]. In particular, alleles of DRB1*08 are particularly difficult to impute correctly, likely due to structural variation and the concomitant loss of SNPs on that haplotype. Finally, in many cases high confidence in imputation results is restricted to the first field of resolution for a given HLA allele; consequently, important variation with respect to disease may not be revealed at this lower resolution.
Box 2. Imputation of HLA from SNP data.
Imputation is a statistical process that infers genotypes that have not been observed experimentally, by exploiting known patterns of LD between genetic markers [107, 108]. Several methods for imputation of HLA alleles from SNP data have been developed [109–111], and each is reliant on a set or sets of “training” data to establish the relationship between SNPs on common GWAS platforms and classically genotyped HLA alleles. After the imputation algorithm has been trained, it is applied to SNP genotype data and the likely HLA genotypes are for each individual are inferred, and associated likelihood or confidence estimates are generally reported.
Additionally, while results from SNP-based methods may appear to fine-map risk variants to specific amino acid residues, due to the nature and extent of LD in the MHC region coupled with the extensive history of gene conversion and recombination event that characterize the relationships between alleles [112], any superficial association with specific residues oftentimes is simply a recapitulation of allelic associations identified through standard genotyping methods. For example, a specific variant may appear to be more closely associated with disease than other amino acid residues, but that may be attributed to the fact that it specifically marks the predisposing allele. There will often not be sufficient power to detect an association at other functional residues because of extensive sharing with other, non-associated alleles. For example, while fine-mapping SNP associations in DRB1, the effect attributable to DRB1*15:01 remained even after sequentially controlling for each of the most significant individual amino acid associations [54]. Given that the allele is the functional unit under observation, it is not surprising that there is limited success in parsing these associations to individual or even small groups of amino acids.
5. HLA associations in other neurological diseases
By far the most well characterized association of HLA variation with neurological disease has been in MS, however there are important parallels observed in other neurological disease that may indicate some common pathogenic mechanisms or pathways. While other neurological diseases have been explored less fully for association with HLA, there exists strong evidence that variation in the region contributes to disease risk in particular disease, with some overlap with HLA associations MS. HLA has been implicated in NMO, where several studies in Asians reported HLA-DRB1*16:02/DPB1*05:01 and HLA-DRB1*09:01 as risk and protective alleles respectively [32, 44, 56, 113, 114]. Interestingly, DRB1*16:02 is closely related to *15:01 and part of the DR2 serogroup. Studies of NMO in other populations, including admixed Brazilians and French Afro-Caribbean, reported a higher frequency of HLA-DRB1*03 [115], another allele identified in MS. However, all studies have been based on relatively small datasets. Likewise in myasthenia gravis [MG], the ancestral DRB1*03:01- bearing haplotype, A1-B8-DR3, has shown the most consistent association with the early onset form of the disease. However, in a recent SNP-based analysis of the extended MHC locus, there was a paucity of significant signals in the class II region; the strongest imputed associated HLA allele was HLA-C*07:01. The strongest association in a recently reported early onset MG GWAS was observed also in the HLA class I region [116]. The role of HLA genes in Parkinson’s disease [PD], first reported in the late 70’s [117], was confirmed in recently reported GWASs [118–120]. SNP-imputed HLA analysis recorded positive associations with the HLA-B*07:02, DRB1*15:01, DQB1*06:02 haplotype, mirroring the association in MS; risk was inversely correlated with DRB1*04:04 [103]. A recent GWAS meta-analysis including 13,708 cases and 95,282 controls identified secondary independent risk variants within the MHC [121]. Finally, reports of HLA association in schizophrenia date over four decades [122, 123] and re-emerged with recent GWAS [124–127]. Some studies showed higher frequencies for HLA-DRB1*01:01 and DRB1*13 in schizophrenic patients. In one study using SNP imputation, the most significant finding was with HLA-C*01:02, whereas DRB1*03:01 and HLA-B*08:01 were protective [128]. Most significantly with respect to MS, an observation of genetic pleiotropy between these two diseases has been observed, primarily driven by association within the MHC [129].
6. New horizons in MS immunogenetics
6.1. Killer-immunoglobulin-like receptors (KIR) in MS
Killer-immunoglobulin-like receptors (KIR) are highly polymorphic receptors expressed on natural killer (NK) cells (Box 3). While clear association with variation in HLA has been established in MS, there has been limited examination of the role of NK cells or their receptors, including KIR. However, because HLA class I molecules serve as the primary ligand for many KIR, it is likely that the association signals observed for many diseases is related to KIR function. In MS, alleles of HLA-A, -B and –C that are known to serve as ligands for KIR have been implicated in disease. KIR may also play a role in other neurological disease such as myasthenia gravis [101, 116] and schizophrenia [128], where there is also evidence for class I involvement at HLA-B and –C. Given that KIR may also be expressed by CD4 T-cells [130] it is conceivable that KIR diversity can influence specific antibody production and thus also explain some class II associations in MS. Importantly, both underlying immunoregulatory dysfunction and inflammatory processes have been proposed for MS [131]. This view is clearly supported by the observation of disease association with HLA variation.
Box 3. Killer immunoglobulin-like receptors (KIR).
KIR are receptors expressed on natural killer (NK) cells, where they regulate cell killing and cytokine response [132]. NK mediated immunity is the first line of defense against viruses and tumors, and the balance between inhibitory and activating receptors and their HLA ligands mediates NK responsiveness [133, 134]. Located on human chromosome 19q13.4, the KIR gene complex displays considerable heterogeneity in gene content both within and between populations. KIR haplotypes contain from 4–14 genes and, based on this genomic structure, are categorized into two groups, termed A and B [135]; the A haplotype is represented by a single configuration of seven genes that express mainly inhibitory KIR, and all other configurations are termed B haplotypes (Figure 3). Despite the large number of unique haplotypes characterized, a few relatively common haplotypes often account for greater than 90% of the KIR haplotypic variation observed within a specific population, and are observed across major ethnic groups [136, 137].
Specific KIR molecules recognize one or more of four epitopes of HLA class I molecules (Figure 4). KIR3DL2KIR2DS2 and KIR2DS4 recognize a subset of HLA-A allotypes carrying the A3/11 epitope [e.g. A*11:01]. KIR3DL1/S1 binds subsets of HLA-A and -B allotypes that carry the Bw4 serological epitope, defined by amino acid positions 77–83 [e.g. B*44:02 and A*24:02]. Finally, the inhibitory KIR2DL1 and KIR2DL2/3 and the activating KIR2DS1KIR2DS2 and KIR2DS4 interact differentially with the mutually exclusive C1 or C2 epitopes carried by all HLA-C allotypes, or the small subset of HLA-B molecules that carry the C1 epitope [134, 138, 139]. However, receptor ligand interactions do not adhere strictly to locus-level associations. Each of the four ligand-receptor interactions is diversified by allelic polymorphism of KIR and HLA class I, and by the sequence of the bound peptide [140–143].
Many of the HLA class I associations outlined above have been identified in the context of GWAS, where the strongest association signal consistently maps to the MHC. However, a direct association of disease risk with KIR polymorphism has not been detectable via GWAS, in part due to the paucity of markers in the KIR region on all common GWAS platforms. A lack of suitable reference alignments has historically precluded inclusion of KIR specific SNPs on GWAS platforms, and the extensive variation in gene-content in KIR haplotypes is typically incompatible with standard quality thresholds. Even the Immunochip [144], which is specifically enriched for markers in the KIR chromosomal region, primarily identifies noncoding variants on the common A [inhibitory] haplotype [144]. Thus, most KIR associations reported to-date in immune-mediated diseases [145], including MS [146, 147], have been identified through focused methods that only assess KIR gene-content polymorphism. Genotyping methods that assay gene-content are generally unable to distinguish copy number rather than simple presence/absence. Because copy-number has direct consequence for the immune response [148], reliance on these results may contribute to a loss of power to detect locus-level associations. Further, strong linkage disequilibrium within gene-content haplotypes [149, 150] makes it difficult to distinguish the causal locus.
Several studies of KIR gene-content have in fact suggested a role for KIR loci in MS susceptibility [Table 3]. Absence of the inhibitory KIR2DL3 [151] was shown to be predisposing in MS, implicating either KIR2DL2 [which segregates as the alternate allele of the same locus] or the closely linked KIR2DS2 in disease. In another study, the presence of KIR2DL5 and KIR3DS1 were found to be elevated in MS patients compared to controls [152], while two other studies identified a different telomeric locus, KIR2DS1, as protective [153, 154]. While these results are intriguing, a comprehensive analysis of KIR allele-level variation is required in order to more fully understand the role of these important immune receptors in human disease.
Table 3.
Summary of studies to-date implicating KIR in MS
| KIR allele or locus | Effect | Cases [N] | Reference |
|---|---|---|---|
| KIR2DL3- absence | Predisposing | 321 | Jelcic et al. 2012 |
| KIR2DL5/ KIR3DS1 | Predisposing | 200 | Garcia-Leon et al. 2011 |
| KIR2DS1 | Protective | 121 | Fusco et al. 2010 |
| KIR2DS1 | Protective | 443 | Bettencourt et al. 2014 |
| Bw4 [3DL1 ligand] | Protective | 631 | Lorentzen et al. 2009 |
Further evidence for a role for KIR is provided by class I association with MS [Table 4]. When HLA class I alleles are analyzed with respect to their role as KIR ligands, important associations emerge. In a Norwegian cohort, no association between KIR carrier frequency and MS was observed [146]. However, analysis of HLA class I revealed that when HLA-B alleles were grouped according to KIR ligand status, the Bw4 motif was observed to be protective. Given that in European populations, including the Norwegian sample, the KIR3DL1 carrier frequencies are generally >95%, it is not surprising that only the ligand reveals a statistically significant association. Likewise, the B*4402 association with protection may be viewed in the context of its role as a Bw4+ KIR3DL1 ligand. Finally, the HLA-C*05 association reported by Yeo et al. [102] may be explained by its role as a C2 group ligand for KIR2DL1. While no association in that study was observed when alleles were grouped according to C1 and C2 status, there is ample evidence to suggest that HLA class I alleles, when characterized at high resolution, differ with respect to the nature and intensity of KIR binding and resulting NK cell activation or inhibition [141], suggesting that specific class I alleles may exert a strong impact on NK cell activation, and thus play a role in disease pathogenesis. However, as noted above, HLA-B*44:02 and –C*05 are in LD in European populations, and thus these results may be reflective of a single association. Finally, although it is likely that the frequently observed association of HLA-A*03 with MS is secondary to the class II effect, the presence of this ligand for KIR3DL2 on the extended DRB1*15:01∼DQB1*06:02 predisposing haplotype may have important ramifications for NK cell reactivity in MS.
Table 4.
HLA class I associations in MS and relevant KIR interaction potential
| HLA class I association with MS | KIR ligand | KIR receptor |
|---|---|---|
| HLA-B*44:02 | Bw4 | KIR3DL1S1 |
| HLA-B*37:01 | Bw4 | KIR3DL1S1 |
| HLA-B*38:01 | Bw4 | KIR3DL1S1 |
| HLA-C*07 | C1 | KIR2DL2/3 |
| HLA-C*05 | C2 | KIR2DL1; KIR2DS1 |
| HLA-A*0301 | A3 | KIR3DL2 |
Substantial data implicate NK cells in MS pathogenesis or protection, bolstering the notion that KIR polymorphism may be important in disease susceptibility and/or progression. While the precise role of innate immunity in the pathogenesis of MS is unclear, several lines of evidence indicate that NK cells contribute to disease susceptibility or progression, either indirectly via immunoregulatory activity or directly through cytotoxicity of self-tissues. Studies in EAE suggest a role for NK cells in down-regulation of disease progression [155–157]. At the same time, increased susceptibility and disease severity in EAE has been associated with NK cells in conjunction with particular cytokines [158, 159]. In humans, an immunoregulatory role for NK cells in MS has been observed, resulting in a diminishment of the inflammatory process [160, 161]. In contrast, in vitro studies show that NK cells can directly lyse neural tissue, and may therefore directly contribute to the tissue injury in MS [162, 163], while at the same time reduced capacity for killing may also play a role in disease [164]. It is likely that many seemingly conflicting results thus far may be explained by variation in control of NK cell activation, likely mediated by KIR. Taken together, these data indicate a role for NK cells and KIR in MS and neuroinflammation, as has been proposed for numerous autoimmune and other immune-mediated diseases.
6.2. Examination of non-coding variation in MS
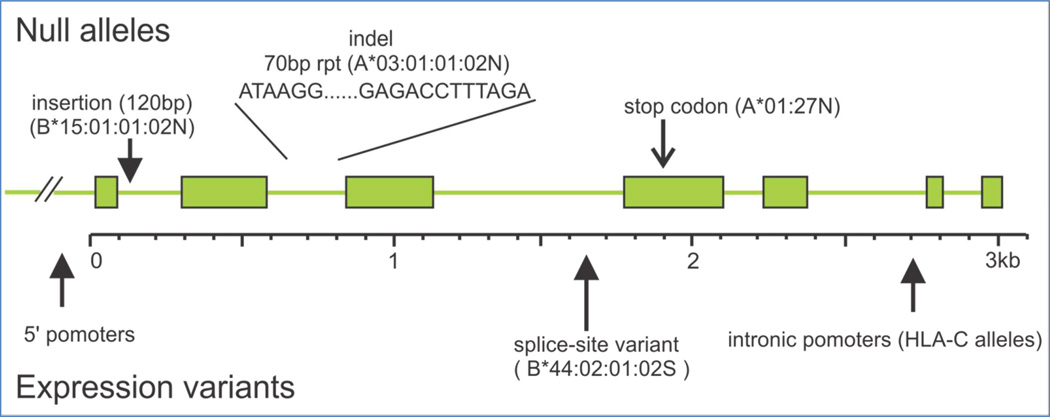
Further investigation into the finer details of HLA variation in MS is promised with the advent of next-generation sequencing (NGS) for the HLA loci. Numerous methods have been developed in the last several years, many of which produce full length genomic sequence, inclusive of all exons, 5’ and 3’ untranslated regions (UTR) as well as intronic sequence [165]. At present, nearly all recognized HLA alleles have been sequenced only through exons 2 (class I and class II) and 3 (class I). While the impact of polymorphism outside of these exons is not fully understood, evidence suggests that additional coding and noncoding variation may be important in the immune response. Many sequence variants have been identified that impact HLA expression or interaction with accessory molecules. One need only consider the many null or expression variants of common alleles to appreciate the potential influence this variation has on immune function (Figure 4). For example, a variant of the MS associated allele HLA-B*44:02 has been described that produces only a soluble, rather than cell surface, molecule; a point mutation at the end of intron 4 alters the exon 5 splice site [166]. However, this polymorphism is not detectable through standard genotyping methods, and thus these alleles are generically genotyped as B*44:02; likewise, the actual population-level frequency of the alternative allele is not known. If in fact the non- surface expressed variant of this allele is common, this could explain the relationship to disease. Similarly, alternative splice sites for DQA1 have been identified through sequence analysis of the 3’ UTR [167]. Likewise, a SNP upstream of HLA-C that was found to be associated with control of HIV infection and cell surface expression of the HLA molecule was shown to be in LD with a 3’ UTR polymorphism that regulates binding of micro RNA, the putative source of the expression variation [168]. This groundbreaking study confirmed that additional polymorphism in the HLA UTRs may play a critical role in disease. Notably, the LD observed between the SNP originally associated with this effect and the 3’ UTR polymorphism has not been found to be consistent across populations [169], further indicating the necessity to directly query these noncoding sequences. In addition to expression variants, polymorphism outside of exons 2 and 3 may impact contact with other receptors or accessory molecules such as CD8 [170]. Finally, sequence variation in the cytoplasmic domain of class I molecules appear to modulate viral down-regulation of HLA [171].
Figure 4.
Noncoding variation in HLA loci can impact expression, through promoter polymorphism [including intronic promoters], introduction of stop codons, via splice site variation, or through sequence disruption
7. Concluding remarks
MS is an example of a multifactorial, complex condition whose understanding has been transformed by advances in genomics, and specifically immunogenomics. Convergent epidemiological and laboratory results are consistent with a polygenic model of inheritance, while the data also supports the long-held view that MS susceptibility rests on the action of polymorphisms common in the population. The incomplete penetrance and moderate individual effect observed for most genetic associations in MS most likely reflect interactions with other genes, post-transcriptional regulatory mechanisms, and significant environmental influences. The strong HLA association with MS, which was first described several decades ago, supports the idea that MS is, at its core, an antigen-specific autoimmune disease. Further investigation into the specific nature of immune system polymorphism in MS will continue to reveal the genetic underpinnings of this debilitating disease. In addition to gene identification, these studies will drive a forceful paradigm shift in the study of MS by allowing a more refined mechanistic representation of disease pathogenesis.
Figure 3.
Structure of the six most common KIR gene content haplotypes observed worldwide. The centromeric [Cen] and telomeric [Tel] motifs are labeled according to their association with the KIR A or B haplotype
Over 100 loci have been firmly associated with susceptibility to MS, whereas the main signal genome-wide maps to the class II region of the human leukocyte antigen (HLA) gene cluster
Despite the remarkable molecular dissection of the HLA region in MS, further studies are needed to generate unifying models to account for the role of the MHC in disease pathogenesis.
New horizons include application of modern sequencing methodologies coupled with state of the art bioinformatics and analytical approaches to understand the role of Killer-immunoglobulin-like receptors (KIR) and non-coding variation in MS.
Acknowledgements
The authors are supported by grants from NINDS, NIAID, NIGMS, NHGRI, and the National Multiple Sclerosis Society. We thank Wesley Marin and Paul J. Norman for assistance with figures. We would like to dedicate this manuscript to our HLA mentors, Glenys Thomson and Chaim Brautbar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Hauser SL, Goodin DS. Multiple sclerosis and other demyelinating diseases. In: Longo DL, Fauci AD, Kasper DL, Hauser SL, Jameson JL, Loscalzo J, editors. Harrison’s Principle of Internal Medicine. 18th Edition. McGraw Hill, NY: 2012. [Google Scholar]
- 3.Lassmann H. Multiple sclerosis: lessons from molecular neuropathology. Exp Neurol. 2014;262(Pt A):2–7. doi: 10.1016/j.expneurol.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Simons M, Misgeld T, Kerschensteiner M. A unified cell biological perspective on axon-myelin injury. J Cell Biol. 2014;206:335–345. doi: 10.1083/jcb.201404154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser SL. Multiple lessons for multiple sclerosis. N Engl J Med. 2008;359:1838–1841. doi: 10.1056/NEJMe0806738. [DOI] [PubMed] [Google Scholar]
- 6.Hauser SL, Chan JR, Oksenberg JR. Multiple sclerosis: Prospects and promise. Annals of Neurology. 2013;74:317–327. doi: 10.1002/ana.24009. [DOI] [PubMed] [Google Scholar]
- 7.Ontaneda D, Fox RJ, Chataway J. Clinical trials in progressive multiple sclerosis: lessons learned and future perspectives. Lancet Neurol. 2015;14:208–223. doi: 10.1016/S1474-4422(14)70264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugliatti M, Sotgiu S, Rosati G. The worldwide prevalence of multiple sclerosis. Clin Neurol Neurosurg. 2002;104:182–191. doi: 10.1016/s0303-8467(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 9.Simpson S, Jr, Blizzard L, Otahal P, Van der Mei I, Taylor B. Latitude is significantly associated with the prevalence of multiple sclerosis: a meta-analysis. J Neurol Neurosurg Psychiatry. 2011;82:1132–1141. doi: 10.1136/jnnp.2011.240432. [DOI] [PubMed] [Google Scholar]
- 10.Orton SM, Herrera BM, Yee IM, Valdar W, Ramagopalan SV, Sadovnick AD, et al. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006;5:932–936. doi: 10.1016/S1474-4422(06)70581-6. [DOI] [PubMed] [Google Scholar]
- 11.Koch-Henriksen N, Sorensen PS. The changing demographic pattern of multiple sclerosis epidemiology. Lancet Neurol. 2010;9:520–532. doi: 10.1016/S1474-4422(10)70064-8. [DOI] [PubMed] [Google Scholar]
- 12.Wallin MT, Page WF, Kurtzke JF. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann Neurol. 2004;55:65–71. doi: 10.1002/ana.10788. [DOI] [PubMed] [Google Scholar]
- 13.Wallin MT, Culpepper WJ, Coffman P, Pulaski S, Maloni H, Mahan CM, et al. The Gulf War era multiple sclerosis cohort: age and incidence rates by race, sex and service. Brain. 2012;135:1778–1785. doi: 10.1093/brain/aws099. [DOI] [PubMed] [Google Scholar]
- 14.Langer-Gould A, Brara SM, Beaber BE, Zhang JL. Incidence of multiple sclerosis in multiple racial and ethnic groups. Neurology. 2013;80:1734–1739. doi: 10.1212/WNL.0b013e3182918cc2. [DOI] [PubMed] [Google Scholar]
- 15.Cree BA, Khan O, Bourdette D, Goodin DS, Cohen JA, Marrie RA, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63:2039–2045. doi: 10.1212/01.wnl.0000145762.60562.5d. [DOI] [PubMed] [Google Scholar]
- 16.Cree BA, Reich DE, Khan O, De Jager PL, Nakashima I, Takahashi T, et al. Modification of Multiple Sclerosis Phenotypes by African Ancestry at HLA. Arch Neurol. 2009;66:226–233. doi: 10.1001/archneurol.2008.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadovnick AD, Baird PA. The familial nature of multiple sclerosis: age-corrected empiric recurrence risks for children and siblings of patients. Neurology. 1988;38:990–991. doi: 10.1212/wnl.38.6.990. [DOI] [PubMed] [Google Scholar]
- 18.Robertson NP, Fraser M, Deans J, Clayton D, Walker N, Compston DA. Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain. 1996;119(Pt 2):449–455. doi: 10.1093/brain/119.2.449. [DOI] [PubMed] [Google Scholar]
- 19.Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100:12877–12882. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebers GC, Sadovnick AD, Risch NJ. A genetic basis for familial aggregation in multiple sclerosis. Nature. 1995;377:150–151. doi: 10.1038/377150a0. [DOI] [PubMed] [Google Scholar]
- 21.Ebers GC, Yee IM, Sadovnick AD, Duquette P. Conjugal multiple sclerosis: population-based prevalence recurrence risks in offspring Canadian Collaborative Study Group. Ann Neurol. 2000;48:927–931. [PubMed] [Google Scholar]
- 22.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosisPart I: the role of infection. Ann Neurol. 2007;61:288–299. doi: 10.1002/ana.21117. [DOI] [PubMed] [Google Scholar]
- 23.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis Part II: Noninfectious factors. Ann Neurol. 2007;61:504–513. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 24.Sawcer S, Franklin RJ, Ban M. Multiple sclerosis genetics. Lancet Neurol. 2014;13:700–709. doi: 10.1016/S1474-4422(14)70041-9. [DOI] [PubMed] [Google Scholar]
- 25.Bertrams J, Kuwert E, Liedtke U. HLA antigens and multiple sclerosis. Tissue Antigens. 1972;2:405–408. doi: 10.1111/j.1399-0039.1972.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 26.Maito S, Manerow N, Mickey MR, Terasaki PI. Multiple sclerosis: association with HL-A3. Tissue Antigens. 1972;2:1–4. doi: 10.1111/j.1399-0039.1972.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 27.Vandiedonck C, Knight JC. The human Major Histocompatibility Complex as a paradigm in genomics research. Brief Funct Genomic Proteomic. 2009 doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SG. IMGT/HLA database--a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 29.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Jersild C, Fog T, Hansen GS, Thomsen M, Svejgaard A, Dupont B. Histocompatibility determinants in multiple sclerosis. Lancet. 1973;(ii):1221–1225. doi: 10.1016/s0140-6736(73)90970-7. [DOI] [PubMed] [Google Scholar]
- 31.Winchester R, Ebers G, Fu SM, Espinosa L, Zabriskie J, Kunkel HG. B-cell alloantigen Ag 7a in multiple sclerosis. Lancet. 1975;2:814. doi: 10.1016/s0140-6736(75)80033-x. [DOI] [PubMed] [Google Scholar]
- 32.Naito S, Namerow N, Mickey MR, Terasaki PI. Multiple sclerosis: association with HL-A3. Tissue antigens. 1972;2:1–4. doi: 10.1111/j.1399-0039.1972.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 33.Compston DA, Batchelor JR, McDonald WI. B-lymphocyte alloantigens associated with multiple sclerosis. Lancet. 1976;2:1261–1265. doi: 10.1016/s0140-6736(76)92027-4. [DOI] [PubMed] [Google Scholar]
- 34.Hauser SL, Fleinschnick E, Weiner HL, Marcus D, Awdeh Z, Yunis EJ, et al. Extended major histocompatibility complex haplotypes in patients with multiple sclerosis. Neurol. 1989;39:275–277. doi: 10.1212/wnl.39.2.275. [DOI] [PubMed] [Google Scholar]
- 35.Haines JL, Terwedow HA, Burgess K, Pericak-Vance MA, Rimmler JB, Martin ER, et al. Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. Hum Mol Genet. 1998;7:1229–1234. doi: 10.1093/hmg/7.8.1229. [DOI] [PubMed] [Google Scholar]
- 36.Terasaki PI. Selection of organ donors. N Engl J Med. 1969;280:1304. doi: 10.1056/NEJM196906052802322. [DOI] [PubMed] [Google Scholar]
- 37.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nature Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 38.de Bakker PI, Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet. 2012;21:R29–R36. doi: 10.1093/hmg/dds384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olerup O, Hillert J. HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue antigens. 1991;38:1–15. doi: 10.1111/j.1399-0039.1991.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 40.Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, Schmidt S, et al. Genetic basis for clinical expression in multiple sclerosis. Brain. 2002;125:150–158. doi: 10.1093/brain/awf009. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am J Epidemiol. 2007;165:1097–1109. doi: 10.1093/aje/kwk118. [DOI] [PubMed] [Google Scholar]
- 42.Consortium TIMSGCtWTCC. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet. 2003;72:710–716. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 45.Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A. 2003;100:12390–12395. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Heide A, Verduijn W, Haasnoot GW, Drabbels JJ, Lammers GJ, Claas FH. HLA dosage effect in narcolepsy with cataplexy. Immunogenetics. 2015;67:1–6. doi: 10.1007/s00251-014-0808-z. [DOI] [PubMed] [Google Scholar]
- 47.Mackie SL, Taylor JC, Martin SG, Consortium Y, Consortium U, Wordsworth P, et al. A spectrum of susceptibility to rheumatoid arthritis within HLA-DRB1: stratification by autoantibody status in a large UK population. Genes Immun. 2012;13:120–128. doi: 10.1038/gene.2011.60. [DOI] [PubMed] [Google Scholar]
- 48.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harbor perspectives in medicine. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, et al. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 50.Kwon OJ, Karni A, Israel S, Brautbar C, Amar A, Meiner Z, et al. HLA class II susceptibility to multiple sclerosis among Ashkenazi and non-Ashkenazi Jews. Arch Neurol. 1999;56:555–560. doi: 10.1001/archneur.56.5.555. [DOI] [PubMed] [Google Scholar]
- 51.Hensiek AE, Sawcer SJ, Feakes R, Deans J, Mander A, Akesson E, et al. HLA-DR 15 is associated with female sex and younger age at diagnosis in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2002;72:184–187. doi: 10.1136/jnnp.72.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Celius EG, Harbo HF, Egeland T, Vartdal F, Vandivik B, Spurkland A. Sex and age at diagnosis are correlated with the HLA-DR2, DQ6 haplotype in multiple sclerosis. J Neurol Sci. 2000;178:132–135. doi: 10.1016/s0022-510x(00)00389-0. [DOI] [PubMed] [Google Scholar]
- 53.Okuda DT, Srinivasan R, Oksenberg JR, Goodin DS, Baranzini SE, Beheshtian A, et al. Genotype-Phenotype correlations in multiple sclerosis: HLA genes influence disease severity inferred by 1HMR spectroscopy and MRI measures. Brain. 2009;132:250–259. doi: 10.1093/brain/awn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patsopoulos NA, Barcellos LF, Hintzen RQ, Schaefer C, van Duijn CM, Noble JA, et al. Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects. PLoS genetics. 2013;9:e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marrosu MG, Murru R, Murru MR, Costa G, Zavattari P, Whalen M, et al. Dissection of the HLA association with multiple sclerosis in the founder isolated population of Sardinia. Hum Mol Genet. 2001;10:2907–2916. doi: 10.1093/hmg/10.25.2907. [DOI] [PubMed] [Google Scholar]
- 56.Thomson G, Valdes AM, Noble JA, Kockum I, Grote MN, Najman J, et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue antigens. 2007;70:110–127. doi: 10.1111/j.1399-0039.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 57.Nepom GT, Hansen JA, Nepom BS. The molecular basis for HLA class II associations with rheumatoid arthritis. J Clin Immunol. 1987;7:1–7. doi: 10.1007/BF00915418. [DOI] [PubMed] [Google Scholar]
- 58.Kooy-Winkelaar Y, van Lummel M, Moustakas AK, Schweizer J, Mearin ML, Mulder CJ, et al. Gluten-specific T cells cross-react between HLA-DQ8 and the HLA-DQ2alpha/DQ8beta transdimer. J Immunol. 2011;187:5123–5129. doi: 10.4049/jimmunol.1101179. [DOI] [PubMed] [Google Scholar]
- 59.Matsuki K, Grumet FC, Lin X, Gelb M, Guilleminault C, Dement WC, et al. DQ (rather than DR) gene marks susceptibility to narcolepsy. Lancet. 1992;339:1052. doi: 10.1016/0140-6736(92)90571-j. [DOI] [PubMed] [Google Scholar]
- 60.Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quelvennec E, Bera O, Cabre P, Alizadeh M, Smadja D, Jugde F, et al. Genetic and functional studies in multiple sclerosis patients from Martinique attest for a specific and direct role of the HLA-DR locus in the syndrome. Tissue Antigens. 2003;61:166–171. doi: 10.1046/j.0001-2815.2002.00008.x. [DOI] [PubMed] [Google Scholar]
- 62.Lincoln MR, Ramagopalan SV, Chao MJ, Herrera BM, Deluca GC, Orton SM, et al. Epistasis among HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci determines multiple sclerosis susceptibility. Proc Natl Acad Sci U S A. 2009;106:7542–7547. doi: 10.1073/pnas.0812664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshimura S, Isobe N, Yonekawa T, Matsushita T, Masaki K, Sato S, et al. Genetic and infectious profiles of Japanese multiple sclerosis patients. PLoS One. 2012;7:e48592. doi: 10.1371/journal.pone.0048592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuoka T, Matsushita T, Osoegawa M, Kawano Y, Minohara M, Mihara F, et al. Association of the HLA-DRB1 alleles with characteristic MRI features of Asian multiple sclerosis. Mult Scler. 2008;14:1181–1190. doi: 10.1177/1352458508097818. [DOI] [PubMed] [Google Scholar]
- 65.Marrosu MG, Muntoni F, Murru MR. Sardinian multiple sclerosis is associated with HLA-DR4: A serological and molecular analysis. Neurology. 1988;38:1749–1753. doi: 10.1212/wnl.38.11.1749. [DOI] [PubMed] [Google Scholar]
- 66.Brassat D, Salemi G, Barcellos L, McNeill G, Proia P, Hauser SL, et al. The HLA locus and multiple sclerosis in Sicily. Neurol. 2005;64:361–363. doi: 10.1212/01.WNL.0000149765.71212.0A. [DOI] [PubMed] [Google Scholar]
- 67.Isobe N, Gourraud PA, Harbo HF, Caillier SJ, Santaniello A, Khankhanian P, et al. Genetic risk variants in African Americans with multiple sclerosis. Neurology. 2013;81:219–227. doi: 10.1212/WNL.0b013e31829bfe2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Modin H, Olsson W, Hillert J, Masterman T. Modes of action of HLA-DR susceptibility specificities in multiple sclerosis. Am J Hum Genet. 2004;74:1321–1322. doi: 10.1086/420977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marrosu MG, Murru MR, Costa G, Cucca F, Sotgiu S, Rosati G, et al. Multiple sclerosis in Sardinia is associated and in linkage disequilibrium with HLA-DR3 and -DR4 alleles. Am J Hum Genet. 1997;61:454–457. doi: 10.1016/S0002-9297(07)64074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karni A, Kohn Y, Safirman C, Abramsky O, Barcellos L, Oksenberg JR, et al. Evidence for the genetic role of human leukocyte antigens in low frequency DRB1*1501 multiple sclerosis patients in Israel. Mult Scler. 1999;5:410–415. doi: 10.1177/135245859900500i607. [DOI] [PubMed] [Google Scholar]
- 72.Cocco E, Sardu C, Pieroni E, Valentini M, Murru R, Costa G, et al. HLA-DRB1-DQB1 haplotypes confer susceptibility and resistance to multiple sclerosis in Sardinia. PLoS One. 2012;7:e33972. doi: 10.1371/journal.pone.0033972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amirzargar A, Mytilineos J, Yousefipour A, Farjadian S, Scherer S, Opelz G, et al. HLA class II associated genetic susceptibility in Iranian multiple sclerosis patients. Eur J immunogenet. 1998;25:297–301. doi: 10.1046/j.1365-2370.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 74.Brum DG, Barreira AA, Louzada-Junior P, Mendes-Junior CT, Donadi EA. Association of the HLA-DRB1*15 allele group and the DRB1*1501 and DRB1*1503 alleles with multiple sclerosis in White and Mulatto samples from Brazil. J Neuroimmunol. 2007;189:118–124. doi: 10.1016/j.jneuroim.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 75.Sollid LM, Pos W, Wucherpfennig KW. Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr Opin Immunol. 2014;31:24–30. doi: 10.1016/j.coi.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis Rheum. 2010;62:1781–1791. doi: 10.1002/art.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verreck FA, Termijtelen A, Koning F. HLA-DR beta chain residue 86 controls DR alpha beta dimer stability. Eur J Immunol. 1993;23:1346–1350. doi: 10.1002/eji.1830230624. [DOI] [PubMed] [Google Scholar]
- 78.Finn TP, Jones RE, Rich C, Dahan R, Link J, David CS, et al. HLA-DRB1*1501 risk association in multiple sclerosis may not be related to presentation of myelin epitopes. J Neurosci Res. 2004;78:100–114. doi: 10.1002/jnr.20227. [DOI] [PubMed] [Google Scholar]
- 79.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- 81.Caillier SJ, Briggs F, Cree BA, Baranzini SE, Fernandez-Vina M, Ramsay PP, et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol. 2008;181:5473–5480. doi: 10.4049/jimmunol.181.8.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574–7. doi: 10.1038/nature05133. [DOI] [PubMed] [Google Scholar]
- 84.Field J, Browning SR, Johnson LJ, Danoy P, Varney MD, Tait BD, et al. A polymorphism in the HLA-DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS ONE. 2010;5:e13454. doi: 10.1371/journal.pone.0013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dekker JW, Easteal S, Jakobsen IB, Gao X, Stewart GJ, Buhler MM, et al. HLA-DPB1 alleles correlate with risk for multiple sclerosis in Caucasoid and Cantonese patients lacking the high-risk DQB1*0602 allele. Tissue antigens. 1993;41:31–36. doi: 10.1111/j.1399-0039.1993.tb01974.x. [DOI] [PubMed] [Google Scholar]
- 86.Fukazawa T, Kikuchi S, Miyagishi R, Miyazaki Y, Yabe I, Hamada T, et al. HLA-dPB1*0501 is not uniquely associated with opticospinal multiple sclerosis in Japanese patients Important role of DPB1*0301. Mult Scler. 2006;12:19–23. doi: 10.1191/135248506ms1252oa. [DOI] [PubMed] [Google Scholar]
- 87.Marrosu MG, Cocco E, Costa G, Murru MR, Mancosu C, Murru R, et al. Interaction of loci within the HLA region influences multiple sclerosis course in the Sardinian population. J Neurol. 2006;253:208–213. doi: 10.1007/s00415-005-0957-y. [DOI] [PubMed] [Google Scholar]
- 88.Wu XM, Wang C, Zhang KN, Lin AY, Kira J, Hu GZ, et al. Association of susceptibility to multiple sclerosis in Southern Han Chinese with HLA-DRB1, -DPB1 alleles and DRB1-DPB1 haplotypes: distinct from other populations. Mult Scler. 2009;15:1422–1430. doi: 10.1177/1352458509345905. [DOI] [PubMed] [Google Scholar]
- 89.Matsushita T, Matsuoka T, Isobe N, Kawano Y, Minohara M, Shi N, et al. Association of the HLA-DPB1*0501 allele with anti-aquaporin-4 antibody positivity in Japanese patients with idiopathic central nervous system demyelinating disorders. Tissue antigens. 2009;73:171–176. doi: 10.1111/j.1399-0039.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 90.Ramagopalan SV, Anderson C, Sadovnick AD, Ebers GC. Genomewide study of multiple sclerosis. N Engl J Med. 2007;357:2199–2200. doi: 10.1056/NEJMc072836. author reply 200-1. [DOI] [PubMed] [Google Scholar]
- 91.Kaimen-Maciel DR, Reiche EM, Borelli SD, Morimoto HK, Melo FC, Lopes J, et al. HLA-DRB1* allele-associated genetic susceptibility and protection against multiple sclerosis in Brazilian patients. Molecular medicine reports. 2009;2:993–998. doi: 10.3892/mmr_00000204. [DOI] [PubMed] [Google Scholar]
- 92.Ramagopalan SV, Morris AP, Dyment DA, Herrera BM, DeLuca GC, Lincoln MR, et al. The inheritance of resistance alleles in multiple sclerosis. PLoS genetics. 2007;3:1607–1613. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laaksonen M, Pastinen T, Sjoroos M, Kuokkanen S, Ruutiainen J, Sumelahti ML, et al. HLA class II associated risk and protection against multiple sclerosis-a Finnish family study. J Neuroimmunol. 2002;122:140–145. doi: 10.1016/s0165-5728(01)00456-8. [DOI] [PubMed] [Google Scholar]
- 94.Tsai S, Santamaria P. MHC Class II Polymorphisms, Autoreactive T-Cells, and Autoimmunity. Front Immunol. 2013;4:321. doi: 10.3389/fimmu.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harbo HF, Lie BA, Sawcer S, Celius EG, Dai KZ, Oturai A, et al. Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue antigens. 2004;63:237–247. doi: 10.1111/j.0001-2815.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 96.Fogdell-Hahn A, Ligers A, Gronning M, Hillert J, Olerup O. Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens. 2000;55:140–148. doi: 10.1034/j.1399-0039.2000.550205.x. [DOI] [PubMed] [Google Scholar]
- 97.Chao MJ, Barnardo MC, Lui GZ, Lincoln MR, Ramagopalan SV, Herrera BM, et al. Transmission of class I/II multi-locus MHC haplotypes and multiple sclerosis susceptibility: accounting for linkage disequilibrium. Hum Mol Genet. 2007;16:1951–1958. doi: 10.1093/hmg/ddm142. [DOI] [PubMed] [Google Scholar]
- 98.Brynedal B, Duvefelt K, Jonasdottir G, Roos IM, Akesson E, Palmgren J, et al. HLA-A confers an HLA-DRB1 independent influence on the risk of multiple sclerosis. PLoS ONE. 2007;2:e664. doi: 10.1371/journal.pone.0000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bergamaschi L, Leone MA, Fasano ME, Guerini FR, Ferrante D, Bolognesi E, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun. 2010;11:173–180. doi: 10.1038/gene.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harbo HF, Isobe N, Berg-Hansen P, Bos SD, Caillier SJ, Gustavsen MW, et al. Oligoclonal bands and age at onset correlate with genetic risk score in multiple sclerosis. Multiple sclerosis. 2013 doi: 10.1177/1352458513506503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yeo TW, De Jager PL, Gregory SG, Barcellos LF, Walton A, Goris A, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol. 2007;61:228–236. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wissemann WT, Hill-Burns EM, Zabetian CP, Factor SA, Patsopoulos N, Hoglund B, et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. American journal of human genetics. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vlachopoulou E, Lahtela E, Wennerstrom A, Havulinna AS, Salo P, Perola M, et al. Evaluation of HLA-DRB1 imputation using a Finnish dataset. Tissue antigens. 2014;83:350–355. doi: 10.1111/tan.12343. [DOI] [PubMed] [Google Scholar]
- 105.Hsieh AR, Chang SW, Chen PL, Chu CC, Hsiao CL, Yang WS, et al. Predicting HLA genotypes using unphased and flanking single-nucleotide polymorphisms in Han Chinese population. BMC Genomics. 2014;15:81. doi: 10.1186/1471-2164-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pillai NE, Okada Y, Saw WY, Ong RT, Wang X, Tantoso E, et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet. 2014;23:4443–4451. doi: 10.1093/hmg/ddu149. [DOI] [PubMed] [Google Scholar]
- 107.Halperin E, Stephan DA. SNP imputation in association studies. Nature biotechnology. 2009;27:349–351. doi: 10.1038/nbt0409-349. [DOI] [PubMed] [Google Scholar]
- 108.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS genetics. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dilthey A, Leslie S, Moutsianas L, Shen J, Cox C, Nelson MR, et al. Multi-population classical HLA type imputation. PLoS computational biology. 2013;9:e1002877. doi: 10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, et al. HIBAG--HLA genotype imputation with attribute bagging. The pharmacogenomics journal. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics (Oxford, England) 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Erlich HA, Gyllensten UB. Shared epitopes among HLA class II alleles: gene conversion, common ancestry and balancing selection. Immunol Today. 1991;12:411–414. doi: 10.1016/0167-5699(91)90143-H. [DOI] [PubMed] [Google Scholar]
- 113.Karinen H, Karkkainen P, Pihlajamaki J, Janatuinen E, Heikkinen M, Julkunen R, et al. Gene dose effect of the DQB1*0201 allele contributes to severity of coeliac disease. Scandinavian journal of gastroenterology. 2006;41:191–199. doi: 10.1080/00365520500206277. [DOI] [PubMed] [Google Scholar]
- 114.Masterman T, Ligers A, Olsson T, Andersson M, Olerup O, Hillert J. HLA-DR15 is associated with lower age at onset in multiple sclerosis. Ann Neurol. 2000;48:211–219. [PubMed] [Google Scholar]
- 115.Deschamps R, Paturel L, Jeannin S, Chausson N, Olindo S, Bera O, et al. Different HLA class II (DRB1 and DQB1) alleles determine either susceptibility or resistance to NMO and multiple sclerosis among the French Afro-Caribbean population. Multiple sclerosis. 2011;17:24–31. doi: 10.1177/1352458510382810. [DOI] [PubMed] [Google Scholar]
- 116.Gregersen PK, Kosoy R, Lee AT, Lamb J, Sussman J, McKee D, et al. Risk for myasthenia gravis maps to a (151) Pro-->Ala change in TNIP1 and to human leukocyte antigen-B*08. Annals of neurology. 2012;72:927–935. doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Emile J, Truelle JL, Pouplard A, Hurez D. [Association of Parkinson’s disease with HLA-B17 and B18 antigens] Nouv Presse Med. 1977;6:4144. [PubMed] [Google Scholar]
- 118.Hamza TH, Payami H. The heritability of risk and age at onset of Parkinson’s disease after accounting for known genetic risk factors. Journal of human genetics. 2010;55:241–243. doi: 10.1038/jhg.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ahmed I, Tamouza R, Delord M, Krishnamoorthy R, Tzourio C, Mulot C, et al. Association between Parkinson’s disease and the HLA-DRB1 locus. Mov Disord. 2012;27:1104–1110. doi: 10.1002/mds.25035. [DOI] [PubMed] [Google Scholar]
- 121.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nature genetics. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Eberhard G, Franzen G, Low B. Schizophrenia susceptibility and HL-A antigen. Neuropsychobiology. 1975;1:211–217. doi: 10.1159/000117496. [DOI] [PubMed] [Google Scholar]
- 123.Amar A, Zohar M, Tiwari J, Brautbar C. HLA and schizophrenia in Israel. Isr J Med Sci. 1988;24:28–31. [PubMed] [Google Scholar]
- 124.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, Zhao Z. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS computational biology. 2012;8:e1002587. doi: 10.1371/journal.pcbi.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Molecular psychiatry. 2014 doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.2. ISGCatWTCCC. Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biological psychiatry. 2012;72:620–628. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2015;20:207–214. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van Bergen J, Kooy-Winkelaar EM, van Dongen H, van Gaalen FA, Thompson A, Huizinga TW, et al. Functional killer Ig-like receptors on human memory CD4+ T cells specific for cytomegalovirus. J Immunol. 2009;182:4175–4182. doi: 10.4049/jimmunol.0800455. [DOI] [PubMed] [Google Scholar]
- 131.Hartung HP, Aktas O, Menge T, Kieseier BC. Immune regulation of multiple sclerosis. Handbook of clinical neurology. 2014;122:3–14. doi: 10.1016/B978-0-444-52001-2.00001-7. [DOI] [PubMed] [Google Scholar]
- 132.Biron CA. Activation and function of natural killer cell responses during viral infections. Curr Opin Immunol. 1997;9:24–34. doi: 10.1016/s0952-7915(97)80155-0. [DOI] [PubMed] [Google Scholar]
- 133.Bashirova AA, Martin MP, McVicar DW, Carrington M. The Killer Immunoglobulin-like Receptor Gene Cluster: Tuning the Genome for Defense. Annu Rev Genomics Hum Genet. 2006 doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 134.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nature reviews Immunology. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parham P. Influence of KIR diversity on human immunity. Adv Exp Med Biol. 2005;560:47–50. doi: 10.1007/0-387-24180-9_6. [DOI] [PubMed] [Google Scholar]
- 136.JA Hollenbach IN, Ladner MB, Single RM. EATrachtenberg Killer Cell Immunoglobulin-like Receptor (KIR) Gene-Content Variation in the HGDP-CEPH Populations Immunogenetics. 2012 doi: 10.1007/s00251-012-0629-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, et al. 16(th) IHIW: Population Global Distribution of Killer Immunoglobulin-like Receptor (KIR) and Ligands. International journal of immunogenetics. 2013;40:39–45. doi: 10.1111/iji.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Colonna M, Moretta A, Vely F, Vivier E. A high-resolution view of NK-cell receptors: structure and function. Immunol Today. 2000;21:428–431. doi: 10.1016/s0167-5699(00)01697-2. [DOI] [PubMed] [Google Scholar]
- 139.Moretta L, Bottino C, Pende D, Castriconi R, Mingari MC, Moretta A. Surface NK receptors and their ligands on tumor cells. Semin Immunol. 2006;18:151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 140.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. Journal of immunology. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 142.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. Journal of immunology. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 143.Cassidy SA, Cheent KS, Khakoo SI. Effects of Peptide on NK cell-mediated MHC I recognition. Front Immunol. 2014;5:133. doi: 10.3389/fimmu.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis research & therapy. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Seminars in immunology. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lorentzen AR, Karlsen TH, Olsson M, Smestad C, Mero IL, Woldseth B, et al. Killer immunoglobulin-like receptor ligand HLA-Bw4 protects against multiple sclerosis. Ann Neurol. 2009;65:658–666. doi: 10.1002/ana.21695. [DOI] [PubMed] [Google Scholar]
- 147.Kaur G, Trowsdale J, Fugger L. Natural killer cells and their receptors in multiple sclerosis. Brain. 2013;136:2657–2676. doi: 10.1093/brain/aws159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, et al. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gourraud PA, Gagne K, Bignon JD, Cambon-Thomsen A, Middleton D. Preliminary analysis of a KIR haplotype estimation algorithm: a simulation study. Tissue antigens. 2007;69(Suppl 1):96–100. doi: 10.1111/j.1399-0039.2006.762_4.x. [DOI] [PubMed] [Google Scholar]
- 150.Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jelcic I, Hsu KC, Kakalacheva K, Breiden P, Dupont B, Uhrberg M, et al. Killer immunoglobulin-like receptor locus polymorphisms in multiple sclerosis. Mult Scler. 2012;18:951–958. doi: 10.1177/1352458511431726. [DOI] [PubMed] [Google Scholar]
- 152.Garcia-Leon JA, Pinto-Medel MJ, Garcia-Trujillo L, Lopez-Gomez C, Oliver-Martos B, Prat-Arrojo I, et al. Killer cell immunoglobulin-like receptor genes in Spanish multiple sclerosis patients. Mol Immunol. 2011;48:1896–1902. doi: 10.1016/j.molimm.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 153.Bettencourt A, Silva AM, Carvalho C, Leal B, Santos E, Costa PP, et al. The role of KIR2DS1 in multiple sclerosis--KIR in Portuguese MS patients. J Neuroimmunol. 2014;269:52–55. doi: 10.1016/j.jneuroim.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 154.Fusco C, Guerini FR, Nocera G, Ventrella G, Caputo D, Valentino MA, et al. KIRs and their HLA ligands in remitting-relapsing multiple sclerosis. J Neuroimmunol. 2010;229:232–237. doi: 10.1016/j.jneuroim.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 155.Xu W, Fazekas G, Hara H, Tabira T. Mechanism of natural killer (NK) cell regulatory role in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;163:24–30. doi: 10.1016/j.jneuroim.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 156.Huang D, Shi FD, Jung S, Pien GC, Wang J, Salazar-Mather TP, et al. The neuronal chemokine CX3CL1/fractalkine selectively recruits NK cells that modify experimental autoimmune encephalomyelitis within the central nervous system. FASEB J. 2006;20:896–905. doi: 10.1096/fj.05-5465com. [DOI] [PubMed] [Google Scholar]
- 157.Hao J, Liu R, Piao W, Zhou Q, Vollmer TL, Campagnolo DI, et al. Central nervous system (CNS)-resident natural killer cells suppress Th17 responses and CNS autoimmune pathology. J Exp Med. 2010;207:1907–1921. doi: 10.1084/jem.20092749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shi FD, Takeda K, Akira S, Sarvetnick N, Ljunggren HG. IL-18 directs autoreactive T cells and promotes autodestruction in the central nervous system via induction of IFN-gamma by NK cells. J Immunol. 2000;165:3099–3104. doi: 10.4049/jimmunol.165.6.3099. [DOI] [PubMed] [Google Scholar]
- 159.Vollmer TL, Liu R, Price M, Rhodes S, La Cava A, Shi FD. Differential effects of IL-21 during initiation and progression of autoimmunity against neuroantigen. J Immunol. 2005;174:2696–2701. doi: 10.4049/jimmunol.174.5.2696. [DOI] [PubMed] [Google Scholar]
- 160.Takahashi K, Miyake S, Kondo T, Terao K, Hatakenaka M, Hashimoto S, et al. Natural killer type 2 bias in remission of multiple sclerosis. J Clin Invest. 2001;107:R23–R29. doi: 10.1172/JCI11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Takahashi K, Aranami T, Endoh M, Miyake S, Yamamura T. The regulatory role of natural killer cells in multiple sclerosis. Brain. 2004;127:1917–1927. doi: 10.1093/brain/awh219. [DOI] [PubMed] [Google Scholar]
- 162.Backstrom E, Chambers BJ, Kristensson K, Ljunggren HG. Direct NK cell-mediated lysis of syngenic dorsal root ganglia neurons in vitro. J Immunol. 2000;165:4895–4900. doi: 10.4049/jimmunol.165.9.4895. [DOI] [PubMed] [Google Scholar]
- 163.Backstrom E, Chambers BJ, Ho EL, Naidenko OV, Mariotti R, Fremont DH, et al. Natural killer cell-mediated lysis of dorsal root ganglia neurons via RAE1/NKG2D interactions. Eur J Immunol. 2003;33:92–100. doi: 10.1002/immu.200390012. [DOI] [PubMed] [Google Scholar]
- 164.Hauser SL, Ault KA, Levin MJ, Garovoy MR, Weiner HL. Natural killer cell activity in multiple sclerosis. J Immunol. 1981;127:1114–1147. [PubMed] [Google Scholar]
- 165.De Santis D, Dinauer D, Duke J, Erlich HA, Holcomb CL, Lind C, et al. 16(th) IHIW : review of HLA typing by NGS. Int J Immunogenet. 2013;40:72–76. doi: 10.1111/iji.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dubois V, Tiercy JM, Labonne MP, Dormoy A, Gebuhrer L. A new HLA-B44 allele (B*44020102S) with a splicing mutation leading to a complete deletion of exon 5. Tissue Antigens. 2004;63:173–180. doi: 10.1111/j.1399-0039.2004.00134.x. [DOI] [PubMed] [Google Scholar]
- 167.Hoarau JJ, Cesari M, Caillens H, Cadet F, Pabion M. HLA DQA1 genes generate multiple transcripts by alternative splicing and polyadenylation of the 3’ untranslated region. Tissue antigens. 2004;63:58–71. doi: 10.1111/j.1399-0039.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 168.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gentle NL, Paximadis M, Puren A, Tiemessen CT. Genetic variability in markers of HLA-C expression in two diverse South African populations. PLoS One. 2013;8:e67780. doi: 10.1371/journal.pone.0067780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Salter RD, Benjamin RJ, Wesley PK, Buxton SE, Garrett TP, Clayberger C, et al. A binding site for the T-cell co-receptor CD8 on the alpha 3 domain of HLA-A2. Nature. 1990;345:41–46. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- 171.Rajapaksa US, Li D, Peng YC, McMichael AJ, Dong T, Xu XN. HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc Natl Acad Sci U S A. 2012;109:13353–13358. doi: 10.1073/pnas.1204199109. [DOI] [PMC free article] [PubMed] [Google Scholar]